Abstract
Background
Non-tuberculous mycobacteria (NTM) are a group of bacteria that cause rare lung infections and are increasingly recognized as causative agents of opportunistic and device-associated infections in humans. In Gabon, there is a lack of data on NTM species identification and drug susceptibility. The aim of this study was to identify the frequency of NTM species and their genotypic susceptibility pattern to commonly used antibiotics for NTM infections in Gabon.
Methods
A cross-sectional study was conducted at the CERMEL TB laboratory from January 2020 to December 2022, NTM subspecies identification and drug susceptibility testing to macrolides and aminoglycosides were performed using the genotype NTM-DR kit.
Results
The study found that out of 524 culture-positive specimens, 146 (28%) were NTM, with the predominant group being Mycobacterium avium complex (MAC) and Mycobacterium abscessus complex (MABC). All MAC isolates were fully susceptible to macrolides and aminoglycosides, while five MABC isolates carried mutations indicative of reduced susceptibility to macrolide and aminoglycoside drugs.
Conclusions
These findings suggest that clinicians may use macrolides and aminoglycosides to manage NTM infections caused by MAC, but further investigation is required to determine MABC drug susceptibility.
Similar content being viewed by others
Background
Non-tuberculous mycobacteria (NTM) are mycobacteria other than M. tuberculosis and M. leprae. They are environmental pathogens predominantly found in water, soil, dust and animal sources [1]. In the past, NTM infections did not attract much attention since they did not exhibit human-to-human transmission. Very often, isolates were considered as contaminants by laboratories, and were therefore overlooked [2, 3].
Moreover, their epidemiology has been difficult to establish because reporting of NTM infections was not mandatory in most parts of the world [4]. Advanced diagnostic laboratory techniques, as well as increased clinicians’ awareness certainly contribute to a recent increase in worldwide reporting of their incidence, which is estimated to be between 1.0 and 1.8 per 100,000 people per year [5]. However, one determining factor behind the evolution of the epidemiology of NTMs is the emergence of the human immunodeficiency virus. In fact, people living with HIV, cystic fibrosis (CF), bronchiectasis, emphysema and chronic obstructive pulmonary disease (COPD) constitute the high-risk population for NTM infections [3, 5,6,7].
There are more than 170 NTM species identified to date, but only a few species are believed to be opportunistic pathogens and responsible for human infections. The most-common of these species are M. avium complex (MAC), M. abscessus complex (MABC), M. kansasii, M. fortuitum, M. chelonae, M. szulgai, M. triviale and M. scrofulaceum.
Infections caused by NTM are diverse and present in both immunocompromised and immunocompetent patients. Bone infections, skin infections and disseminated disease have been described; 90% of which are chronic lung infections in the same way as tuberculosis [8]. In fact, clinically, it is difficult to discern the symptoms of NTM lung disease and tuberculosis. This can lead to incorrect diagnoses in the absence of microbiological identification. Moreover, NTM are resistant to several antimycobacterial agents; hence the low efficacy of the latter [9, 10].
In order to inform clinical decision-making to optimise treatment efficacy, as the intrinsic antibiotic resistance profile is species-specific [5, 11], species identification must precede sensitivity profile determination. Macrolides and aminoglycosides have been considered first-choice molecules in the management of NTM infection according to the American Thoracic Society/Infectious Diseases Society of America (ATS/IDSA) [5].
In high-income countries, the management of NTM can be considered routine [5]. However, in the sub-Saharan region, there is very little data on NTM management and drug susceptibility profiles, as very few laboratories perform these tests. Most studies are focused on the prevalence of pulmonary NTM. In 2017, a systematic review and meta-analysis of 37 articles on NTM in the South-of the-Sahara region revealed a prevalence of 7.5% of pulmonary NTM, with MAC as the predominant species [12].
Like the other sub-Saharan countries, Gabon has some data on the isolation of NTM. Indeed, a study conducted by our group in 2022 showed a proportion of 10% of NTM in patients presumed to have tuberculosis in the Lambaréné region, with MAC being the most-common species [13]. Whilst antimicrobial susceptibility information is essential for clinicians to select appropriate treatment regimens, drug susceptibility testing for NTM species has not been widely conducted in Gabon.
Thus, the objective of this study was to identify the species and drug susceptibility profiles of NTM commonly isolated at the Tuberculosis Reference Laboratory of the Lambaréné Medical Research Center (CERMEL).
Methods
Study site
This study was carried out at the CERMEL TB laboratory, which is the national TB reference laboratory of Gabon. The laboratory receives samples from laboratories in peripheral areas for routine surveillance of drug-resistant tuberculosis. In addition, the laboratory performs susceptibility testing (Line Probe Assay and conventional DST) and monitors MDR-TB patients from the whole country using a culture technique [14,15,16].
Study design
This was a prospective cross-sectional study of isolates from positive cultures of presumptive TB patients identified as NTM species, from January 2020 to December 2022. A limited number of specimens collected in 2020 in the framework of an earlier-reported study were also included in this analysis [13].
Sampling
Two sputum samples of each presumptive TB patient were collected over two consecutive days. Specimens that were positive after culture were specified as NTM using Genotype CM/AS version 2.0 (Bruker Hain Lifescience, Nehren, Germany). Only specimen from patients with species found to be identical in each of the culture isolates were eligible for susceptibility testing.
Data collection
All data were collected from patients using a structured questionnaire and on the Laboratory test request form. Demographic data (age, sex), clinical information (HIV status, history of TB treatment, clinical symptoms), AFB smear results, and radiological results were collected. Study data were managed using REDCap (Research Electronic Data Capture) [17, 18].
Extraction and species identification
DNA was extracted from a positive mycobacterial culture using the Genolyse kit (Hain Lifescience, Nehren, Germany). The GenoType CM/AS kit (Hain Lifescience, Nehren, Germany) was used to prepare the master mix for polymerase chain reaction (PCR) amplification of the speciation-determining region using biotinylated primers present in the kit. After amplification, the labelled PCR products are hybridised with specific oligonucleotide probes immobilised on a strip. The post-hybridisation reaction results in the appearance of coloured bands on the band at the probe binding site and can be observed with the naked eye. Labelled hybrids captured and detected by colorimetric development can be used to detect different species. All the assays were performed according to the manufacturer's instructions [19].
Genotypic drug susceptibility testing of NTM (GenoType NTM-DR test)
In vitro antimicrobial susceptibility by the broth microdilution testing of NTM was not performed within the realm of this study. We performed the genotypic method with the GenoType NTM-DR assay, a qualitative in vitro test assay based on PCR and DNA strip technology. Mycobacterial DNA is extracted from cultivated material, specifically amplified via PCR, and detected on membrane strips using reverse hybridisation and an enzymatic colour reaction. The Genotype NTM-DR assay permits the simultaneous genetic detection of several relevant NTM: M. avium, M intracellulare, M chimaera, M. chelonae and the M. abscessus complex (M. abscessus subsp. abscessus, M. abscessus subsp. massilliense and M. abscessus subsp. bolletii); their resistance to aminoglycosides (kanamycin, amikacin, gentamicin) via the detection of the most relevant mutations of the rrs gene; and their resistance to macrolides (clarithromycin, azithromycin) via the detection of the most relevant mutations of the rrl gene. Additionally, the erm(41) gene is analysed for the identification of macrolide resistance in members of the M. abscessus complex. The gene is divided into two probes; the erm(41) C28 probe and the erm(41) T28 probe. When the erm(41) C28 probe is positive, this indicates that the strain tested is susceptible to macrolides (except for strains with the additional rrl mutation). When the erm(41) T28 probe is positive, this indicates that the strain tested is resistant to macrolides, and the rrl gene is also examined for the detection of resistance to macrolides (clarithromycin or azithromycin). The rrs gene is examined for polymorphisms indicative of resistance to aminoglycosides (kanamycin, amikacin, gentamicin).
Data analysis
Data analysis was performed using R version 4.0.1; the dplyr package was used for calculus methods. Chi-square tests were performed to establish a relationship between the categorical or binary variables. The factors associated with positive NTM status were quantified by logistic regression. The model building process followed a backward stepwise strategy whereby univariable analysis of all variables preceded the multivariable analysis. A variable was retained in the model if it was significant (p < 0.05). Two-sided p-values of 0.05 or less were considered statistically significant.
Study results
NTM species identification
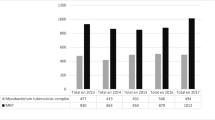
The specimens in this study came from 524 culture positive samples among presumptive TB patients, including 29 fast-growing samples collected in 2020 from a previous study [13]. A total of 146 isolates (27.9%) were identified as NTM between February 2020 to November 2022. Of the 146 isolates confirmed as NTM, 142 (97.3%) were from sputum, while four (2.2%) were from urine samples. Four species (complexes) were identified among the isolates, and the predominant group was M. avium complex (MAC 80/146; 54.8%; of which M. intracellulare 53/146 (36.3%) and M. avium 27/146 (18. 5%)). M. abscessus complex (MABC) amounted to 38/146 (26.0%) of the isolates. However, in 11 cases (7.5%), there were two species detected. GenoType Mycobacterium NTM-DR could identify the subspecies of MABC M. abscessus subsp. abscessus (20 isolates), M. abscessus subsp. bollettii (8), and M. abscessus subsp. massiliense (10). The isolated species are listed in Table 1.
Patient characteristics
The average age of study participants was 46.8 (SD:14) years; 88/142 (62.0%) of the patients were male; 47/142 (33.1%) patients were HIV-co-infected. Regarding treatment history, 32/142 (22. 5%) of the NTM patients isolates had previously been treated for TB. Among the positive NTM isolates, only 34/142 (23.9%) of the smears were positive (Table 2). The clinical characteristics of NTM infections of male and female patients are compared in Table 3. The results show that the proportion of cough and hemoptysis in male patients were significantly higher than in female patients (93% versus 36%, p < 0.030; 37% vs 20%, p < 0.040, respectively).
Genotypic drug susceptibility testing of NTM species
Of the 146 NTM isolates, 135 were tested for antimicrobial susceptibility to macrolides (azithromycin, clarithromycin and capreomycin) and aminoglycosides (kanamycin, amikacin, and gentamycin) using GenoType NTM-DR. The other 11 species were not tested due to the multiple NTM species identified in the same isolate. Of the 38 isolates of the subspecies of the MABC complex, five isolates of the subspecies M. abscessus subsp. abscessus were found to be resistant to macrolides, with a mutation present in T28 in the erm probe (41), and a mutation in the rrl gene. Resistance to aminoglycosides was also found by a mutation in the rrs gene. The five strains that presented the erm(41) mutation also carried rrl and rrs mutations. However, no resistance-conferring mutations were detected for either MAC complex or M. chelonae (Table 4).
Discussion
This study identified NTM species in presumptive tuberculosis patients in Gabon and determined their drug susceptibility profile using the LPA GenoType Mycobacterium NTM-DR test. The study revealed that M. intracellulare was the most-commonly occurring species, as observed previously in a study done in the same region, and also demonstrated in a study involving several countries in Sub-Saharan Africa [12, 13]. However, the MABC was the second most-common species isolated, which is contrary to the previous study where M. fortuitum was the second most-frequently found species. This describes the diversity of NTM species circulating among patients presumed to have tuberculosis in the Lambaréné region. As well, it has been documented that 25 to 60% of patients with a positive respiratory specimen for microbiological, respiratory signs and radiographic criteria have NTM pulmonary disease, and that due to its variable virulence, it is important to identify the species of NTM for better patient management [20].
In this study, results show that NTM isolates were more prevalent in male patients (88/142; 62%) as compared to female patients. This is in line with studies performed in Europe with 70%, and Tanzania 66%, which have suggested NTM pulmonary isolates to be originating mostly from men [21, 22]. However, some studies have indicated that pulmonary disease from NTM affects more women than men [23,24,25]. The findings from our study can be explained by the fact that our study population is based on cases of presumptive tuberculosis patients, and that in most studies on tuberculosis the male gender is the most-affected by the disease [22, 26]. Also, having a history of tuberculosis for pulmonary tuberculosis has been reported to be associated with NTM pulmonary isolates [13]. In this study, about 25% of the patients with NTM pulmonary isolates had a history of confirmed tuberculosis. Radiologically, the pulmonary presentation of NTM in our study was characterized by the presence of infiltrations and cavitations, which are also radiographic features similar to those of tuberculosis [27].
According to the Clinical Laboratory Standards Institute (CLSI) recommendations, drug susceptibility testing of NTM is usually done by the broth microdilution method as the gold standard. However, this method is time-consuming, given the delay due to the slow mycobacterial growth, as it is the case in M. avium [11]. In our study, 5/38 isolates of the MABC group were resistant to macrolides, with a mutation in the erm(41) gene at position T28 and mutation in the rrl gene in M. abscessus subsp. abscessus. These findings are similar to those of Maya et al., which is due to the fact that M. abscessus subsp. abscessus and M. abscessus subsp. bolettii macrolide resistance is identified by polymorphisms at position 28 in the erm(41) gene (cytosine is replaced by thymine leading to resistance) and mutations at positions 2058/2059 (adenine is replaced by cytosine leading to resistance) in the rrl gene [22, 28, 29]. However, other species of this group such as M. massiliense have a non-functional erm(41) gene, do not have the target base at position 28 which results from a large deletion of a non-functional 276 bp, and therefore do not exhibit resistance to macrolides. To determine NTM macrolide resistance, it is necessary to check whether there is a mutation in the rrl gene [30, 31]. Five isolates of M. abscessus subsp. abscessus were resistant to aminoglycosides through the rrs gene. Management of MABC group NTM that are resistant to macrolides and aminoglycosides should be performed according to guidelines [32]. In our study, no mutation was detected in the MAC group (M. avium, M. intracellulare) indicating that MAC is sensitive to macrolides and aminoglycosides; in line with a study conducted in Ghana showing sensitivity of all MAC isolates to macrolides and aminoglycosides drugs [33].
This study has limitations. Firstly, the presence of two species of NTM in an isolate does not allow the genotypic susceptibility test to be carried out for those type of isolates, and the genotype can only be used for the species indicated in the kit, while the reference method (microdilution) can be used as a complement for the other species. Secondly, although macrolides and aminoglycosides are the first-line molecules for NTM lung disease, other molecules that can be combined with the therapeutic regimens have not been taken into account by the test used.
Conclusions
Our study identified NTM species and tested drug susceptibility in routine culture isolates among tuberculosis patients in Lambaréné, Gabon. The study confirmed that the predominant NTM species was M. intracellulare, and that all MAC species were sensitive to macrolides and aminoglycosides. On the other hand, five MABC group isolates linked to the subspecies M. abscessus subsp. abscessus showed genotypical resistance to macrolides and aminoglycosides. Our research shows that NTM are present among presumptive TB cases in our setting, hence National Tuberculosis Control Programs should consider screening for NTM among presumptive TB cases. Knowledge of the NTM infection status and NTM drug susceptibility pattern is essential for correct diagnosis and selection of appropriate treatment regimens and should guide clinical decision-making.
Availability of data and materials
All data generated and analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- AFB:
-
Acid Fast Bacilli
- ATS:
-
American Thoracic Society
- CERMEL:
-
Centre de Recherche Médicales de Lambaréné
- CLSI:
-
Clinical Laboratory Standards Institute DNA: deoxyribonucleic acid
- DST:
-
Drug Sensitivity Testing
- HIV:
-
Human Immunodeficiency Virus
- IDSA:
-
Infectious Diseases Society of America
- LPA:
-
Line Probe Assay
- MABC:
-
Mycobacterium abscessus Complex
- MAC:
-
Mycobacterium avium Complex
- MDR-TB:
-
Multidrug-resistant tuberculosis
- NTM:
-
Non-tuberculous mycobacteria
- PCR:
-
Polymerase chain reaction
- REDCap:
-
Research Electronic Data Capture
- SD:
-
Standard Deviation
- TB:
-
Tuberculosis
References
Falkinham JO. Environmental sources of nontuberculous mycobacteria. Clin Chest Med. 2015;36:35–41.
Harris KA, Underwood A, Kenna DTD, Brooks A, Kavaliunaite E, Kapatai G, Tewolde R, Aurora P, Dixon G. Whole-genome sequencing and epidemiological analysis do not provide evidence for cross-transmission of mycobacterium abscessus in a cohort of pediatric cystic fibrosis patients. Clin Infect Dis. 2015;60:1007–16.
Honda JR, Knight V, Chan ED. Pathogenesis and risk factors for nontuberculous mycobacterial lung disease. Clin Chest Med. 2015;36:1–11.
Zhang H, Luo M, Zhang K, Yang X, Hu K, Fu Z, Zhang L, Wu P, Wan D, Han M, Wang X. Species identification and antimicrobial susceptibility testing of non-tuberculous mycobacteria isolated in Chongqing, Southwest China. Epidemiol Infect. 2020;149:e7.
Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, Holland SM, Horsburgh R, Huitt G, Iademarco MF, Iseman M, Olivier K, Ruoss S, Von Reyn CF, Wallace RJ, Winthrop K. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med. 2007;175:367–416.
Thomson RM, Carter R, Tolson C, Coulter C, Huygens F, Hargreaves M. Factors associated with the isolation of Nontuberculous mycobacteria (NTM) from a large municipal water system in Brisbane, Australia. BMC Microbiol. 2013;13:89.
Donohue MJ. Epidemiological risk factors and the geographical distribution of eight Mycobacterium species. BMC Infect Dis. 2021;21:1–11.
Santos A, Carneiro S, Silva A, Gomes JP, Macedo R. Nontuberculous Mycobacteria in Portugal: trends from the last decade. Pulmonology. 2022. https://doi.org/10.1016/J.PULMOE.2022.01.011.
Kasperbauer SH, De Groote MA. The treatment of rapidly growing mycobacterial infections. Clin Chest Med. 2015;36:67–78.
Wang D, Lin W, Cheng H, Bao X, Xu D, Liang S, Jiang Y, Wang C. Clinical Characteristics and Antimicrobial Susceptibility of Mycobacterium intracellulare and Mycobacterium abscessus Pulmonary Diseases: a retrospective study. Can J Infect Dis Med Microbiol = J Can des Mal Infect la Microbiol Médicale. 2022;2022:2642200.
Mougari F, Loiseau J, Veziris N, Bernard C, Bercot B, Sougakoff W, Jarlier V, Raskine L, Cambau E, Aubry A, Brossier F, Chauffour A, Lecorche E, Reibel F, Robert J. Evaluation of the new GenoType NTM-DR kit for the molecular detection of antimicrobial resistance in non-tuberculous mycobacteria. J Antimicrob Chemother. 2017;72:1669–77.
Okoi C, Anderson STB, Antonio M, Mulwa SN, Gehre F, Adetifa IMO. Non-tuberculous Mycobacteria isolated from pulmonary samples in sub-Saharan Africa - a systematic review and meta analyses. Sci Rep. 2017;7:1–12.
Epola Dibamba Ndanga M, Achimi Agbo Abdul JBP, Edoa JR, Chester Mevyann R, Adegbite BR, Mfoumbi A, Mebiame Biyogho C, Beh Mba R, Mahoumbou J, McCall MBB, Grobusch MP, Adegnika AA, Alabi AS. Non-tuberculous mycobacteria isolation from presumptive tuberculosis patients in Lambaréné, Gabon. Trop Med Int Heal. 2022;27:438–44.
Alabi AS, Traoré AN, Loembe MM, Ateba-Ngoa U, Frank M, Adegnika AA, Lell B, Mahoumbou J, Köhler C, Kremsner PG, Grobusch MP. Enhanced laboratory capacity development: a boost for effective tuberculosis control in resource-limited settings. Int J Infect Dis. 2017;56:81–4.
Mekota AM, Gillespie SH, Hoelscher M, Diacon AH, Dawson R, Churchyard G, Sanne I, Minja L, Kibiki G, Maboko L, Lakhi S, Joloba M, Alabi A, Kirenga B, McHugh TD, Grobusch MP, Boeree MJ. Building sustainable clinical trial sites in Sub-Saharan Africa through networking, infrastructure improvement, training and conducting clinical studies: the PanACEA approach. Acta Trop. 2023;238:106776.
Ramharter M, Agnandji ST, Adegnika AA, Lell B, Mombo-Ngoma G, Grobusch MP, McCall M, Muranaka R, Kreidenweiss A, Velavan TP, Esen M, Schaumburg F, Alabi A, Druml C, Mordmüller B, Köhler C, Kremsner PG. Development of sustainable research excellence with a global perspective on infectious diseases: Centre de Recherches Médicales de Lambaréné (CERMEL) Gabon. Wien Klin Wochenschr. 2021;133:500–8.
Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42:377–81.
Harris PA, Taylor R, Minor BL, Elliott V, Fernandez M, O’Neal L, McLeod L, Delacqua G, Delacqua F, Kirby J, Duda SN. The REDCap consortium: building an international community of software platform partners. J Biomed Inform. 2019;95: 103208.
Hain LifeScience. GenoType Mycobacterium CM instructions for use. 2014.
Stout JE, Koh WJ, Yew WW. Update on pulmonary disease due to non-tuberculous mycobacteria. Int J Infect Dis. 2016;45:123–34.
Van Ingen J, Bendien SA, De Lange WCM, Hoefsloot W, Dekhuijzen PNR, Boeree MJ, Van Soolingen D. Clinical relevance of non-tuberculous mycobacteria isolated in the Nijmegen-Arnhem region The Netherlands. Thorax. 2009;64:502–6.
Maya TG, Komba EV, Mensah GI, Mbelele PM, Mpagama SG, Mfinanga SG, Addo KK, Kazwala RR. Drug susceptibility profiles and factors associated with non-tuberculous mycobacteria species circulating among patients diagnosed with pulmonary tuberculosis in Tanzania. PLoS ONE. 2022;17: e0265358.
Park SC, Kang MJ, Han CH, Lee SM, Kim CJ, Lee MJ, Kang YA. Prevalence, incidence, and mortality of nontuberculous mycobacterial infection in Korea: a nationwide population-based study. BMC Pulmon Med. 2019;19:140.
Dodd PJ, Looker C, Plumb ID, Bond V, Schaap A, Shanaube K, Muyoyeta M, Vynnycky E, Godfrey-Faussett P, Corbett EL, Beyers N, Ayles H, White RG. Age- and sex-specific social contact patterns and incidence of Mycobacterium tuberculosis infection. Am J Epidemiol. 2016;183:156–66.
Kim RD, Greenberg DE, Ehrmantraut ME, Guide SV, Ding L, Shea Y, Brown MR, Chernick M, Steagall WK, Glasgow CG, Lin JP, Jolley C, Sorbara L, Raffeld M, Hill S, Avila N, Sachdev V, Barnhart LA, et al. Pulmonary nontuberculous mycobacterial disease: prospective study of a distinct preexisting syndrome. Am J Respir Crit Care Med. 2008;178:1066.
De Castro DB, De Seixas Maciel EMG, Sadahiro M, Pinto RC, De Albuquerque BC, Braga JU. Tuberculosis incidence inequalities and its social determinants in Manaus from 2007 to 2016. Int J Equity Health. 2018;17:187.
Sharma S, Upadhyay V. Epidemiology, diagnosis & treatment of non-tuberculous mycobacterial diseases. Indian J Med Res. 2020;152:185–226.
Huh HJ, Kim SY, Shim HJ, Kim DH, Yoo IY, Kang OK, Ki CS, Shin SY, Jhun BW, Shin SJ, Daley CL, Koh WJ, Yong Lee N. GenoType NTM-DR performance evaluation for identification of mycobacterium avium complex and mycobacterium abscessus and determination of clarithromycin and amikacin resistance. J Clin Microbiol. 2019;57:e00516–19.
Nash KA, Brown-Elliott AB, Wallace RJ. A novel gene, erm(41), confers inducible macrolide resistance to clinical isolates of Mycobacterium abscessus but is absent from Mycobacterium chelonae. Antimicrob Agents Chemother. 2009;53:1367–76.
Kim SY, Kim CK, Bae IK, Jeong SH, Yim JJ, Jung JY, Park MS, Kim YS, Kim SK, Chang J, Kang YA. The drug susceptibility profile and inducible resistance to macrolides of Mycobacterium abscessus and Mycobacterium massiliense in Korea. Diagn Microbiol Infect Dis. 2015;81:107–11.
Rubio M, March F, Garrigó M, Moreno C, Español M, Coll P. Inducible and acquired clarithromycin resistance in the mycobacterium abscessus complex. PLoS ONE. 2015;10: e0140166.
Haworth CS, Banks J, Capstick T, Fisher AJ, Gorsuch T, Laurenson IF, Leitch A, Loebinger MR, Milburn HJ, Nightingale M, Ormerod P, Shingadia D, Smith D, Whitehead N, Wilson R, Floto RA. British Thoracic Society guidelines for the management of non-tuberculous mycobacterial pulmonary disease (NTM-PD). 2017.
Addo KK, Addo SO, Mensah GI, Mosi L, Bonsu FA. Genotyping and drug susceptibility testing of mycobacterial isolates from population-based tuberculosis prevalence survey in Ghana. BMC Infect Dis. 2017;17:1–7.
Acknowledgements
We are grateful to the fieldworkers and TB nurses of the tuberculosis research team at CERMEL, for their hard work.
Funding
The longitudinal routine data collection from which this study is deducted is supported by funding from WHO AFRO/TDR/EDCTP Small Grant Project (2019/893805) and Central Africa Network on Tuberculosis, HIV/AIDS and Malaria (CANTAM), which is a network of excellence supported by The European & Developing Countries Clinical Trials Partnership (EDCTP). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
Conceptualisation, M.E.D.N., J.B.P.A.A.A., J.R.E., B.R.A., A.A.A.; and A.S.A.; methodology, M.E.D.N., G.A.R.M.I., J.M., S.M., N.M.R., Data analysis, M.E.D.N., and J.R.E.; validation, J.B.P.A.A.A., M.E.D.N., and A.A.A.; Specimen collection, M.E.D.N., R.C.M.; A.M. and C.M.B.; writing—original draft preparation, M.E.D.N.; funding acquisition, A.A.A., B.L., P.G.K and M.P.G. All authors have contributed to, and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study protocol was approved by the Institutional Ethics Committee of the Centre de Recherches Médicales de Lambaréné (CERMEL) under the number CEI-014-2019. Written informed consent was obtained from all volunteers prior to study, study identification numbers and laboratory serial numbers were used instead of patient numbers. names so that the identity of each participant remains completely anonymous.
All work on human tissue samples, and human data reporting in the manuscript were performed in accordance with relevant guidelines and regulations such as the Declaration of Helsinki.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Ndanga, M.E.D., Abdul, J.B.P.A.A., Edoa, J.R. et al. Species identification and drug susceptibility testing of non-tuberculous mycobacteria by Line Probe Assay in Lambaréné, Gabon—a cross-sectional study. BMC Infect Dis 23, 651 (2023). https://doi.org/10.1186/s12879-023-08617-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-023-08617-x