Abstract
Background
In 2018 Pakistan initiated its national antimicrobial resistance (AMR) surveillance aligned with Global Antimicrobial Surveillance System (GLASS). To complement this surveillance, we conducted a situational analysis of AMR rates among GLASS organisms in the country. Data from published studies and from antibiograms was compared and role of antibiograms as potential contributors to national AMR surveillance explored.
Methods
AMR rates for GLASS specified pathogen/antimicrobials combination from Pakistan were reviewed. Data sources included published studies (2006–2018) providing AMR rates from Pakistan (n = 54) as well as antibiograms (2011–2018) available on the Pakistan Antimicrobial Resistance Network (PARN) website. Resistance rates were categorized as follows: Very low: 0–10%, Low: 11–30%, Moderate: 30–50% and High: > 50%.
Results
Published data from hospital and community/laboratory-based studies report resistance rates of > 50% and 30–50% respectively to 3rd generation cephalosporins, fluoroquinolones and cotrimoxazole amongst Klebsiella pneumoniae and Escherichia coli. Carbapenem resistance rates amongst these organisms remained below 30%. High (> 50%) resistance was reported in Acinetobacter species to aminoglycosides and carbapenems among hospitalized patients. The evolution of ceftriaxone resistant Salmonella Typhi and Shigella species is reported. The data showed > 50% to fluoroquinolones amongst Neisseria gonorrhoeae and the spread of methicillin resistant Staphylococcus aureus (< 30%; 2008) to (> 50%; 2010) in hospital settings. Resistance reported in published studies aligned well with antibiogram data. The latter also captured a clear picture of evolution of resistance over the study period.
Conclusion
Both published studies as well antibiograms suggest high rates of AMR in Pakistan. Antibiogram data demonstrating steady increase in AMR highlight its potential role towards supplementing national AMR surveillance efforts particularly in settings where reach of national surveillance may be limited.
Similar content being viewed by others
Background
Antimicrobial resistance (AMR) recognized as a natural evolutionary process is facilitated by the genomic plasticity of microorganisms [1]. In recent times, this process has been greatly accelerated by the increased exposure of microorganisms to antimicrobial agents. Excessive usage of antimicrobial drugs in human and animal populations as well in agriculture contribute to their increasing availability in the environment placing a strong selective pressure within microorganisms and resulting in the development of antimicrobial resistance [2]. At a global level, the emergence and spread of antimicrobial resistance is enhanced by poor prescribing practices, counterfeit drugs, and poor infection control practices. Travel and trade can contribute to this spread; the spread of NDM-1, from Indian sub-continent region to Europe and United States highlights the global nature of this public health disaster [3]. Reports of high levels of antibacterial resistance [4,5,6,7] from hospitals as well as the community [8, 9] in Pakistan, the fifth most populous country globally [10], places an additional burden on an overstressed and under resourced health care system. Circulating mobile genetic elements in bacterial species [5, 7, 11] that can transfer multidrug resistance genes within and between species have also been reported. Further, the detection of extensively drug resistant (XDR) Salmonella Typhi from Hyderabad, Pakistan [12] reinforces concerns over the complex interaction between environment and anthropogenic activities in contributing towards emerging AMR.
Hence, following the Global Action Plan to tackle antimicrobial resistance at the 68th World Health Assembly, 2015, Global Antimicrobial Surveillance System (GLASS) was established [13] to collate antimicrobial resistance data at a global level.
Responding to the Global Action Plan and reports of increasing resistance in Pakistan, the National Strategic Framework for Containment of Antimicrobial Resistance, was translated into the National Action Plan of Pakistan for Antimicrobial Resistance and launched in 2018, initiating national AMR surveillance system (PASS) aligned with GLASS in 2018 [9]. To complement PASS, which collects data from 2018 onward, we present an analysis of the available information on AMR in the country from 2006 to 2018 based on literature review of published studies on available antibiograms. Resistance patterns over the course of the study period are presented.
Methods
Data sources
The situational analysis was conducted using two methods: literature review of published studies and review of data available on the Pakistan Antimicrobial Resistance Network (PARN) [14] website.
Review of published literature
Literature search was performed for studies reporting antimicrobial susceptibility rates of GLASS specified microorganisms (GLASS) [13]: Klebsiella pneumoniae, Escherichia coli, Acinetobacter baumannii, Salmonella Typhi, Shigella species, Neisseria gonorrhoeae, Staphylococcus aureus and Streptococcus pneumoniae from Pakistan.
Search strategy
Studies and reports were identified by searching electronic database Medline (PubMed) peer reviewed literature. Key terms used in the search are presented in Additional file 1.
Selection criteria
English language articles published January 01, 2006–January 31, 2018 were included. Systematic reviews, case reports, novel antibacterial therapeutics, articles not supported by quality control methodologies, studies focusing on vaccination program outcomes and in-data-review were excluded.
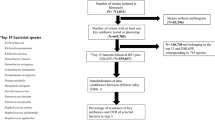
Hospital- as well as community-based studies for both adult and paediatric populations were included (Fig. 1).
Search methodology. Algorithm showing search methodology and reasons for exclusion; 53 studies selected from a total of 117 studies identified using the search strategies (Additional file 1). GLASS Global Antimicrobial Surveillance System, ARG antimicrobial resistance genes, QC quality control, RCT randomized control trial, MDR multi drug resistant
The papers retrieved were independently reviewed by two authors (DKS and RH). Relevant studies based on selection criteria were included. Discrepancies arising between the reviewers were resolved by consensus discussions between the two reviewers.
Outcomes of interest
Data from the reviewed literature was categorized under the following headings in MS Excel: author, journal, year published, year of study, location, organism names, isolate numbers, site of infection, hospital/community, age group, antibiotics tested, percent resistance, multi drug resistance [15], clinical outcomes of treatment based on mortality linked to the infectious episode, methods provided for susceptibility testing, statistical analysis, quality assurance and use of Clinical Laboratory Standards Institute (CLSI) guidelines, key findings and limitations.
Antibiogram data
Information on antimicrobial resistance included in antibiograms published on the PARN website was reviewed. To address the gaps and strengthen laboratory capacity for AMR surveillance PARN [14]; a collaborative initiative between public and private health organizations from Pakistan was established in 2006. It is an electronic platform that aims to share data and information relating to antimicrobial resistance in Pakistan. As part of these activities, PARN publishes antibiograms that are contributed voluntarily by individual laboratories. Based on their capacity and interest, contribution by the individual laboratories has varied from year to year. However, cumulatively, such information is considered an effective resource providing Level 2 evidence i.e., laboratory-based information in the absence of standardized AMR surveillance processes [16] for gauging antimicrobial resistance trends in the country. Data from antibiograms available for the years 2011–2018 was included. The antibiogram data available for the study period and included in this analysis were from the following institutions: Aga Khan University (2011–2018), Patel Hospital Karachi (2011–2012), Tabba Hospital Karachi (2011–2012), Indus Hospital Karachi (2011–2012), Dr Ziauddin Hospital (2011), Shifa Hospital Islamabad (2015), Jinnah Hospital Karachi (2014–2018), Civil Hospital Karachi (2016–2018), Shaukat Khanum Memorial Cancer Hospital and Research Centre, Lahore (2017).
Data analysis
The pathogens/antimicrobial based on GLASS specified bug/drug combination (GLASS) [13] investigated for resistance from both published reports and from the PARN website are shown in Table 1.
In order to minimize the effect of heterogeneity arising from different settings in which studies were conducted, the reviewed literature was divided into the following groups:
-
1.
Hospital based studies including Intensive Care Units (ICUs), Medical Intensive Care Units (MICUs), and medical wards.
-
2.
Laboratory based surveillance studies, community studies.
-
3.
Hospital and community-based studies specifically reporting antimicrobial resistance rates in neonates and children.
-
4.
Antimicrobial Resistance data based on published antibiograms.
Resistance rates reported in published studies were included in groups 1–3 while data from antibiograms was included in group 4.
Median percentage resistance along with their confidence intervals were determined for studies/antibiograms published in the same year using STATA/MP 13[17] by calculating the 50th centile resistance rates with 95% CIs. Where susceptibility rates were given, resistance rates were calculated by subtracting the percentage susceptibility rates provided from 100.
Resistance rates were categorized from very low to high as follows:
Very low: 0–10%, Low: 11–30%, Moderate: 30–50% and High: ≥ 50%
Results
PubMed search returned a total of 287 articles. A total of 53 studies were selected for this review (Fig. 1).
While the majority (40/53) of studies included data from both adults and children, this data was not disaggregated by age (Tables 2, and 3, Additional Files 2 and 3). One study [18] disaggregated data by age and sex but did not report the denominators (entire population sampled). A few studies (n = 12) focused only on the paediatric age group, and data from these are described separately (Table 4, Additional file 4). The majority of the studies included resistance data from hospitalized patients, with only 16 studies reporting community level data. These are included in Table 3.
Antibiogram data available were not further classified by the setting (hospital vs community), site of infection, or age. However, overall resistance rates presented in the antibiograms (Table 5, Additional file 5) for K. pneumoniae, E. coli and Acinetobacter species, Shigella species, N. gonorrhoeae as well as S. aureus approached those in reported studies (Tables 2, 3 and 4).
Additional files 2, 3, 4 and 5 complement Tables 2, 3, 4 and 5. These additional files provide resistance rates reported from each selected literature studies and published antibiogram.
Klebsiella pneumoniae and Escherichia coli
Hospital-based studies consistently indicated high rates of resistance to 3rd generation cephalosporins, fluoroquinolones and cotrimoxazole amongst both K. pneumoniae and E. coli (Table 2). Meanwhile, moderate to high resistance to fluoroquinolones and co-trimoxazole and increasing resistance to 3rd generation cephalosporins is reported from laboratory surveillance, from community-based studies (2012–2018) (Table 3) and in the antibiogram data (Table 5). Information on resistant K. pneumoniae amongst the paediatric population is sparse. However, published data reporting resistance in this age group is available for E. coli suggesting moderate to high rates of resistance to 3rd generation cephalosporins, fluoroquinolones and high resistance rates to cotrimoxazole (Table 4). Carbapenem resistance rates of under 30% for both K. pneumoniae and E. coli were reported in published studies as well as in the antibiograms (Tables 2, 3, 4, and 5). With the antibiograms suggesting increasing resistance 2013–2018 (Table 5) and one ICU based study (Table 2) reporting 56% carbapenem resistance amongst their K. pneumoniae isolates [19].
Acinetobacter species
High resistance rates to aminoglycosides (amikacin and gentamicin) and to carbapenems (Tables 2, 4, and 5) is documented for Acinetobacter species.
Salmonella Typhi
Fluoroquinolone resistance rates of over 80% amongst S. Typhi in the country have been documented in published reports since 2014 (Table 3) and in antibiograms since 2012 (Table 5). The emergence of ceftriaxone resistant S. Typhi was captured in both publications as well as in antibiograms (Tables 3 and 5). Laboratory-based studies conducted between 2008 and 2013 documented very low rates of S. Typhi resistant to ceftriaxone [20,21,22]. In December 2016, an outbreak of ceftriaxone resistant S. Typhi or extensively drug resistant (XDR) S. Typhi was reported from the southern province of Sindh, as a result of CTX-M gene acquisition in the widely prevalent fluoroquinolone, ampicillin, cotrimoxazole and chloramphenicol resistant strain of S. Typhi. This outbreak was recorded in antibiogram data in 2017 (Table 5) and published a year later [23, 24].
Shigella species
While strains showing resistance to ceftriaxone have been reported, the rate of such resistance remains low (Table 3). A laboratory-based study conducted in 2006–2007 reported an increase in resistance to ceftriaxone from 2 to 8%, and to ofloxacin from 4.3 to 10.9% over the study period [25]. This study further reported that ceftriaxone resistance was highest amongst Shigella flexineri followed by Shigella sonnei [25]. Similar figures were reported from another study conducted between 2011 and 2013 on isolates (n = 45) from cancer patients documenting resistance rates of 7% for ceftriaxone and 25% for ciprofloxacin [26]. Information from published studies is supported by the antibiogram data reporting low rate of resistance amongst Shigella species to both ceftriaxone and ciprofloxacin (2011–2016). Subsequent data however reports an increase in resistance rates to moderate levels for ceftriaxone (2017–2018) and for ciprofloxacin in 2017 (Table 5).
Neisseria gonorrhoeae
Resistance amongst N. gonorrhoeae to cephalosporins, azithromycin or spectinomycin was not reported in either the published studies or in the antibiograms from 2006 to 2018. However high fluoroquinolone resistance rates were consistently reported (Tables 3 and 5).
Staphylococcus aureus
Heat map for the years 2007–2014 (Table 2) shows an increase in prevalence of hospital acquired methicillin resistant S. aureus (MRSA) from low (< 30%) in 2008 [27] to moderate levels (> 50%) in 2017 [28]. MRSA rates in community and laboratory surveillance data (Table 3) also remained low to moderate; 12% in 2005–2008 [29] to 28% from 2009 to 2010 [29]. However, antibiogram data show high median MRSA rates (Table 5).
Streptococcus pneumoniae
Published reports from the country for resistance amongst S. pneumoniae strains are scant. However, a study [30] based on data from 2013 to 2014 reported high rates of resistance to penicillin and cotrimoxazole, and moderate resistance to ceftriaxone (Table 3). These findings are in agreement with antibiograms showing increasing resistance to penicillin between 2015 and 2018, and high rate of cotrimoxazole resistance between 2011 and 2018 (Table 5).
Antimicrobial resistance based on patients’ demographics
A few studies [18, 28, 31,32,33] disaggregated frequency of drug resistant isolates based on age and or gender. Jadoon et al. (2015) [31] reported higher frequency of ciprofloxacin resistant E. coli in males (61%) as compared to females (32.8%). Whereas, Ali et al. (2017) [18] reported fluoroquinolone resistant E. coli to be higher in female patients (75–83.3%), aged 1–15 years, compared to male patients (44.4%). In age groups ≥ 16 years similar frequency of fluoroquinolone resistant isolates was reported in both genders. Kalam et al. (2014) [32] reported higher frequency of MDR Gram negative bacteria in males (66%) compared to females (34.2%). Similarly, MRSA were found to be present at a higher frequency in adult males (70%) compared to female patients (30%) [28]. A study conducted in the Pediatric Intensive Care Unit (PICU) found that amongst male patients (aged 1 month–15 years) MDR Gram negative infection occurred most frequently in age group < 1 year (52.7%) [33]. Socioeconomic status of the patients was not reported in the reviewed studies.
Only two studies shared outcome data of patients with drug resistant infection. Kalam et al. (2014) [32] reported deaths in 46.2% of patients with Gram negative bacterial infection. Similarly, 42.9% of critically ill pediatric patients receiving intravenous polymyxin B with MDR Gram negative infection expired [33].
Discussion
Our data provides a window into the gradual emergence and spread of antimicrobial resistance in Pakistan from 2009 to 2018. During this period both published reports as well as laboratory based antibiogram data reveal increasing resistance to antimicrobial agents amongst GLASS priority pathogens in the country. The number of studies reporting resistance rates from children were few, but those available were consistent with findings from adult populations. These findings are also consistent with recent publications reporting increasing AMR in the region [73,74,75,76].
Widespread resistance to fluoroquinolones together with rapid increase in Carbapenem resistant Enterobacteriaceae (CRE) over the past decade is widely reported from South Asia [73, 74, 76,77,78,79]. Colistin is frequently used as a last resort antibiotic in such cases. However, the recommended susceptibility testing method for colistin has recently been revised. Therefore, while high colistin resistance is reported in the literature [75], given the concern that the reported rates may not be based on the recommended methods, colistin resistance data was not included.
The quality and standardization of antimicrobial sensitivity testing methodologies used in laboratories across the country vary. This is likely to be reflected in all AMR related data including from published studies as well as from antibiogram based information. Additionally, antibiograms submitted to PARN website are on a voluntary basis, therefore the data presented is limited by the information available for that year. Frequency of antimicrobial resistance in accordance with gender and age groups was reported in a few studies [18, 28, 31,32,33]. However published literature [80,81,82] indicate that age, gender, comorbid conditions and underlying factors such as: previous use of antibiotics and duration of in-dwelling catheters have a significant role. That for patients with urinary tract infection caused by E. coli, amikacin, nitrofurantoin and colistin, age and gender should be considered while prescribing antibacterials [81]. Similarly, Bruie et al. (2004) [80], found female patients to be at an increased risk of penicillin resistant S. pneumoniae infection. Further, their study indicates that presence of HIV increases the risk of AMR in pneumococcal infections. Such information can provide an evidence-base for physicians to avoid unnecessary antibiotics for critical patients.
Hospital settings including intensive care units (ICU) reported relatively high resistance to fluoroquinolones amongst K. pneumoniae and E. coli, as well as resistance amongst these organisms to cephalosporins and carbapenems with an increase in resistance being reported for K. pneumoniae. These data support published reports [83,84,85] enforcing concern about role of hospitals in contributing to spread of resistance.
The few community-based reports available from the country, endorsed hospital and laboratory-based findings for high AMR rates. These findings are also supported by a systematic review and meta-analysis reporting moderate to high levels of antimicrobial resistance amongst bacterial pathogens associated with community acquired paediatric bloodstream infections in low- and middle-income countries [86] as well as by increasing reports of community acquired AMR globally including from LMIC [23, 87,88,89]. The paucity of community-based AMR reports in our study points not only to a weakness in the AMR data being presented, but also to a dependence on hospital and laboratory-based studies for information on AMR in LMICs and to the challenges of capturing community-based information on AMR in these settings.
Surveillance using GLASS as well as PASS relies on information from select surveillance sites which is then combined as national data. While this system is valuable at the macro level, the combined data overlooks the granularity and local information required by treating physicians. As such antibiograms are a useful means of sharing antimicrobial resistance information at a local level. Therefore, a compilation of antibiogram data may also provide useful information at a national level. As the country moves towards strengthening AMR surveillance, contribution of published antibiograms should be explored towards supplementing national surveillance efforts.
It was encouraging to note that hospital and laboratory antibiogram based information from the PARN website agreed with, and complemented published reports well suggesting that such information constitutes an important source of AMR data, which needs to be harnessed, and utilized in national AMR analyses. We observed rare discrepancies between published data and antibiograms viz in the case of S. aureus. Higher median resistance in S. aureus in antibiograms is likely driven by a predominance of hospital and ICU data.
More importantly such antibiogram data also correlated well with the AMR information from Pakistan included in the GLASS report 2017–2018 [90]. This is not unexpected as GLASS data reported from many LMICs is primarily reliant on laboratory-based data as well.
National AMR surveillance efforts which provide information to the GLASS platform currently focuses on collecting resistance data on select organisms from specified enrolled sites. Such efforts while essential, have limitations; they include only select laboratories connected to the surveillance system, information supplied is limited to specific pathogens included in GLASS. As such localized resistance patterns and geographic distribution are difficult to assess. We therefore propose that national level data, such as that collected for GLASS, be supplemented with individual hospital antibiograms based information to inform sub-national AMR rates. This concept is similar to that of ResistanceMap [91]; an open-access online resource reporting resistance data from 66 countries from 1999 to 2017, and also to Resistancebank [92]; an online repository created in 2019 for surveillance data on animal antimicrobial resistance.
Based on our findings we propose similar initiatives at a national level offering a central platform for sharing antibiograms, allowing comparison of susceptibility rates at sub-national levels to create opportunities for information sharing, monitoring of AMR, and focused control efforts.
Conclusion
Data from both published studies and from antibiograms were complementary in showing high resistance to most antibacterials studied. These included third generation cephalosporins and carbapanems. Gaps in literature including paucity of information on AMR amongst peadiatric and community based populations, as well as a lack of studies exploring the association between patient demographic, underlying co-morbidities and resistance are highlighted by our study. We further show that antibiograms are a valuable tool to aid physicians in understanding resistance rates locally towards improving prescribing practices.
Availability of data and materials
All data generated or analysed during this study are provided in the manuscript and supplementary information files. PARN is openly available to the public. The papers can be accessed through the journal links on PubMed. Most of these articles are open access. Resistance rates can be also accessed through the referenced studies available on PubMed (https://pubmed.ncbi.nlm.nih.gov/). Antibiogram data can be accessed through the following URL: http://parn.org.pk/antimicrobial-data/.
Abbreviations
- AMR:
-
Antimicrobial resistance
- ARG:
-
Antimicrobial resistance genes
- CLSI:
-
Clinical Laboratory Standards Institute
- CRE:
-
Carbapenem resistant Enterobacteriaceae
- GDP:
-
Gross Domestic Product
- GLASS:
-
Global Antimicrobial Surveillance System
- PASS:
-
Pakistan AMR surveillance system
- NDM:
-
New Delhi Metallo-betalactamase
- XDR:
-
Extensively drug resistant
- MDR:
-
Multi drug resistant
- QC:
-
Quality control
- RCT:
-
Randomized control trial
- ICU:
-
Intensive Care Unit
- MICU:
-
Medical Intensive Care Unit
- MRSA:
-
Methicillin Resistant Staphylococcus aureus
- PICU:
-
Pediatric Intensive Care Unit
- LMIC:
-
Low Middle Income Countries
References
Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010;74(3):417–33.
Fodor A, Abate BA, Deak P, Fodor L, Gyenge E, Klein MG, Koncz Z, Muvevi J, Otvos L, Szekely G, et al. Multidrug resistance (MDR) and collateral sensitivity in bacteria, with special attention to genetic and evolutionary aspects and to the perspectives of antimicrobial peptides—a review. Pathogens. 2020;9(7):522.
Johnson AP, Woodford N. Global spread of antibiotic resistance: the example of New Delhi metallo-beta-lactamase (NDM)-mediated carbapenem resistance. J Med Microbiol. 2013;62(Pt 4):499–513.
Javaid N, Sultana Q, Rasool K, Gandra S, Ahmad F, Chaudhary SU, Mirza S. Trends in antimicrobial resistance amongst pathogens isolated from blood and cerebrospinal fluid cultures in Pakistan (2011–2015): a retrospective cross-sectional study. PLoS ONE. 2021;16(4):e0250226.
Hameed F, Khan MA, Muhammad H, Sarwar T, Bilal H, Rehman TU. Plasmid-mediated mcr-1 gene in Acinetobacter baumannii and Pseudomonas aeruginosa: first report from Pakistan. Rev Soc Bras Med Trop. 2019;52:e20190237.
Imtiaz W, Syed Z, Rafaque Z, Andrews SC, Dasti JI. Analysis of antibiotic resistance and virulence traits (genetic and phenotypic) in Klebsiella pneumoniae clinical isolates from Pakistan: identification of significant levels of carbapenem and colistin resistance. Infect Drug Resist. 2021;14:227–36.
Bilal H, Hameed F, Khan MA, Khan S, Yang X, Rehman TU. Detection of mcr-1 gene in extended-spectrum β-lactamase-producing Klebsiella pneumoniae from human urine samples in Pakistan. Jundishapur J Microbiol. 2020;13(4):e96646.
GARP. Situational analysis report on antimicrobial resistance in Pakistan. Findings and recommendations for antibiotics use and resistance. CDDEP; 2018.
NIH. Pakistan antimicrobial resistance surveillance system surveillance report: surveillance report 2017–2018. In: edited by Anjum J, Nisar M. 1st edn. Islamabad: National Institute of Health Sciences; 2018. Available from: https://www.nih.org.pk/wp-content/uploads/2020/03/Report_2017-2018-NIH-Final.pdf. Last Accessed 4 Apr 2021.
Pakistan Population 2021 (Live) https://worldpopulationreview.com/countries/pakistan-population.
Bilal H, Zhang G, Rehman T, Han J, Khan S, Shafiq M, Yang X, Yan Z, Yang X. First report of blaNDM-1 bearing IncX3 plasmid in clinically isolated ST11 Klebsiella pneumoniae from Pakistan. Microorganisms. 2021;9(5):951.
Klemm EJ, Wong VK, Dougan G. Emergence of dominant multidrug-resistant bacterial clades: lessons from history and whole-genome sequencing. Proc Natl Acad Sci USA. 2018;115(51):12872–7.
W.H.O. Global antimicrobial resistance surveillance system manual for early implementation. Geneva: W.H.O; 2015. Available from: https://apps.who.int/iris/handle/10665/188783. Last Accessed 4 Apr 2021.
Pakistan Antimicrobial Resistance Network (PARN). Available from: http://parn.org.pk/antimicrobial-data/. Last Accessed 4 Apr 2021.
Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–81.
Seale AC, Gordon NC, Islam J, Peacock SJ, Scott JAG. AMR surveillance in low and middle-income settings—a roadmap for participation in the Global Antimicrobial Surveillance System (GLASS). Wellcome Open Res. 2017;2:92.
StataCorp. Stata statistical software: release 13. College Station, TX: StataCorp LP; 2013.
Ali I, Shabbir M, Noor Ul I. Antibiotics susceptibility patterns of uropathogenic E. coli with special reference to fluoroquinolones in different age and gender groups. J Pak Med Assoc. 2017;67(8):1161–5.
Qadeer A, Akhtar A, Ain QU, Saadat S, Mansoor S, Assad S, Ishtiaq W, Ilyas A, Khan AY, Ajam Y. Antibiogram of medical intensive care unit at tertiary care hospital setting of Pakistan. Cureus. 2016;8(9):e809.
Bushra R, Sial AA, Rizvi M, Shafiq Y, Aslam N, Bano N. Report: sensitivity pattern of ceftriaxone against different clinical isolates. Pak J Pharm Sci. 2016;29(1):249–53.
Abdullah FE, Haider F, Fatima K, Irfan S, Iqbal MS. Enteric fever in Karachi: current antibiotic susceptibility of Salmonellae isolates. J Coll Physicians Surg Pak. 2012;22(3):147–50.
Qamar FN, Azmatullah A, Kazi AM, Khan E, Zaidi AK. A three-year review of antimicrobial resistance of Salmonella enterica serovars Typhi and Paratyphi A in Pakistan. J Infect Dev Ctries. 2014;8(8):981–6.
Klemm EJ, Shakoor S, Page AJ, Qamar FN, Judge K, Saeed DK, Wong VK, Dallman TJ, Nair S, Baker S, et al. Emergence of an extensively drug-resistant Salmonella enterica Serovar Typhi clone harboring a promiscuous plasmid encoding resistance to fluoroquinolones and third-generation cephalosporins. mBio. 2018. https://doi.org/10.1128/mBio.00105-18.
Qamar FN, Yousafzai MT, Khalid M, Kazi AM, Lohana H, Karim S, Khan A, Hotwani A, Qureshi S, Kabir F, et al. Outbreak investigation of ceftriaxone-resistant Salmonella enterica serotype Typhi and its risk factors among the general population in Hyderabad, Pakistan: a matched case-control study. Lancet Infect Dis. 2018;18(12):1368–76.
Khan E, Jabeen K, Ejaz M, Siddiqui J, Shezad MF, Zafar A. Trends in antimicrobial resistance in Shigella species in Karachi, Pakistan. J Infect Dev Ctries. 2009;3(10):798–802.
Raza A, Mahboob A, Nizammudin S, Nazeer SH, Sultan F. Shigella infections: a two year experience in cancer patients. J Pak Med Assoc. 2016;66(1):37–9.
Malik N, Butt T, Arfan ul B. Frequency and antimicrobial susceptibility pattern of methicillin resistant Staphylococcus aureus. J Coll Physicians Surg Pak. 2009;19(5):287–90.
Siddiqui T, Muhammad IN, Khan MN, Naz S, Bashir L, Sarosh N, Masood R, Ali A, Fatima S, Naqvi T. MRSA: prevalence and susceptibility pattern in health care setups of Karachi. Pak J Pharm Sci. 2017;30(6(Supplementary)):2417–21.
Ahmad MK, Asrar A. Prevalence of methicillin resistant Staphylococcus aureus in pyogenic community and hospital acquired skin and soft tissues infections. J Pak Med Assoc. 2014;64(8):892–5.
Shah SN, Ullah B, Basit A, Begum A, Tabassum A, Zafar S, Saleha S. Prevalence and susceptibility patterns of bacteria causing respiratory tract infections in North Waziristan, Pakistan. Pak J Pharm Sci. 2016;29(2 Suppl):701–6.
Jadoon RJ, Jalal-ud-Din M, Khan SAE. E. coli resistance to ciprofloxacin and common associated factors. J Coll Physicians Surg Pak. 2015;25(11):824–7.
Kalam K, Qamar F, Kumar S, Ali S, Baqi S. Risk factors for carbapenem resistant bacteraemia and mortality due to gram negative bacteraemia in a developing country. J Pak Med Assoc. 2014;64(5):530–6.
Siddiqui NU, Qamar FN, Jurair H, Haque A. Multi-drug resistant gram negative infections and use of intravenous polymyxin B in critically ill children of developing country: retrospective cohort study. BMC Infect Dis. 2014;14:626.
Khan MS, Siddiqui SZ, Haider S, Zafar A, Zafar F, Khan RN, Afshan K, Jabeen A, Khan MS, Hasan R. Infection control education: impact on ventilator-associated pneumonia rates in a public sector intensive care unit in Pakistan. Trans R Soc Trop Med Hyg. 2009;103(8):807–11.
Akhtar N. Hospital acquired infections in a medical intensive care unit. J Coll Physicians Surg Pak. 2010;20(6):386–90.
Khan E, Ejaz M, Zafar A, Jabeen K, Shakoor S, Inayat R, Hasan R. Increased isolation of ESBL producing Klebsiella pneumoniae with emergence of carbapenem resistant isolates in Pakistan: report from a tertiary care hospital. J Pak Med Assoc. 2010;60(3):186–90.
Miyan Z, Fawwad A, Sabir R, Basit A. Microbiological pattern of diabetic foot infections at a tertiary care center in a developing country. J Pak Med Assoc. 2017;67(5):665–9.
Saeed MA, Haque A, Ali A, Mohsin M, Bashir S, Tariq A, Afzal A, Iftikhar T, Sarwar Y. A profile of drug resistance genes and integrons in E. coli causing surgical wound infections in the Faisalabad region of Pakistan. J Antibiot (Tokyo). 2009;62(6):319–23.
Jan WA, Khan MS, Jehanzeb M, Muazam. Surgical site infection and pattern of antibiotic use in a tertiary care hospital in Peshawar. JPMA. 2010;22(3):141–5.
Parveen A, Sultan F, Raza A, Zafar W, Nizamuddin S, Mahboob A, Saleem S, Nazeer SH. Bacteraemia caused by Escherichia coli in cancer patients at a specialist center in Pakistan. J Pak Med Assoc. 2015;65(12):1271–6.
Saad U, Anwar S, Kahara UZ, Siddiqui M, Hina H. Antimicrobial susceptibility of intra-abdominal infection isolates from a tertiary care hospital in Karachi. J Ayub Med Coll Abbottabad. 2016;28(3):568–71.
Shabbir S, Jamil S, Hafiz S. Pattern of polymicrobial isolates and antimicrobial susceptibility from blood. J Coll Physicians Surg Pak. 2016;26(7):585–8.
Jadoon SA, Ahmed A, Irshad R. Spectrum of bacterial culture and drug sensitivity vs resistance in uncomplicated urinary tract infection. J Ayub Med Coll Abbottabad. 2018;30(3):432–8.
Zahid KF, Hafeez H, Afzal A. Bacterial spectrum and susceptibility patterns of pathogens in adult febrile neutropenic patients: a comparison between two time periods. J Ayub Med Coll Abbottabad. 2009;21(4):146–9.
Hasan B, Perveen K, Olsen B, Zahra R. Emergence of carbapenem-resistant Acinetobacter baumannii in hospitals in Pakistan. J Med Microbiol. 2014;63(Pt 1):50–5.
Tahseen U, Talib MT. Acinetobacter infections as an emerging threat in intensive care units. J Ayub Med Coll Abbottabad. 2015;27(1):113–6.
Ahmed A, Hussain S, Ijaz T, Hashemy I. Susceptibility of methicillin-resistant Staphylococcus aureus and enterococci to teicoplanin in Pakistan: the MRSET study. JPMA. 2014;64:256–9.
Afridi FI, Zeb M, Hussain A, Farooqi BJ, Murtuza G. Inducible clindamycin resistance in Staphylococcus species. J Coll Physicians Surg Pak. 2014;24(7):481–4.
Liaqat F, Sheikh AA, Nazir J, Hussain T, Rabbani M, Shaheen AY, Muhammad J. Report-isolation identification and control of vancomycin resistant Staphylococcus aureus. Pak J Pharm Sci. 2015;28(3):997–1004.
Rafiq MS, Rafiq MI, Khan T, Rafiq M, Khan MM. Effectiveness of simple control measures on methicillin-resistant Staphylococcus aureus infection status and characteristics with susceptibility patterns in a teaching hospital in Peshawar. J Pak Med Assoc. 2015;65:915–20.
Ahmad S, Bacha N, Bakht J, Ahmed J. Characterization of pathogens involved in ventilator associated pneumonia in surgical and medical intensive care units—a single center experience. Pak J Pharm Sci. 2017;30(6):2091–9.
Abdullah FE, Memon AA, Bandukda MY, Jamil M. Increasing ciprofloxacin resistance of isolates from infected urines of a cross-section of patients in Karachi. BMC Res Notes. 2012;5:696.
Abdullah FE, Mushtaq A, Irshad M, Rauf H, Afzal N, Rasheed A. Current efficacy of antibiotics against Klebsiella isolates from urine samples—a multi-centric experience in Karachi. Pak J Pharm Sci. 2013;26(1):11–5.
Fatima S, Muhammad IN, Usman S, Jamil S, Khan MN, Khan SI. Incidence of multidrug resistance and extended-spectrum beta-lactamase expression in community-acquired urinary tract infection among different age groups of patients. Indian J Pharmacol. 2018;50(2):69–74.
Ahmad W, Jamshed F, Ahmad W. Freequncy of Escherichia coli in patients with community acquired urinary tract infection and their resistance pattern against some commonly used anti bacterials. J Ayub Med Coll Abbottabad. 2015;27(2):333–7.
Zehra NM, Irfan F, Mirza IA, Imtiaz A, Nadeem S, Hameed F. Current trends of antimicrobial susceptibility of typhoidal Salmonellae isolated at Tertiary Care Hospital. J Coll Physicians Surg Pak. 2017;27(11):690–2.
Qamar FN, Yousafzai MT, Sultana S, Baig A, Shakoor S, Hirani F, Wassay A, Khushboo S, Mehmood J, Freeman A, et al. A retrospective study of laboratory-based enteric fever surveillance, Pakistan, 2012–2014. J Infect Dis. 2018;218(suppl_4):S201-205.
Nizamuddin S, Jabeen K, Zafar A. Evaluation of predominant Neisseria gonorrhoeae strain types and its correlation with fluoroquinolone resistance in Pakistan. J Pak Med Assoc. 2011;61(5):446–9.
Jabeen K, Nizamuddin S, Irfan S, Khan E, Malik F, Zafar A. Increasing trend of resistance to penicillin, tetracycline, and fluoroquinolone resistance in Neisseria gonorrhoeae from Pakistan (1992–2009). J Trop Med. 2011;2011:960501.
Sethi SGD, Bala M, Dorji D, Ibrahim M, Kausar JK, Unemo M. Antimicrobial susceptibility and genetic characteristics of Neisseria gonorrhoeae isolates from India, Pakistan and Bhutan in 2007–2011. BMC Infect Dis. 2013;13:35–9.
Jabeen K, Bhawan Mal P, Khan E, Chandio S, Jacobsson S, Unemo M. Antimicrobial resistance and Neisseria gonorrhoeae multiantigen sequence typing (NG-MAST) genotypes in N. gonorrhoeae during 2012–2014 in Karachi, Pakistan. BMC Infect Dis. 2016;16:353.
Saleem AF, Qamar FN, Shahzad H, Qadir M, Zaidi AK. Trends in antibiotic susceptibility and incidence of late-onset Klebsiella pneumoniae neonatal sepsis over a six-year period in a neonatal intensive care unit in Karachi, Pakistan. Int J Infect Dis. 2013;17(11):e961–5.
Muhammad Z, Ahmed A, Hayat U, Wazir MS, Rafiyatullah, Waqas H. Neonatal sepsis: causative bacteria and their resistance to antibiotics. J Ayub Med Coll Abbottabad. 2010;22(4):33–36.
Najeeb S, Gillani S, Rizvi SK, Ullah R, ur Rehman A. Causative bacteria and antibiotic resistance in neonatal sepsis. J Ayub Med Coll Abbottabad. 2012;24(3–4):131–34.
Ullah O, Khan A, Ambreen A, Ahmad I, Akhtar T, Gandapor AJ, Khan AM. Antibiotic sensitivity pattern of bacterial isolates of neonatal septicemia in Peshawar, Pakistan. Arch Iran Med. 2016;19(12):866–9.
Khalil U, Younus M, Asghar N, Siddiqui F, Gomez-Duarte OG, Wren BW, Bokhari H. Phenotypic and genotypic characterization of enteroaggregative Escherichia coli isolates from pediatric population in Pakistan. APMIS. 2016;124(10):872–80.
Younas M, Siddiqui F, Noreen Z, Bokhari SS, Gomez-Duarte OG, Wren BW, Bokhari H. Characterization of enteropathogenic Escherichia coli of clinical origin from the pediatric population in Pakistan. Trans R Soc Trop Med Hyg. 2016;110(7):414–20.
Anwar M, Ejaz H, Zafar A, Hamid H. Phenotypic detection of metallo-beta-lactamases in carbapenem resistant Acinetobacter baumannii isolated from pediatric patients in Pakistan. J Pathog. 2016;2016:8603964.
Indhar F, Durrani MA, Bux A, Sohail M. Carbapenemases among Acinetobacter species isolated from NICU of a tertairy care hospital in Karachi. J Pak Med Assoc. 2017;67(10):1547–51.
Khan MI, Soofi SB, Ochiai LR, Khan MJ, Shah Muhammad Sahito SM, Mohammad Atif Habib MA, Puri MK, von Seidlein L, Park JK, Young YA, Ali M, Nizami SQ, Acosta CJ, Sack RB, Clemens JD, Bhutta ZA. Epidemiology, clinical presentation, and patterns of drug resistance of Salmonella Typhi in Karachi, Pakistan. J Infect Dev Ctries. 2012;6(10):704–14.
Owais A, Sultana S, Zaman U, Rizvi A, Zaidi AK. Incidence of typhoid bacteremia in infants and young children in southern coastal Pakistan. Pediatr Infect Dis J. 2010;29(11):1035–9.
Mir F, Tikmani SS, Shakoor S, Warraich HJ, Sultana S, Ali SA, Zaidi AK. Incidence and etiology of omphalitis in Pakistan: a community-based cohort study. J Infect Dev Ctries. 2011;5(12):828–33.
Malchione MD, Torres LM, Hartley DM, Koch M, Goodman JL. Carbapenem and colistin resistance in Enterobacteriaceae in southeast Asia: review and mapping of emerging and overlapping challenges. Int J Antimicrob Agents. 2019;54(4):381–99.
Browne AJ, Kashef Hamadani BH, Kumaran EAP, Rao P, Longbottom J, Harriss E, Moore CE, Dunachie S, Basnyat B, Baker S, et al. Drug-resistant enteric fever worldwide, 1990 to 2018: a systematic review and meta-analysis. BMC Med. 2020;18(1):1.
Pormohammad A, Mehdinejadiani K, Gholizadeh P, Nasiri MJ, Mohtavinejad N, Dadashi M, Karimaei S, Safari H, Azimi T. Global prevalence of colistin resistance in clinical isolates of Acinetobacter baumannii: a systematic review and meta-analysis. Microb Pathog. 2020;139:103887.
Chaurasia S, Sivanandan S, Agarwal R, Ellis S, Sharland M, Sankar MJ. Neonatal sepsis in South Asia: huge burden and spiralling antimicrobial resistance. BMJ. 2019;364:k5314.
Britto CD, Dyson ZA, Mathias S, Bosco A, Dougan G, Jose S, Nagaraj S, Holt KE, Pollard AJ. Persistent circulation of a fluoroquinolone-resistant Salmonella enterica Typhi clone in the Indian subcontinent. J Antimicrob Chemother. 2020;75(2):337–41.
Chung The H, Boinett C, Pham Thanh D, Jenkins C, Weill FX, Howden BP, Valcanis M, De Lappe N, Cormican M, Wangchuk S, et al. Dissecting the molecular evolution of fluoroquinolone-resistant Shigella sonnei. Nat Commun. 2019;10(1):4828.
Ingle DJ, Levine MM, Kotloff KL, Holt KE, Robins-Browne RM. Dynamics of antimicrobial resistance in intestinal Escherichia coli from children in community settings in South Asia and sub-Saharan Africa. Nat Microbiol. 2018;3(9):1063–73.
Buie KA, Klugman KP, von Gottberg A, Perovic O, Karstaedt A, Crewe-Brown HH, Madhi SA, Feldman C. Gender as a risk factor for both antibiotic resistance and infection with pediatric serogroups/serotypes, in HIV-infected and -uninfected adults with pneumococcal bacteremia. J Infect Dis. 2004;189(11):1996–2000.
Hossain A, Hossain SA, Fatema AN, Wahab A, Alam MM, Islam MN, Hossain MZ, Ahsan GU. Age and gender-specific antibiotic resistance patterns among Bangladeshi patients with urinary tract infection caused by Escherichia coli. Heliyon. 2020;6(6):e04161.
Lee DS, Choe HS, Kim HY, Yoo JM, Bae WJ, Cho YH, Kim SW, Han CH, Bae SR, Jang H, et al. Role of age and sex in determining antibiotic resistance in febrile urinary tract infections. Int J Infect Dis. 2016;51:89–96.
Manandhar S, Zellweger RM, Maharjan N, Dongol S, Prajapati KG, Thwaites G, Basnyat B, Dixit SM, Baker S, Karkey A. A high prevalence of multi-drug resistant Gram-negative bacilli in a Nepali tertiary care hospital and associated widespread distribution of Extended-Spectrum Beta-Lactamase (ESBL) and carbapenemase-encoding genes. Ann Clin Microbiol Antimicrob. 2020;19(1):48.
Roberts T, Limmathurotsakul D, Turner P, Day NPJ, Vandepitte WP, Cooper BS. Antimicrobial-resistant Gram-negative colonization in infants from a neonatal intensive care unit in Thailand. J Hosp Infect. 2019;103(2):151–5.
Shrestha R, Koju P, Xinliang Liu, Maharjan B, Madhup S, Shah P, Hao L. Health care associated infection and trend of antimicrobial resistance in tertiary care hospital—a study in low income setting. Kathmandu Univ Med J. 2019;68(4):329–35.
Droz N, Hsia Y, Ellis S, Dramowski A, Sharland M, Basmaci R. Bacterial pathogens and resistance causing community acquired paediatric bloodstream infections in low- and middle-income countries: a systematic review and meta-analysis. Antimicrob Resist Infect Control. 2019;8:207.
Mohamed ES, Khairy RMM, Abdelrahim SS. Prevalence and molecular characteristics of ESBL and AmpC beta-lactamase producing Enterobacteriaceae strains isolated from UTIs in Egypt. Antimicrob Resist Infect Control. 2020;9(1):198.
Steinig EJ, Duchene S, Robinson DA, Monecke S, Yokoyama M, Laabei M, Slickers P, Andersson P, Williamson D, Kearns A, et al. Evolution and global transmission of a multidrug-resistant, community-associated methicillin-resistant Staphylococcus aureus Lineage from the Indian Subcontinent. mBio. 2019. https://doi.org/10.1128/mBio.01105-19.
George CRR, Enriquez RP, Gatus BJ, Whiley DM, Lo YR, Ishikawa N, Wi T, Lahra MM. Systematic review and survey of Neisseria gonorrhoeae ceftriaxone and azithromycin susceptibility data in the Asia Pacific, 2011 to 2016. PLoS ONE. 2019;14(4):e0213312.
W.H.O. GLASS early implementation report: 2017–2018. Geneva: World Health Organization; 2019. Available from: https://apps.who.int/iris/bitstream/handle/10665/279656/9789241515061-eng.pdf. Last Accessed 4 Apr 2021.
CDDEP. Antibiotic resistance. Resistance Map. Available from: https://resistancemap.cddep.org/DRI.php. Last Accessed 4 Apr 2021.
Resistancebank. Resistancebank. Available from: https://resistancebank.org/. Last Accessed 4 Apr 2021.
Acknowledgements
We would like to thank Mr. Faisal Malik for his help with editing the tables, Dr Kauser Jabeen for her insight during the preliminary review, and Khalid Wahab for assistance with lab antibiogram data. We would also like to thank Health Security Partners, USA, for partial salary support for Dania K. Saeed.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
Study conception and design: RH, SS. Literature review: RH, and DKS. Writing, Data Analysis, and Review: RH, SS, JF, and DKS. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1.
Terms used for searching peer reviewed literature from electronic database Medline (PubMed).
Additional file 2.
Antimicrobial resistance rates (shown as percent resistance) reported in hospital-based studies (2006–2017).
Additional file 3.
Antimicrobial resistance rates (shown as percent resistance) from laboratory surveillance and community-based literature review (2009–2018).
Additional file 4.
Antimicrobial resistance rates (shown as percent resistance) amongst pediatric population based on literature review (2010–2017).
Additional file 5.
Antimicrobial resistance rates from laboratory based antibiograms (2006–2018).
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Saeed, D.K., Farooqi, J., Shakoor, S. et al. Antimicrobial resistance among GLASS priority pathogens from Pakistan: 2006–2018. BMC Infect Dis 21, 1231 (2021). https://doi.org/10.1186/s12879-021-06795-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-021-06795-0