Abstract
Background
Leishmaniasis is one of the most important infectious diseases affecting the Colombian National Army due to the high number of reported cases and exposure throughout military operations in endemic areas. The main aim of this study was to estimate the geographical distribution along with the genetic diversity and treatment outcome of Leishmania species in Colombian military personnel.
Methods
Skin lesion samples by smear and aspirate were collected in 136 patients having parasitological cutaneous leishmaniasis (CL) diagnosis. DNA was extracted, the nuclear marker heat shock protein 70 (HSP70) was amplified by PCR and sequenced. Leishmania species were identified by BLASTn. The geo-spatial distribution of the identified parasites was determined according to the possible site of infection. Gene tree was constructed by maximum likelihood (ML), diversity indices (π, h) were estimated and haplotype network was constructed under the Templeton-Crandall-Sing algorithm in order to determine the geographic relationships of the genetic variants of Leishmania species circulating in Colombian military population.
Results
The species were identified in 77.94% of the samples, with a predominance of L. braziliensis (65.09%), followed by L. panamensis (31.13%), L. naiffi by the first time reported in Colombia in two patients (1.89%) as well as L. lindenbergi in a single patient (0.945%) with possible infection in the municipality of Miraflores, Guaviare and L. infantum in a single patient (0.945%) notified with CL in the municipality of Tumaco, Nariño. The phylogenetic analysis was consistent according to bootstrap, showing four strongly differentiated clades.
Conclusions
The geo-spatial distribution suggested that L. braziliensis has a greater abundance, while L. panamensis has a greater dispersion. The phylogenetic relationships of Leishmania species in Colombian military personnel was estimated with the confirmation of two new species circulating without prior report in the country and a species with no background for CL in the Colombian army. A substantial genetic diversity of Leishmania braziliensis was defined. This study contributes through the understanding of the molecular epidemiology to the CL transmission in Colombia.
Similar content being viewed by others
Background
The Leishmania parasites are the etiological agents of leishmaniasis, a tropical vector-borne disease (VBD) transmitted to humans through a bite of infected hematophagous female insects of the Psychodidae subfamily [1]. There are approximately 20 Leishmania species that can generate different clinical manifestations such as cutaneous leishmaniasis (CL), mucocutaneous leishmaniasis (MCL) or visceral leishmaniasis (VL) [2, 3]. Seventeen countries of the Americas reported to the Pan-American Health Organization (PAHO) and the World Health Organization (WHO) a total of 989.096 CL and MCL cases from 2001 to 2018 with a decreasing trend with some exceptions. Colombia is the 2nd (13.82%) after Brazil (35.69%) with the highest number of CL and MCL cases as well as the eighth (incidence: 26.17) with the highest disease incidence in the continent according to 2018 data [4]. A total of 6426 cases of leishmaniasis were reported during 2018 in Colombia in its three clinical forms corresponding to 98.3% (6319) CL, 1.4% (90) MCL, and 0.3% (17) VL cases [5]. CL is currently the infectious disease that most affects the Colombian National Army, where 41.774 cases have been reported between 2008 and 2019, followed by 575 cases of MCL in active military personnel, according to data from the National Surveillance System in Public Health (SIVIGILA) and Operational Health department of the Army Health Directorate [6].
Molecular markers such as the heat shock protein gene (HSP70) can be sequenced to identify by barcoding the Leishmania species [7, 8]. Studies conducted in Colombia have shown that in civil population a predominance of L. braziliensis (86.42%) exists, followed by L. panamensis (11.11%) by using of PCR-RFLP of the nuclear marker HSP70 [9], while others found a predominance of L. panamensis (61.3%), followed by L. braziliensis (23.1%) by sequencing of the mitochondrial marker Cytb [7] as well as a predominance of L. panamensis (56.3%) followed by L. braziliensis (36.8%), L guyanensis and L. amazonensis by PCR-RFLP and sequencing of mini-exon and HSP70 markers [10]. Regarding the military population, it has been reported to date the highest predominance of L. braziliensis (95.4%), followed by L. guyanensis (2.3%) throughout PCR-RFLP [11], compared to a predominance of L. braziliensis (61.1%), followed by L. panamensis (33.5%) using Cytb and HSP70 markers sequencing [8].
Colombia is one of the countries with the greatest richness of human pathogenic Leishmania species (nine species) [7, 12, 13], moving the CL prevalence from sylvatic to domestic cycles in most of the country [14], and the military activities in sylvatic areas are considered one of the factors related to changes in the dynamics of disease transmission [8]. Due to variations in the relative abundances of Leishmania species range in different studies, it is worth conducting studies in order to understand the factors that determine the molecular epidemiology of Leishmania parasites in relation to the geographical origins [7, 8]. Therefore, the aim of this study was to determine the geographical distribution and genetic diversity of Leishmania species in military personnel with CL.
Methods
Sampling size
A sample size of 136 patients from leishmaniasis centers in Bonza, Boyacá (89 patients) and health battalion (BASAN) in Bogotá (47 patients) was estimated in Epi Info v7.2.2.6 (https://www.cdc.gov/epiinfo/index.html), with 95% CI and an expected sampling error of 5% based on the average number of patients treated for one year in these military establishments. Two groups of patients were defined: first group made up of those who received first-line treatment with meglumine antimoniate and the second group for those who had presented therapeutic failure with meglumine antimoniate and initiated second-line treatment with pentamidine isothionate. The study had as inclusion criteria patients who participated voluntarily from male military population, older than 18 years old, with a parasitological diagnosis of CL and where lesion size larger than 1 cm in diameter and without clinical evidence of bacterial or fungal infection at the beginning of the study. As an exclusion criterion was considered patients with facial, genitals lesions and / or presenting comorbidities as pathologies where treatment is contraindicated.
Sampling, storing and medical resolution
In the case of smear samples to identify Leishmania species by barcoding, hand ischemia was performed using thumb-index fingers, scraped off the base and center of the ulcer with a sterile lancet [15]. Smear samples obtained were deposited in a screw cap vial with 250 μL of sterile saline solution. It was subsequently stored at − 70 °C until processing.
The clinical-epidemiological record was filled out by all of the patients with variables such as age, number of lesions, disease evolution, internal and external areas of lesions and municipality of possible infection. Once the patients were diagnosed, they were treated with the first line or second line treatment.
DNA extraction, PCR, purification and sequencing
DNA extraction was carried out with the Invisorb® Spin Universal commercial kit - Stratec molecular (Berlin, Germany) according to the manufacturer’s indications. HSP70F (5 ‘AGG TGA AGG CGA CGA ACG 3’) and HSP70R (5 ‘CGC TTG TCC ATC TTT GCG TC 3’) primers were used to amplify a 337 bp partial region of HSP70 [16]. PCR mix for HSP70 consisted Master Mix Green Promega 1X PCR (Madison-WI, USA), 5 μM of each primer and 3 μL of DNA for a final volume of 15 μL. The thermal profile consisted of a pre-denaturation at 94 °C for 5 min, followed by 40 cycles with denaturation at 94 °C for 1 min, annealing at 58 °C for 1 min and elongation at 72 °C for 1 min. Then a final extension at 72 °C for 10 min [16]. All PCR products were verified by 1% agarose gel, stained with Gel Red™ 1000X and run at 80 V for 40 min in a horizontal electrophoresis system with TAE 1X. Finally, PCR products were subsequently purified by ExoSAP-IT and sequenced by the terminal dideoxy method using Big Dye chemistry v 3.1 within an AB3730xI automatic sequencer in Macrogen Korea.
Sequence editing and species identification
HSP70 sequences were edited manually using Geneious Pro 4.8.1 software (http://www.geneious.com) [17]. Sequence alignment was performed under the MUSCLE algorithm [18] and identity of sequences was confirmed with BLASTn in NCBI (http://blast.ncbi.nlm.nih.gov), where the Leishmania species was identified by the similarity between study sequences and others deposited in GenBank. All haplotypes were identified using MacClade software v 4.0.8 [19, 20]. A geo-referenced database was constructed to address the spatial distribution of identified Leishmania species. Sample data belong to the possible site of infection, as reported by the patient. Distribution and frequency of the parasite species was mapped by using ArcGIS10.3 [21].
Phylogenetic analysis
Sequences were obtained from GenBank for the phylogenetic analyses. The following sequences were downloaded for the phylogenetic reconstruction: Leishmania major (FN395023 and HF586344), L. aethiopica (FN395018 and FN395019), L. tropica (FN395025 and FN395026), L donovani (FN395027 and HF586385), L. mexicana (EU599091, HF586401 and FN395038), L. amazonensis (HF586353, LN907831 and MF344846), L. lainsoni (FN395047, FN395048 and LN907839), L. peruviana (EU599089, FN395044 and FR872765), L. guyanensis (FN395052, LN907836 and MG029128) and L. shawi (GU071177 and MG029127) [22,23,24,25,26,27]. Additionally, sequences FN395039, FR715987, GU071173, EU599094, FN395055, HF586359, FN395056, FR872767, MG029126, FN395031, FN395032, HF586350 and MG029124 were used as reference species for L. braziliensis, L. panamensis, L. naiffi, L. lindenbergi and L. infantum respectively [22, 23, 26,27,28,29,30]. Phylogenetic relationships between Leishmania species were inferred by maximum likelihood (ML) with the evolutionary model of Generalized-Time-Reversible γ + Invariant (GTR γ + I) substitution and 1000 bootstrap replicates using the RAxML-HPC BlackBox software on the CIPRES Science Gateway portal (http://www.phylo.org/). Finally, gene tree in a circular polar form was edited in FigTree [31].
Genetic diversity and haplotype network
Nucleotide (π) and haplotype (h) diversity indices were estimated in DNAsp v5.0 [32]. Haplotype network based on HSP70 for Leishmania braziliensis was constructed using the TCS method in Popart software [33] in order to determine nucleotide changes between genetic variants of Leishmania braziliensis in Colombia. All sequences of each haplotype were assigned to a specific biogeographical region for Colombia (Additional file 1).
Statistical analysis
All descriptive and univariate statistical analyzes for patients were developed in IBM SPSS Statistics v25. Non-parametric test was used when the data did not follow Gaussian distribution according to the Kolmogorov-Smirnov test before and after transforming them using base 10 Logarithm. All variables (age, number of lesions, disease evolution, internal and external areas of lesions) were analyzed according to their categorical or quantitative nature and independence of data and were analyzed by the Mann Whitney U test (MWU) and Chi square (X2). The variables analyzed in this study were infectious species (L. braziliensis and L. panamensis), type of treatment (meglumine antimoniate and pentamidine isethionate), internal and external area of lesions, number of lesions, disease evolution (months), age and medical resolution. All tests were two-tailed, and the results were considered significant (*) when p < 0.05 and highly significant (**) when p < 0.01.
Results
Geographical distribution patterns
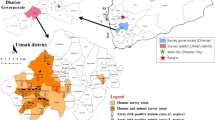
The 77.94% (106/136) of the patient samples in the study were successfully amplified and sequenced (106/136). The species with the highest relative abundance was L. braziliensis with 65.09% (69/106), followed by L. panamensis with 31.13% (33/106), L. naiffi with 1.89% (2/106), L. lindenbergi with 0.945% (1/106) and L. infantum with 0.945% (1/106). L. braziliensis was reported in eight departments with exclusive distribution in the Amazon, Andean and Orinoco regions, while L. panamensis had a wider distribution with report in eleven departments representing four biogeographical regions of Colombia (Amazon, Andean, Caribbean and Pacific) with exception of Insular and Orinoco regions. L. braziliensis and L. panamensis were identified in reported infected samples from departments such as Antioquia, Cundinamarca, Nariño and Norte de Santander. L. naiffi and L. lindenbergi were identified by the first time through sequencing for Colombia in the municipality of Miraflores, Guaviare, being the only department with report of four Leishmania species in this study. Likewise, L. infantum was identified by HSP70 in the BAS131 patient with CL in the municipality of Tumaco, Nariño department (Fig. 1).
Distribution patterns and relative abundance of Leishmania spp. circulating within National Army of Colombia. 65.09% (n = 69) of the barcoding identified species corresponded to L. braziliensis, while 31.13% (n = 33) to L. panamensis, only 1.89% (n = 2) to L. naiffi, 0,945% (n = 1) to L. lindenbergi and 0.945% (n = 1) to L. infantum. Barcoding of the parasite species was carried out by BLASTn with a percentage identity greater than 98% from HSP70 (106/136) sequences obtained by PCR amplification and terminal dideoxy paired end sequencing for skin smear samples. 106 skin smear samples of patients were analyzed within BONZA, Boyacá (n = 74) and BASAN, Bogotá (n = 32). (Own elaboration)
Phylogenetic relationships
Gene tree showed phylogenetic consistency for the barcoding confirmation of Leishmania species circulating in Colombia (Fig. 2). Bootstrap was reported in branches of main Leishmania clades within gene tree based on HSP70 (Fig. 2). Reciprocal monophyly was defined among all clades, as well as total concordance with NCBI BLASTn barcoding. In the gene tree was found BON32 into L. lindenbergi cluster and BON48, BON70, BON71 and BON96 within L. naiffi cluster.
Phylogenetic reconstruction by maximum likelihood of Leishmania spp. under GTR γ + I evolutionary model according HSP70 gene. Phylogeny in a circular polar form shows Bootstrap support in branches with percentages of 100% or > 53%. L. naiffi is highlighted in black, L. lindenbergi in pink, L. braziliensis in red wine, L. panamensis in dark green and L. infantum in yellow. Outgroup: Leishmania major. Sister groups: L. aethiopica, L. tropica, L donovani, L. mexicana, L. amazonensis, L. lainsoni, L. peruviana, L. guyanensis and L. shawi. The numbers in the branches tip are sample codes
Genetic diversity and haplotype network
Two haplotypes were found because of two variable sites, a nucleotide diversity π = 0.110% (+/− 0.0877%) and a haplotype diversity h = 0.111 (+/− 0.0498) for L. braziliensis by HSP70 (NCBI GenBank accession numbers MT543301 and MT543302). No genetic variants were found for L. panamensis, L. naiffi, L. infantum and L. lindenbergi by using HSP70 (NCBI GenBank accession numbers MT543303 – MT543306). All variable sites for HSP70 in L. braziliensis are described in Additional file 2: Table S1. The genetic diversity indices for L. naiffi were not considered due to the low sample size. The haplotype frequency was used in order to do haplotype network according to biogeographical region of possible infection for Colombia (Additional file 2: Table S2). The most frequent haplotype was LbCol01 with greater abundance in the Amazon and Orinoco regions, and present in all eight departments where L. braziliensis was notified (Fig. 3).
Haplotype network reconstruction for L. braziliensis under parsimony criteria with the TCS algorithm according HSP70 (n = 69, 337 bp). The size of the circles indicates the frequency of each haplotype. Black dot indicates the hypothetical ancestral haplotype and the perpendicular lines between the haplotypes refer to the number of nucleotide substitutions between them
Medical resolution, infectious species and clinical-demographic data
The 72.06% (98/136) of the patients were notified with cure according to medical criteria, while 10.29% (14/136) with therapeutic failure and 17.65% (24/136) as without data. This meant an efficacy of the medications during the treatment. Appraising the patients by treatment, 64.29% (63/98) of the patients submitted to the 1st line of meglumine antimoniate had disease healing, while 92.11% (35/38) healed when underwent the 2nd line with pentamidine isethionate (Additional file 3). All patients who had therapeutic failure were referred to a 3rd line treatment according to medical criteria. No significant differences were found in the disease resolution according to the infectious species (L. braziliensis versus L. panamensis) for meglumine antimoniate (Chi2 = 1561; gl = 1), pentamidine isethionate (Chi2 = 1105; gl = 1) and both treatments (Chi2 = 2.47; gl = 1) (Additional file 3). L. naiffi was not considered due to sample size.
Significant differences were found in the medical resolution of lesions (MWU = 381 **) according to patient age, being on average younger those who had therapeutic failure. In contrast, neither lesion areas, nor the disease evolution, nor the number of lesions explained the differences in whether a patient was healed or had a therapeutic failure. However, descriptive statistics such as mean, median and maximum value of the internal and external areas of lesions were higher for those who did not resolve according to medical criteria at the end of treatment (Table 1).
Clinical history of patients with L. naiffi, L. lindenbergi and L. infantum infection
Patient BON33 (L. naiffi) and BON32 (L. lindenbergi) entered the study on August 15, 2017, being both 19 years old professional soldiers, with 2.5 months of evolution disease before starting first-line treatment with meglumine antimoniate and latter with lesion in left hand. Additionally, patient BAS19 (L. naiffi) entered the study on August 3, 2017, as a 21 years old regular soldier, with 3 months of evolution disease before starting first-line treatment with meglumine antimoniate. BON33, BON32 and BAS19 were notified in Miraflores, Guaviare as a potential site of infection. In contrast, the BAS131 (L. infantum) patient participated in the study on March 12, 2019 as a 23 years old professional soldier, with therapeutic failure to meglumine antimoniate and before starting second-line treatment with pentamidine isethionate (Fig. 4).
Lesions in patients with CL caused by L. naiffi and L. infantum. Above is BON33: 19 years old professional soldier with ulcerated lesion on right forearm, possible site of infection in the municipality of Miraflores, Guaviare and with medical resolution as healing after first-line treatment with meglumine antimoniate. In the middle is BAS19: 21 years old regular soldier with ulcerated lesion on right leg, possible site of infection in Miraflores, Guaviare and with medical resolution as healing after first-line treatment with meglumine antimoniate. Below is BAS131: 23 years old professional soldier with plaque-shaped lesion on back, possible site of infection in the municipality of Tumaco, Nariño and with medical resolution as healing at the end of the second-line treatment with pentamidine isethionate
Discussion
The molecular epidemiology of CL in military population is complex due to continuous movement of troops, internal armed conflict and the sociodemographic features this population has which merits to conduct studies understanding the Leishmania species distribution and their potential association with clinical and demographic variables. Herein, we were able to identify several species infecting this vulnerable population (L. braziliensis, L. panamensis, L. infantum, L. naiffi, L. lindenbergi). The infectious species with greater relative abundance was L. braziliensis followed by L. panamensis in accordance with previous reports (Fig. 1) [8, 11]. When comparing our results with Patino et al., 2017, a 5.52% increase in the relative abundance of L. braziliensis and a decrease in species richness were observed. The sample size in our study was approximately half compared to the previous study and for Meta department only 25 patients were sampled. Additionally, some differences in relative abundance and richness of species could also be explained by the limitations of inclusion criteria in this study that suggests the need of a more comprehensive sampling to fully unveil the Leishmania species distribution in this vulnerable population. In terms of genetic diversity, the indices for L. braziliensis according to HSP70 were much lower in this study compared to indices reported in the military population previously [8] and this could be due to the sampling bias of our study as well.
Leishmania naiffi is reported for the first time in Colombia, its main reservoir is the nine-banded armadillo (Dasypus novemcinctus) and cutaneous leishmaniasis cases have been reported to date in humans within large areas of the Brazilian Amazon, French Guiana, Ecuador, Peru and Suriname, as well as in Panama and Martinique [34,35,36,37,38,39]. Its circulation in Colombia was suspected based on HSP70 through PCR-RFLP [9, 40], consequently, this new report for Colombia highlighted its clinical and epidemiological role in areas where its circulation was not previously known. Another L. naiffi reservoir includes animals of rodentia order as Sao Lourenço Punare (Thrichomys Laurentius) and Paraguayan punaré (Thrichomys pachyurus) endemics for Brazil [41, 42], while the main reported vectors have been Lutzomyia squamiventris, Lu. paraensis, Lu. davisi, Lu. hirsuta and Lu. tortura in Brazilian and Ecuadorian Amazon as well as Lu. gomezi and Nyssomyia trapidoi in Panamá [36, 37, 43,44,45,46]. In Colombia, Lu. gomezi, Lu. hirsuta and Lu. davisi have been reported in the Guaviare department [14, 47] explaining a potential transmission cycle for this species in the country. Nevertheless, future studies should consider exploring the transmission cycle of this species and its clinical and biological properties.
Leishmania lindenbergi is also reported as a new species circulating in Colombia. To date L. lindenbergi is endemic for the Amazon region of Brazil and has been reported as the rarest species with a restricted ecological distribution [48]. Nevertheless, it was reported by the first time in Brazilian soldiers performing activities in secondary forests as well as in a woman from the municipality of Bélem, state of Pará and state of Rondônia, Brazil respectively [49]. Nyssomyia antunesi has been defined as the main vector playing a determinant role in the transmission cycle of L. lindenbergi [49]. The patient BON32 reported as the potential place of infection for CL, the municipality of Miraflores, Guaviare. According to previous records of N. antunesi for Colombia, this sandfly species was reported in Miraflores, Guaviare as well as mainly in other departments such as Arauca, Caquetá, Casanare, Guajira, Meta, Vaupés and Vichada belonging to Amazon, Caribbean and Orinoco regions of Colombia [14, 50]. In this regard, this would explain a potential transmission scenario for this species in several Colombian regions suggesting the need to conduct new studies to understand the circulating species in unexplored ecotopes for CL, including its vectors.
Regarding the potential association of Leishmania species and clinical data, we did not find statistically significant associations. As of today, very few studies have identified a specific Leishmania variant with the disease outcome. In our case as most of the samples were L. braziliensis, this decreased the statistical power of the test. Then, a more robust epidemiological study must be in place in the light of trying to find evidence associating Leishmania species causality and disease outcome. Considering L. braziliensis is strongly correlated to MCL in the Americas [51, 52], it was suggested to carry out a clinical and preventive monitoring of patients who were identified with this infectious species and who, according to medical criteria, resulted in therapeutic failure at the end of the study [53]. Similarly, it is important to evaluate in further analyses main factors involved in the therapeutic failure of patients infected with L. panamensis. The therapeutic failure may be related to the genetic variants of Leishmania species [54]. Moreover, 35 cases of MCL were reported in military personnel between 2018 and 2019 [6] with 51.43% that occurred within departments of Meta (7 cases), Guaviare (6 cases), Santander (3 cases), Putumayo (1 case) and Guainía (1 case), where currently are notified all cases with therapeutic failure according to this study. These departments deserve particular studies regarding virulence and drug resistance by the parasite as well as local transmission cycles evaluating vectors and reservoirs [55, 56]. Further, the presence of Leishmania RNA virus (LRV-1) in parasites isolated from MCL patients should be explored as this might be related with the virulence of the parasite.
Conclusions
In conclusion, two new species (L. naiffi and L. lindenbergi) were reported for the first time in Colombia with possible infection in the municipality of Miraflores, Guaviare. L. braziliensis had the greatest relative abundance, while L. panamensis had the greatest dispersion within national territory. This confirms previous estimations of these two species circulating in the country. Our findings suggest the need to study unexplored regions as the Amazon and Orinoco where less sampling has been conducted that allowed us to identify these novel species. In addition, we highlight the complex epidemiological scenario of CL in the Colombian military personnel and the need to establish adequate prevention strategies that help to diminish the incidence of CL in this vulnerable population.
Availability of data and materials
All data sets used or analysed in the current study are included in the supplementary information files of this published article. All genetic sequences derived of current study are deposited and available in GenBank (https://www.ncbi.nlm.nih.gov/genbank/) under accession numbers MT543301, MT543302, MT543303, MT543304, MT543305 and MT543306.
Abbreviations
- CL:
-
Cutaneous leishmaniasis
- DNA:
-
Deoxyribonucleic acid
- HSP70 :
-
Heat shock protein 70
- MCL:
-
Mucocutaneous leishmaniasis
- VBD:
-
Vector borne disease
- VL:
-
Visceral leishmaniasis
- WHO:
-
World health organization
- PAHO:
-
Pan-american health organization
- INS:
-
Instituto nacional de salud
- SIVIGILA:
-
Sistema nacional de vigilancia en salud pública
- BASAN:
-
Batallón de sanidad
- CI:
-
Confidence interval
- EDTA:
-
Ethylenediamine tetraacetic acid
- LRI-DISAN-EJC:
-
Laboratorio de referencia e investigación, dirección de sanidad, ejército
- NNN:
-
Novy-macneal-Nicolle medium
- cm:
-
Centimeter
- μM:
-
Micromolar
- μL:
-
Mcrolitre
- ng:
-
Nanogram
- °C:
-
Degree Celsius
- min:
-
Minutes
- s:
-
Seconds
- bp:
-
Base pairs
- TAE:
-
Tris acetate EDTA buffer
- MUSCLE:
-
Multiple sequence comparison by log-expectation
- BLASTn:
-
Basic local alignment search tool for nucleotide databases using a nucleotide query
- NCBI:
-
National Center for Biotechnology Information
- ML:
-
Maximum likelihood
- GTR γ + I:
-
Generalized time reversible gamma and invariant evolutionary model
- RAxML-HPC:
-
Randomized axelerated maximum likelihood-high performance computing
- CIPRES:
-
Cyberinfrastructure for phylogenetic research
- TCS:
-
Templeton-Crandall-Sing algorithm
- MWU:
-
Mann whitney u test
- BON:
-
Bonza
- BAS:
-
BASAN
- WD:
-
Without data
- PCR-RFLP:
-
Polymerase chain reaction and restriction fragment length polymorphism
- TF:
-
Therapeutic failure
- NUCL:
-
Non-ulcerative cutaneous leishmaniasis
- MLST:
-
Multilocus sequence typing
References
Young DG, Morales A, Kreutzer RD, Alexander JB, Corredor A, Tesh RB, et al. Isolations of Leishmania braziliensis (Kinetoplastida: Trypanosomatidae) from cryopreserved Colombian sand flies (Diptera: Psychodidae). J Med Entomol. 1987;24(5):587–9.
Herwaldt BL. Leishmaniasis. Lancet. 1999;354(9185):1191–9.
Ul BA. Chronology of cutaneous leishmaniasis: An overview of the history of the disease. J Pakistan Assoc Dermatologists. 2006;16(1):24–7.
PAHO/WHO. Leishmaniases - Epidemiological Report of the Americas. No 8. Washington DC; 2019.
INS. Informe de evento leishmaniasis cutanea, mucosa y visceral, Colombia, 2018. Bogotá DC; 2019.
INS. Sistema Nacional de Vigilancia en Salud Pública (SIVIGILA), Instituto Nacional de Salud. Ministerio de Salud y Protección Social. Bogotá DC, Colombia; 2020. [cited 2020 Jan 15]. Available from: http://portalsivigila.ins.gov.co/sivigila/index.php#.
Ramírez JD, Hernández C, León CM, Ayala MS, Flórez C, González C. Taxonomy, diversity, temporal and geographical distribution of Cutaneous Leishmaniasis in Colombia: A retrospective study. Sci Rep. 2016;6(1):28266.
Patino LH, Mendez C, Rodriguez O, Romero Y, Velandia D, Alvarado M, et al. Spatial distribution, Leishmania species and clinical traits of Cutaneous Leishmaniasis cases in the Colombian army. PLoS Negl Trop Dis. 2017;11(8):e0005876.
Montalvo AM, Fraga J, Montano I, Monzote L, Auwera G. Van der, Marín M, et al. Identificación molecular con base en el gen hsp70 de aislamientos clínicos de Leishmania spp. en Colombia. Biomédica. 2016;36(Sup1 SE-Artículos originales).
Ovalle-Bracho C, Londoño-Barbosa D, Salgado-Almario J, González C. Evaluating the spatial distribution of Leishmania parasites in Colombia from clinical samples and human isolates (1999 to 2016). PLoS One. 2019;14(3):e0214124.
Perez-Franco JE, Cruz-Barrera ML, Robayo ML, Lopez MC, Daza CD, Bedoya A, et al. Clinical and Parasitological Features of Patients with American Cutaneous Leishmaniasis that Did Not Respond to Treatment with Meglumine Antimoniate. PLoS Negl Trop Dis. 2016;10(5):e0004739.
Corredor A, Gallego JF, Tesh RB, Morales A, de Carrasquilla CF, Young DG, et al. Epidemiology of Visceral Leishmaniasis in Colombia. Am J Trop Med Hyg. 1989;40(5):480–6.
Ovalle CE, Porras L, Rey M, Ríos M, Camargo YC. Distribución geográfica de especies de Leishmania aisladas de pacientes consultantes al Instituto Nacional de Dermatología Federico Lleras Acosta, E.S.E., 1995-2005. Biomédica. 2006;26(Sup1 SE-Artículos originales).
Ferro C, López M, Fuya P, Lugo L, Cordovez JM, González C. Spatial Distribution of Sand Fly Vectors and Eco-Epidemiology of Cutaneous Leishmaniasis Transmission in Colombia. PLoS One. 2015;10(10):e0139391.
Suárez M, Valencia BM, Jara M, Alba M, Boggild AK, Dujardin J-C, et al. Quantification of Leishmania (Viannia) Kinetoplast DNA in Ulcers of Cutaneous Leishmaniasis Reveals Inter-site and Inter-sampling Variability in Parasite Load. PLoS Negl Trop Dis. 2015;9(7):e0003936.
Hernández C, Alvarez C, González C, Ayala MS, León CM, Ramírez JD. Identification of six New World Leishmania species through the implementation of a High-Resolution Melting (HRM) genotyping assay. Parasit Vectors. 2014;7:501.
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647–9.
Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–7.
Maddison WP, Maddison DR. Interactive analysis of phylogeny and character evolution using the computer program MacClade. Folia Primatol (Basel). 1989;53(1–4):190–202.
Maddison D, Maddison W. MacClade 4: Analysis of phylogeny and character evolution. Sunderland MA: Sinauer Associates Inc Publishers; 2000. p. 492.
ESRI. Environmental Systems Research Institute. Redlands CA: ArcGIS; 2015.
Odiwuor S, Veland N, Maes I, Arévalo J, Dujardin J-C, Van der Auwera G. Evolution of the Leishmania braziliensis species complex from amplified fragment length polymorphisms, and clinical implications. Infect Genet Evol. 2012;12(8):1994–2002.
Van der Auwera G, Maes I, De Doncker S, Ravel C, Cnops L, Van Esbroeck M, et al. Heat-shock protein 70 gene sequencing for Leishmania species typing in European tropical infectious disease clinics. Euro Surveill. 2013;18(30):20543.
Van der Auwera G, Bart A, Chicharro C, Cortes S, Davidsson L, Di Muccio T, et al. Comparison of Leishmania typing results obtained from 16 European clinical laboratories in 2014. Euro Surveill. 2016;21(49):30418.
Gómez-Hernández C, Bento EC, Rezende-Oliveira K, Nascentes GAN, Barbosa CG, Batista LR, et al. Leishmania infection in bats from a non-endemic region of Leishmaniasis in Brazil. Parasitology. 2017;144(14):1980–6.
Espada CR, Ortiz PA, Shaw JJ, Barral AMP, Costa JML, Uliana SRB, et al. Identification of Leishmania (Viannia) species and clinical isolates of Leishmania (Leishmania) amazonensis from Brazil using PCR-RFLP of the heat-shock protein 70 gene reveals some unexpected observations. Diagn Microbiol Infect Dis. 2018;91(4):312–8.
Kato H, Gomez EA, Seki C, Furumoto H, Martini-Robles L, Muzzio J, et al. PCR-RFLP analyses of Leishmania species causing cutaneous and mucocutaneous leishmaniasis revealed distribution of genetically complex strains with hybrid and mito-nuclear discordance in Ecuador. PLoS Negl Trop Dis. 2019;13(5):e0007403.
Adaui V, Castillo D, Zimic M, Gutierrez A, Decuypere S, Vanaerschot M, et al. Comparative Gene Expression Analysis throughout the Life Cycle of Leishmania braziliensis: Diversity of Expression Profiles among Clinical Isolates. PLoS Negl Trop Dis. 2011;5(5):e1021.
Fraga J, Montalvo AM, De Doncker S, Dujardin J-C, Van der Auwera G. Phylogeny of Leishmania species based on the heat-shock protein 70 gene. Infect Genet Evol. 2010;10(2):238–45.
da Silva LA, de Sousa CDS, da Graça GC, Porrozzi R, Cupolillo E. Sequence analysis and PCR-RFLP profiling of the hsp70 gene as a valuable tool for identifying Leishmania species associated with human leishmaniasis in Brazil. Infect Genet Evol. 2010;10(1):77–83.
Rambaut A. FigTree, Institute of Evolutionary Biology University of Edinburgh. United Kingdom; 2009.
Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25(11):1451–2.
Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Mol Ecol. 2000;9(10):1657–9.
Lainson R, Shaw JJ. Leishmania (Viannia) naiffi sp. n., a parasite of the armadillo, Dasypus novemcinctus (L.) in Amazonian Brazil. Ann Parasitol Hum Comp. 1989;64(1):3–9.
Pratlong F, Deniau M, Darie H, Eichenlaub S, Pröll S, Garrabe E, et al. Human cutaneous leishmaniasis caused by Leishmania naiffi is wide-spread in South America. Ann Trop Med Parasitol. 2002;96(8):781–5.
Kato H, Gomez EA, Yamamoto Y, Calvopiña M, Guevara AG, Marco JD, et al. Natural infection of Lutzomyia tortura with Leishmania (Viannia) naiffi in an Amazonian area of Ecuador. Am J Trop Med Hyg. 2008;79(3):438–40.
Azpurua J, De La Cruz D, Valderama A, Windsor D. Lutzomyia Sand Fly Diversity and Rates of Infection by Wolbachia and an Exotic Leishmania Species on Barro Colorado Island, Panama. PLoS Negl Trop Dis. 2010;4(3):e627.
van Thiel P-PAM, van Gool T, Kager PA, Bart A. First cases of cutaneous leishmaniasis caused by Leishmania (Viannia) naiffi infection in Surinam. Am J Trop Med Hyg. 2010;82(4):588–90.
Fraga J, Montalvo AM, Van der Auwera G, Maes I, Dujardin J-C, Requena JM. Evolution and species discrimination according to the Leishmania heat-shock protein 20 gene. Infect Genet Evol. 2013;18:229–37.
Montalvo AM, Fraga J, Tirado D, Blandón G, Alba A, Van der Auwera G, et al. Detection and identification of Leishmania spp.: application of two hsp70-based PCR-RFLP protocols to clinical samples from the New World. Parasitol Res. 2017;116(7):1843–8.
Cássia-Pires R, Boité MC, D’Andrea PS, Herrera HM, Cupolillo E, Jansen AM, et al. Distinct Leishmania Species Infecting Wild Caviomorph Rodents (Rodentia: Hystricognathi) from Brazil. PLoS Negl Trop Dis. 2014;8(12):e3389.
Roque ALR, Jansen AM. Wild and synanthropic reservoirs of Leishmania species in the Americas. Int J Parasitol Parasites Wildl. 2014;3(3):251–62.
Naiff RD, Freitas RA, Naiff MF, Arias JR, Barrett TV, Momen H, et al. Epidemiological and nosological aspects of Leishmania naiffi Lainson & Shaw, 1989. Mem Inst Oswaldo Cruz. 1991;86(3):317–21.
Grimaldi G Jr, Momen H, Naiff RD, McMahon-Pratt D, Barrett TV. Characterization and classification of leishmanial parasites from humans, wild mammals, and sand flies in the Amazon region of Brazil. Am J Trop Med Hyg. 1991;44(6):645–61.
Gil LHS, Basano SA, Souza AA, Silva MGS, Barata I, Ishikawa EA, et al. Recent observations on the sand fly (Diptera: Psychodidae) fauna of the State of Rondônia, Western Amazônia, Brazil: the importance of Psychdopygus davisi as a vector of zoonotic cutaneous leishmaniasis, vol. 98: Memórias do Instituto Oswaldo Cruz . scielo; 2003. p. 751–5.
Kato H, Calvopiña M, Criollo H, Hashiguchi Y. First human cases of Leishmania (Viannia) naiffi infection in Ecuador and identification of its suspected vector species. Acta Trop. 2013;128(3):710–3.
Bejarano EE, Estrada LG. FAMILY PSYCHODIDAE. Zootaxa. 2016;4122(1):187–238.
Cantanhêde LM, Mattos CB, de Souza RC, Filgueira CPB, da Silva Júnior CF, Limeira C, et al. First report of Leishmania (Viannia) lindenbergi causing tegumentary leishmaniasis in the Brazilian western Amazon region TT - Premier signalement de Leishmania (Viannia) lindenbergi causant une leishmaniose tégumentaire dans la région amazonienne de l’o. Parasite. 2019;26:30.
Silveira FT, Ishikawa EAY, De Souza AAA, Lainson R. An outbreak of cutaneous leishmaniasis among soldiers in Belém, Pará State, Brazil, caused by Leishmania (Viannia) lindenbergi n. sp. A new leishmanial parasite of man in the Amazon region. Parasite. 2002;9(1):43–50.
Trujillo AV, Reina AEG, Orjuela AG, Suarez EP, Palomares JE, Alvarez LSB. Seasonal variation and natural infection of Lutzomyia antunesi (Diptera: Psychodidae: Phlebotominae), an endemic species in the Orinoquia region of Colombia, vol. 108: Memórias do Instituto Oswaldo Cruz . scielo; 2013. p. 463–9.
Schwartz E, Hatz C, Blum J. New world cutaneous leishmaniasis in travellers. Lancet Infect Dis. 2006;6(6):342–9.
Cincurá C, de Lima CMF, Machado PRL, Oliveira-Filho J, Glesby MJ, Lessa MM, et al. Mucosal leishmaniasis: A Retrospective Study of 327 Cases from an Endemic Area of Leishmania (Viannia) braziliensis. Am J Trop Med Hyg. 2017;97(3):761–6.
Méndez-Bejarano, C. P., Correa-Cárdenas, C. A., Pérez-Rico, J. J., Romero-Barbosa, Y. A., & Rodríguez-Angarita, O. Parasitic load of Leishmania spp. in Colombian military personnel with cutaneous leishmaniasis: a case study. Revista Científica General José María Córdova. 2020;18(29):237–66.
Patino LH, Imamura H, Cruz-Saavedra L, Pavia P, Muskus C, Méndez C, et al. Major changes in chromosomal somy, gene expression and gene dosage driven by Sb(III) in Leishmania braziliensis and Leishmania panamensis. Sci Rep. 2019;9(1):9485.
WHO Expert Committee on the Control of the Organization World Health. Control of the leishmaniases: report of a meeting of the WHO Expert Commitee on the Control of Leishmaniases, Geneva, 22-26 March 2010. Geneva PP - Geneva: World Health Organization; 2010. (WHO technical report series ; 949).
Marin J, Urrea D, Muskus C, Echeverry MC, Mejía AM, Triana O. Curvas de fusión de regiones genómicas específicas: una herramienta prometedora para el diagnóstico y tipificación de las especies causantes de la leishmaniasis cutánea en Colombia. Biomedica. 2017;37(4):538–47.
Acknowledgments
We thank Dirección de Ciencia y Tecnología (DITEC) of the Colombian National Army for its support in the processes, resource management and permanent advice. We also thank the Rehabilitation Center for Leishmaniasis of the Health Battalion in Bogotá and the National Rehabilitation Center for Leishmaniasis in Duitama, Boyacá for their collaboration, willingness and monitoring of patients.
Funding
This study was funded by Dirección de Ciencia y Tecnología (DITEC) of the Comando de Educación y Doctrina (CEDOC), Colombia National Army for the execution of the research project, through royalties for agreements with hydrocarbon companies such as Drummond Ltd., Ecopetrol, Hocol and ODL. Additionally, the authors thank BioMed Central Accounts Team for Publication Fee Assistance number 6340041182029152 of USD 1,717.50 (75% discount). The commercial funders had no role in the study design, data collection, data analysis, decision to publish and in writing the manuscript.
Author information
Authors and Affiliations
Contributions
MCD, CM, JDR conceived and designed the study. MCD, YR, MTA and CM managed the funding acquisition and formulated the needs plan for research project subjected to current study. JP, MCD, YR, OR and CM participated in the collection and storage of samples. CC provided clinical support in the sampling and clinical monitoring of patients. CACC, JP, JDR, LHP, MCD and OR conducted the molecular tests and bioinformatics analyses. CACC, JP, YR, CM, JDR, LHP and OCB analysed the data. CACC, JDR, JP and CM wrote the original draft of the manuscript. JP, JDR, LHP, MCD, YR, OCB, OR, MTA, CC and CM reviewed and edited the written manuscript. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The article 11 of 8430 resolution from 1993 of the Health Ministry allowed to this study to be classified as a minimum risk investigation. The Research Ethics Committee of the Central Military Hospital, Bogotá approved the research project subjected to this publication under Act No. 2043 of March 22, 2017.
The handling of all the data of each patient that participated voluntarily, the clinical-epidemiological record and the results, were carried out under the strictest standards of confidentiality, prior authorization of the patient throughout the application of an informed consent. This prior informed consent was signed and used according to 2378 Colombian resolution of 2008 on Good Clinical Practices.
Consent for publication
Not Applicable.
Competing interests
The authors declare that there are no potential competing interests related to the current study.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1
Biogeographical regions within Colombia. (Own elaboration)
Additional file 2
Variable sites and haplotype frequency in Leishmania species by possible site of infection.
Additional file 3.
Demographic, clinical and epidemiological data of patients
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Correa-Cárdenas, C.A., Pérez, J., Patino, L.H. et al. Distribution, treatment outcome and genetic diversity of Leishmania species in military personnel from Colombia with cutaneous leishmaniasis. BMC Infect Dis 20, 938 (2020). https://doi.org/10.1186/s12879-020-05529-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-020-05529-y