Abstract
Background
Antimicrobial resistance has been declared by the World Health Organization as a threat to the public health. The aim of this study was to analyze antimicrobial resistance patterns of the common pathogens occurring at the Bugando Medical Centre (BMC), Mwanza, Tanzania to provide data for antimicrobial stewardship programmes.
Methods
A total of 3330 microbiological culture results scripts representing non-repetitive specimens reported between June 2013 and May 2015 were retrieved and analyzed for pathogens and their susceptibility patterns using STATA-11 software.
Results
Out of 3330 specimens, 439 (13.2%) had positive culture. Staphylococcus aureus (n = 100; 22.8%), Klebsiella pneumoniae (n = 65; 14.8%) and Escherichia coli (n = 41; 9.3%) were the most frequently isolated bacteria. Of 78 Staphylococcus aureus tested, 27 (34.6%) were found to be methicillin resistant Staphylococcus aureus (MRSA). Rates of resistance of Klebsiella pneumoniae and Escherichia coli isolates to third generation cephalosporins were 38.5% (25/65) and 29.3% (12/41) respectively. Staphylococcus aureus and Klesbiella pneumoniae were commonly isolated from bloodstream infections while Escherichia coli and Pseudomonas aeruginosa were the predominant isolates from urinary tract and wounds infections respectively. Of 23 Salmonella species isolated, 22 (95%) were recovered from the blood. Nine of the 23 Salmonella species isolates (39%) were found to be resistant to third generation cephalosporins. The resistance rate of gram-negative bacteria to third generation cephalosporins increased from 26.5% in 2014 to 57.9% in 2015 (p = 0.004) while the rate of MRSA decreased from 41.2% in 2013 to 9.5% in 2015 (p = 0.016). Multidrug-resistant gram-negative isolates were commonly isolated from Intensive Care Units and it was noted that, the majority of invasive infections were due to gram-negative bacteria.
Conclusion
There is an increase in proportion of gram-negative isolates resistant to third generation cephalosporins. The diversity of potential pathogens resistant to commonly prescribed antibiotics underscores the importance of sustained and standardized antimicrobial resistance surveillance and antibiotic stewardship programmes in developing countries.
Similar content being viewed by others
Background
Antimicrobial resistance (AMR) has reached a critical point and is now becoming a global threat to the public health worldwide. The World Health Organization (WHO) has declared AMR a public health threat and has urged different countries to develop action plan to combat the problem [1]. Knowledge on the epidemiology of healthcare-associated infections (HCAIs) and the antimicrobial susceptibility patterns of the isolates are crucial towards guiding empirical treatment [2]. The spectra of bacteria causing infections and their susceptibility pattern have been found to vary from one setting to another; a fact which highlights the importance of having local surveillance data for planning and implementing infection prevention and control (IPC) measures [3–5]. High income countries including the United States, have managed to decrease the burden of HCAIs by 30% through effective implementation surveillance systems [6]. A study which was conducted in a teaching hospital in Nigeria, showed the reduction of HCAIs rates from 5.8 to 2.8% in 2003 and 2006 respectively through the implementation of an effective infection control program [7]. In fighting emerging infections, surveillance data regarding AMR in Europe have been made available starting at regional to international levels via databases to ease communication between scientists [2, 8]. In Africa there are limited surveillance data irrespective of an upsurge of HCAIs and AMR clones circulating both in hospitals and communities [9–11]. At the BMC hospital in Tanzania the rate of extended spectrum beta-lactamases (ESBL) producing Escherichia coli increased from 25 to 50%, and that of MRSA increased from 16 to 44% between 2009 and 2014 [12–14]. Preliminary findings show significant variation of the epidemiology and evolution of these strains as evidenced by multiple clones of Escherichia coli carrying bla CTX-M-15 [15], causing sepsis among neonates and a new ST1797 MRSA causing wound infections [16]. Infections due to multidrug-resistant isolates have been found to be associated with increased morbidity and mortality [17, 18]. The emerging of these multidrug-resistant clones coupled with the introduction of expensive and last resort antibiotics such as carbapenems and vancomycin pose a great challenge in combating AMR at BMC and in developing countries at large. This analysis was done to assess the pattern of bacterial isolates and their susceptibility patterns from various specimens received at BMC microbiological laboratory between June 2013 and May 2015. These data are crucial in rationalizing empirical treatment and set measures for IPC, surveillance and policy change in Tanzania and in other developing countries.
Methods
Between June 2013 and May 2015 a retrospective analysis of BMC microbiological culture results was done. BMC is a tertiary, consultant and teaching hospital located in Northwestern Tanzania. It serves a population of about 13 million people from eight regions. A total of 3387 laboratory culture results were retrieved during the observational period. Out of 3387 laboratory results, 87 scripts were excluded from the analysis due to incomplete filling of the microbiological laboratory form. Missing particulars included age, clinical history, antibiotic use, specimen type and ward number.
Microbiological analysis
BMC clinical microbiology laboratory is accredited according to ISO 15189 by Southern African Development Community Accreditation Service (SADCAS) accreditation body with registration number MD 002. It participates in bacteriology external quality assurance coordinated by a reference laboratory in South Africa (WHO/NICD).
Clinicians were responsible for decisions regarding taking specimens for microbiological culture based on the clinical assessment of the patients. Specimens were processed following the BMC microbiology laboratory standard operating procedures (SOPs). Contaminants were defined as isolated bacteria that were more likely to be normal flora depending on the type and site where the specimen was taken. Identification of bacteria was carried out by conventional biochemical methods [19]. In case of ambiguity, analytical profile index (API) 20E or 20NE (bioMérieux, France) was used to confirm identification. Antimicrobial susceptibility testing was performed using the disk-diffusion method and interpreted according to the Clinical Laboratory Standards Institute guidelines (CLSI) [20]. Gram-negative bacteria resistant to either ceftriaxone (30 μg) or cefotaxime (30 μg) or both were termed as resistant to third generation cephalosporins. Intermediate susceptibility results were regarded as resistant in the analysis. MRSA was detected using cefoxitin (30 μg) disk and the isolate with zone of inhibition of ≤ 21 mm was confirmed phenotypically to be MRSA.
Quality control
During the observation period, Staphylococcus aureus ATCC 25923, Escherichia coli ATCC 25922 and Klebsiella pneumoniae ATCC 13883 were used as quality control strains. In addition, in-house molecular typed MRSA strain and Escherichia coli ATCC 35218 were also used as positive controls for MRSA and third generation cephalosporin resistant organisms respectively.
Data analysis
In the analysis of these data, children were defined as ≤ 12 years old and adults > 12 years old according to BMC admission criteria. Types of infections analyzed included; blood stream (sepsis, bacteremia), pneumonia, urinary tract, upper respiratory tract, surgical site, skin and soft tissue (abscess, pyoderma, pyomyositis, ulcer and wound), gastro-intestinal, central nervous system and cardiovascular system [21].
All demographic and clinical data as indicated in BMC microbiological request/report form were extracted and entered in Excel spreadsheet. Data were analyzed using STATA version 11 (STATA Corp LP, USA). Categorical variables were summarized as proportions and were analyzed using the Pearson’s Chi-Square test or Fisher’s exact to test statistical differences among the various groups. Odds ratios with their respective 95% confidence interval (CI) were calculated to measure the strength of associations. The p-value of less than 0.05 was considered statistically significant.
Results
Type of specimens
During the study period (June 2013 to May 2015), a total of 3330 non-repetitive specimens were analyzed; blood 2238 (67.2%), urine 307 (9.2%), cerebrospinal fluid 303 (9.1%), pus swabs 206 (6.2%), aspirates 148 (4.4%), sputum 86 (2.6%) and stool 42 (1.3%). Of the analyzed specimens, 971 (29.2%), 1888 (56.7%) and 471 (14.1%) were from year 2013, 2014 and 2015 respectively. The paediatric population contributed 67% (651/971), 47.9% (904/1888) and 74.7% (352/471) of the total specimens analyzed in 2013, 2014 and 2015 respectively.
Culture results
Out of 3330 specimens processed during the observation period, 439 (13.2%) were found to be culture positive and 140 (4.2%) had contamination. Contamination rates were significantly more in pediatric wards than in other wards (106/1907; 5.6% vs. 34/1393; 2.4%, p < 0.001). Positive culture rates were 17.8% (173/971), 11.8% (223/1888) and 9.1% (43/471) in 2013, 2014 and 2015 respectively.
Antibiotic use before specimen collection
Out of 3330 microbiological request/report forms, 1257 (37.8%) reported the patients to have had an exposure to antibiotics before the specimens were taken. Types of antibiotics documented were penicillins (22.2%, n = 279), gentamicin (16.15%, n = 203), ceftriaxone or cefotaxime (16.1%, n = 202), meropenem (0.9%, n = 12), ciprofloxacin (2.2%, n = 28) and unspecified (58.6%, n = 736).
Pathogens and their susceptibility pattern
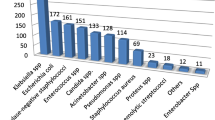
Out of 439 positive cultures, most frequently isolated pathogens were Staphylococcus aureus (22.8%, n = 100), Klebsiella pneumoniae (14.8%, n = 65) and Escherichia coli (9.3%, n = 41) Table 1. Majority of Staphylococcus aureus (61%, n = 61) and Klebsiella pneumoniae (70.8%, n = 46) isolates were from paediatric population (Paediatric, PREM/NICU and PICU) while the majority of Escherichia coli isolates (68.3%, n = 28) were from adults (Table 2). Staphylococcus aureus and Klebsiella pneumoniae were the most frequent isolates recovered from blood (3.6%, n = 80/2238 and 1.9%, n = 42/2238, Table 1). Escherichia coli (7.2%; n = 22/307) dominated the urinary tract infections.
Of 41 Escherichia coli isolates, 19 were tested for the susceptibility to third generation cephalosporins of which, 12 (63.2%) were found to be resistant. Out of 22 Escherichia coli isolates from urinary tract infections, 20 (90.9%) were susceptible to nitrofurantoin. Twenty four Escherichia coli isolates were tested for the susceptibility to ciprofloxacin; 16 (66.7%) were found to be resistant. Regarding 65 Klebsiella pneumoniae isolated, 31 were tested for the susceptibility to third generation cephalosporins of which, 25 (80.6%) were found to be resistant whereas of 61 Klebsiella pneumoniae isolates tested for gentamicin, 51 (83.6%) were found to be resistant. All 48 Klebsiella pneumoniae isolates tested for the susceptibility to imipenem were found to be susceptible.
The proportion of Salmonella species among positive cultures was 5.2% (n = 23/439) of which 95.7% were from blood (n = 22), Table 1. The majority of Salmonella species (82.6%, n = 19) were isolated from paediatric wards (Table 2). Out of 23 Salmonella species, 16 were tested for the susceptibility to third generation cephalosporins of which, 9(56.3%) were found to be resistant. All 15 Salmonella species tested against ciprofloxacin were found to be susceptible.
Most of the Pseudomonas aeruginosa isolates (58.3%, n = 14/24) were recovered from wound swabs. Of 24 Pseudomonas aeruginosa isolates, 16 were tested for the susceptibility to the third generation cephalosporins of which, 14(87.5%) were found to be resistant. A total of 20/22 (90.9%) Pseudomonas aeruginosa were found to be susceptible to amikacin.
Methicillin-resistant Staphylococcus aureus (MRSA) trend
Out of 100 Staphylococcus aureus isolates, 78 isolates were tested for cefoxitin of which, 27 (34.6%) were found to be MRSA. Of 27 MRSA isolates, 62.9% (n = 17) were from the paediatric population. The MRSA proportion kept on fluctuating during the observation period; it was 41.2% (n = 14/34), 47.8% (n = 11/23) and 9.5% (n = 2/21) in 2013, 2014 and 2015 respectively (p = 0.016). MRSA isolates were significantly more isolated from sterile parts of the body (blood, aspirates and cerebrospinal fluid) than from non-sterile (urine, pus and wound swabs) (32.5 vs. 5%; p = 0.01) (Fig. 1).
Resistance to third generation cephalosporins
The overall proportion of gram-negative bacteria resistant to third generation cephalosporins was 35.9% (112/312). The majority (59.8%, n = 67/112) of these isolates were from children. When distributed by years of the observation, the isolation trend was 44.9% (n = 57/127), 26.5% (n = 44/166) and 57.9% (n = 11/19) in 2013, 2014 and 2015 respectively. The gram-negative bacteria resistant to third generation cephalosporins proportion was significantly higher in isolates from Intensive Care Units (ICUs) (46.9%) than in other wards (31.8%); OR = 1.89, 95% CI (1.1–3.3), p = 0.01. In addition, the isolation rate of the gram-negative bacteria resistant to third generation cephalosporins was higher from sterile sites (41.5%) than non-sterile sites (30.9%), p = 0.051 (Fig. 1).
Discussion
In the present analysis, a total of 3387 specimens that were sent for culture and sensitivity between June 2013 and May 2015, the number seem to be little in a hospital with a bed capacity of 900. Relying on the National or local treatment guidelines which emphasize more on empirical treatment might have contributed to low number of specimens received in the laboratory [22]. Deployment and adherence to antimicrobial therapy guidelines and policies using evidence-based generated data might improve the current situation as previously shown in Botswana [23]. Blood specimens contributed to more than half of specimens processed and the majority were from paediatric wards. This could be explained by the local practice in these wards that emphasize on the need of blood culture to all children presenting with fever.
In the present analysis, Staphylococcus aureus and Klebsiella pneumoniae were the predominant organisms causing bloodstream infections from pediatric wards. These findings are in agreement with a prospective study which was conducted at a national hospital in Tanzania among neonates with fever which found Staphylococcus aureus to be the predominant pathogen (54.5%) followed by Klesbiella pneumoniae (32.7%) [24]. Similar findings were observed at BMC in 2010 [12], indicating the persistence of pattern in pediatric wards especially in neonatal units.
With regard to urinary tract infections in this analysis, Escherichia coli was found to be the most frequently isolated pathogen. A similar finding was observed in previous studies done in the same setting [25, 26]. Escherichia coli isolates from urine demonstrated high rates of resistance to ciprofloxacin and third generation cephalosporins than to nitrofurantoin. The rate of resistance of nitrofurantoin has remained low all years in the present study. Despite being cheap, this antibiotic is not commonly self-prescribed. Ciprofloxacin and third generation cephalosporins are commonly used at BMC than nitrofurantoin and they are also commonly self-prescribed in the community. The overuse of ciprofloxacin and cephalosporins might be the most probable cause of higher resistance rates observed.
As in previous study [27], the majority of Salmonella species isolates were isolated from blood in this study and all of the tested Salmonella species isolates were susceptible to ciprofloxacin. This finding is similar to a study done in Ghana, which observed Salmonella typhi to be the most prevalent pathogen isolated from bloodstream infection and all isolates were susceptible to ciprofloxacin [27]. The observation of 100% susceptibility of Salmonella species isolates to ciprofloxacin in our setting could be explained by the non-use of this antimicrobial agent in paediatric wards. This can be evidenced by the fact that the majority of these isolates were from the paediatric wards. This is further supported by the previous study which observed 8% of Salmonella species isolates from adults to be resistant to ciprofloxacin [28]. In this study, a significant proportion of Salmonella species isolates were resistant to third generation cephalosporins. Similar findings have been observed in Europe from people who travelled from Asian countries [29, 30] and in Kenya [31]. However, these findings are in contrast with the previous study conducted three years ago in the same hospital which observed all Salmonella species to be susceptible to ceftriaxone [28]; suggesting the presence of emerging multidrug-resistant Salmonella species in Northwestern Tanzania.
As in previous study [14], most of the Pseudomonas aeruginosa isolates in this study were recovered from wound swabs and more than 50% were resistant to third generation cephalosporins. The majority of Pseudomonas aeruginosa isolates in this study were sensitive to amikacin, as previously observed [32]. This could be explained by the rare use this aminoglycoside in our setting and other setting in low and middle-income countries.
The general proportion of MRSA in this study was found to be in agreement with MRSA proportion observed in other studies done in the same setting in 2010 and 2014 [12, 14]. However, during the observation period a significant decrease was observed in 2015, this could be explained by the ongoing hand washing campaigns and improvement of IPC programs in various BMC wards coupled by increased awareness of the emergence of multidrug-resistant strains among healthcare workers. The latter measures have been shown to reduce MRSA outbreaks [33]. In the present study, MRSA isolates were significantly found to cause invasive infections. Invasive MRSA infections might lead to increased morbidity and mortality of the infected patients as previously observed [12].
Although the overall trend of the proportions of gram-negative bacteria resistant to third generation cephalosporins appeared to decrease in 2014 in comparison to 2013, the significant upsurge of these notorious strains was observed in 2015. The high resistance rates observed in ICU patients could be explained by the fact that patients in ICU tend to stay longer in the hospital and this has been found to increase risk infections due to multidrug-resistant pathogens [34].
Conclusions
The observed high proportion of gram-negative isolates being resistant to third generation cephalosporins is alarming and calls for a surveillance of HCAIs and community infections to establish the source and transmission pathways. The emergence of Salmonella species isolates resistant to third generation cephalosporins is worrisome because of increased infections of Salmonella species observed in HIV infected patients which is endemic in low-income countries.
Despite the good service offered by BMC microbiology laboratory shortcomings have been observed which need rectifications. Monitoring and proper screening of isolates resistant to third generation cephalosporins to know the mechanisms of resistance responsible should be done. Consistency in antimicrobials tested for the isolates should be observed to establish a homogeneous database. Adherence to CLSI guidelines in processing microbiological isolates should be mandatory. It is a high time to consider having the local antimicrobial guide using local generated data to guide treatment. There is need to perform molecular characterization of common isolates involved in HCAIs in developing countries so that evidence based data can be used to improve the IPC in our setting.
Study limitations
The analysis based on getting information from microbiological request/report forms therefore it was difficult to tell whether the infection originated from the community or it was healthcare-associated. Antimicrobial susceptibility testing depended on the availability of antimicrobial discs at that particular time. This has resulted to inconsistent bacterial antibiogram patterns and difficulties in reporting resistance rates of the most frequent isolates. Antimicrobial agents were tested against wrong isolates as per CLSI guidelines e.g. ceftriaxone was tested against Pseudomonas aeruginosa isolates. Moreover, specific tests like inducible clindamycin resistance (D-test) and ESBL testing were not done. Other important epidemiological information such as patient’s outcome, duration of hospital stay etc. were not reported. Lastly, there might be sampling bias in this analysis because there were no guidelines for systematic specimen collection for microbiological culture.
Abbreviations
- AMR:
-
Antimicrobial resistance
- API:
-
Analytical profile index
- BMC:
-
Bugando Medical Centre
- CLSI:
-
Clinical laboratory standards institute
- ESBL:
-
Extended spectrum beta lactamase
- HCAIs:
-
Healthcare-associated infections
- IPC:
-
Infection prevention and control
- MRSA:
-
Methicillin-resistant Staphylococcus aureus
- NICD:
-
National institute for communicable diseases, a division of the national health laboratory service, South Africa
- NICU:
-
Neonatal intensive care unit
- PICU:
-
Paediatric intensive care unit
- PREM:
-
Premature unit
- SADCAS:
-
Southern African development community accreditation service
- WHO:
-
World Health Organization
References
WHO. Antimicrobial resistance: global report on surveillance. World Health Organization; 2014. https://scholar.google.de/scholar?hl=en&q=Antimicrobial+resistance%3A+global+report+on+surveillance&btnG=&as_sdt=1%2C5&as_sdtp=.
De Kraker M, Jarlier V, Monen J, Heuer O, Van De Sande N, Grundmann H. The changing epidemiology of bacteraemias in Europe: trends from the European Antimicrobial Resistance Surveillance System. Clin Microbiol Infect. 2013;19(9):860–8.
Forster AJ, Oake N, Roth V, Suh KN, Majewski J, Leeder C, van Walraven C. Patient-level factors associated with methicillin-resistant Staphylococcus aureus carriage at hospital admission: a systematic review. Am J Infect Control. 2013;41(3):214–20.
Allegranzi B, Pittet D. Role of hand hygiene in healthcare-associated infection prevention. J Hosp Infect. 2009;73(4):305–15.
Loveday H, Wilson J, Kerr K, Pitchers R, Walker J, Browne J. Association between healthcare water systems and Pseudomonas aeruginosa infections: a rapid systematic review. J Hosp Infect. 2014;86(1):7–15.
Allegranzi B, Pittet D. Preventing infections acquired during health-care delivery. Lancet. 2008;372(9651):1719–20.
Abubakar S. Implementation of an Infection Control Programme in Kano, Northern Nigeria. Int J Infect Control. 2007;3(2). https://scholar.google.de/scholar?q=Implementation+of+an+Infection+Control+Programme+in+Kano%2C+Northern+Nigeria&btnG=&hl=en&as_sdt=0%2C5.
Rosenthal VD, Bijie H, Maki DG, Mehta Y, Apisarnthanarak A, Medeiros EA, Leblebicioglu H, Fisher D, Álvarez-Moreno C, Khader IA. International Nosocomial Infection Control Consortium (INICC) report, data summary of 36 countries, for 2004–2009. Am J Infect Control. 2012;40(5):396–407.
Lubell Y, Ashley EA, Turner C, Turner P, White NJ. Susceptibility of community-acquired pathogens to antibiotics in Africa and Asia in neonates–an alarmingly short review. Tropical Med Int Health. 2011;16(2):145–51.
Vlieghe E, Phoba M, Tamfun JM, Jacobs J. Antibiotic resistance among bacterial pathogens in Central Africa: a review of the published literature between 1955 and 2008. Int J Antimicrob Agents. 2009;34(4):295–303.
Mshana SE, Matee M, Rweyemamu M. Antimicrobial resistance in human and animal pathogens in Zambia, Democratic Republic of Congo, Mozambique and Tanzania: an urgent need of a sustainable surveillance system. Ann Clin Microbiol Antimicrob. 2013;12:28.
Kayange N, Kamugisha E, Mwizamholya DL, Jeremiah S, Mshana SE. Predictors of positive blood culture and deaths among neonates with suspected neonatal sepsis in a tertiary hospital, Mwanza-Tanzania. BMC Pediatr. 2010;10(1):39.
Mshana SE, Kamugisha E, Mirambo M, Chakraborty T, Lyamuya EF. Prevalence of multiresistant gram-negative organisms in a tertiary hospital in Mwanza, Tanzania. BMC Res Notes. 2009;2(1):49.
Moremi N, Mushi MF, Fidelis M, Chalya P, Mirambo M, Mshana SE. Predominance of multi-resistant gram-negative bacteria colonizing chronic lower limb ulcers (CLLUs) at Bugando Medical Center. BMC Res Notes. 2014;7(1):211.
Mshana S, Imirzalioglu C, Hain T, Domann E, Lyamuya E, Chakraborty T. Multiple ST clonal complexes, with a predominance of ST131, of Escherichia coli harbouring blaCTX-M-15 in a tertiary hospital in Tanzania. Clin Microbiol Infect. 2011;17(8):1279–82.
Moremi N, Mshana SE, Kamugisha E, Kataraihya J, Tappe D, Vogel U, Lyamuya EF, Claus H. Predominance of methicillin resistant Staphylococcus aureus-ST88 and new ST1797 causing wound infection and abscesses. J Infect Dev Ctries. 2012;6(08):620–5.
Mshana SE, Hain T, Domann E, Lyamuya EF, Chakraborty T, Imirzalioglu C. Predominance of Klebsiella pneumoniae ST14 carrying CTX-M-15 causing neonatal sepsis in Tanzania. BMC Infect Dis. 2013;13(1):466.
Mshana SE, Gerwing L, Minde M, Hain T, Domann E, Lyamuya E, Chakraborty T, Imirzalioglu C. Outbreak of a novel Enterobacter sp. carrying bla CTX-M-15 in a neonatal unit of a tertiary care hospital in Tanzania. Int J Antimicrob Agents. 2011;38(3):265–9.
Gerhardt P, Murray R, Costilow R, Nester EW, Wood WA, Krieg N, Phillips GB. Manual of methods for general bacteriology. 1981.
Wayne P. CLSI document M02-A11 and M07-A9. Clinical and Laboratory Standards Institute. 2012
CDC. CDC/NHSN surveillance definitions for specific types of infections. CDC Atlanta; 2014. https://scholar.google.de/scholar?q=CDC%2FNHSN+surveillance+definitions+for+specific+types+of+infections.+CDC+Atlanta%3B+2014.&btnG=&hl=en&as_sdt=0%2C5.
Welfare MHS. Standard Treatment Guidelines and National Essential Medicines List. 2013.
BMoH. Botswana Antimicrobial Therapy Guide. Botswana: Ministry of Health; 2012.
Mhada TV, Fredrick F, Matee MI, Massawe A. Neonatal sepsis at Muhimbili National Hospital, Dar es Salaam, Tanzania; aetiology, antimicrobial sensitivity pattern and clinical outcome. BMC Public Health. 2012;12(1):904.
Festo E, Kidenya BR, Hokororo A, Mshana SE. Predictors of urinary tract infection among febrile children attending at Bugando Medical Centre, Northwestern Tanzania. Archives Clin Microbiol. 2011;2(5). https://scholar.google.de/scholar?q=Predictors+of+urinary+tract+infection+among+febrile+children+attending+at+Bugando+Medical+Centre%2C+Northwestern+Tanzania&btnG=&hl=en&as_sdt=0%2C5.
Msaki BP, Mshana SE, Hokororo A, Mazigo HD, Morona D. Prevalence and predictors of urinary tract infection and severe malaria among febrile children attending Makongoro health centre in Mwanza city, North-Western Tanzania. Archives Public Health. 2012;70(1):1–8.
Groß U, Amuzu SK, De Ciman R, Kassimova I, Groß L, Rabsch W, Rosenberg U, Schulze M, Stich A, Zimmermann O. Bacteremia and antimicrobial drug resistance over time, Ghana. Emerg Infect Dis. 2011;17(10):1879.
Meremo A, Mshana SE, Kidenya BR, Kabangila R, Peck R, Kataraihya JB. High prevalence of Non–typhoid salmonella bacteraemia among febrile HIV adult patients admitted at a tertiary Hospital, North-Western Tanzania. Int Arch Med. 2012;5(1):28.
Al Naiemi N, Zwart B, Rijnsburger MC, Roosendaal R, Debets-Ossenkopp YJ, Mulder JA, Fijen CA, Maten W, Vandenbroucke-Grauls CM, Savelkoul PH. Extended-spectrum-beta-lactamase production in a Salmonella enterica serotype Typhi strain from the Philippines. J Clin Microbiol. 2008;46(8):2794–5.
González-López JJ, Piedra-Carrasco N, Salvador F, Rodríguez V, Sánchez-Montalvá A, Planes AM, Molina I, Larrosa MN. ESBL-producing Salmonella enterica serovar typhi in traveler returning from Guatemala to Spain. Emerg Infect Dis. 2014;20(11):1918.
Kariuki S, Okoro C, Kiiru J, Njoroge S, Omuse G, Langridge G, Kingsley RA, Dougan G, Revathi G. Ceftriaxone-Resistant Salmonella enterica Serotype Typhimurium Sequence Type 313 from Kenyan Patients Is Associated with the blaCTX-M-15 Gene on a Novel IncHI2 Plasmid. Antimicrob Agents Chemother. 2015;59(6):3133–9.
Richard P, Le Floch R, Chamoux C, Pannier M, Espaze E, Richt H. Pseudomonas aeruginosa outbreak in a burn unit: role of antimicrobials in the emergence of multiply resistant strains. J Infect Dis. 1994;170(2):377–83.
Rampling A, Wiseman S, Davis L, Hyett A, Walbridge A, Payne G, Cornaby A. Evidence that hospital hygiene is important in the control of methicillin-resistant Staphylococcus aureus. J Hosp Infect. 2001;49(2):109–16.
Fridkin SK. Increasing prevalence of antimicrobial resistance in intensive care units. Crit Care Med. 2001;29(4):N64–8.
Acknowledgements
The authors thank Lisa Gerwing-Adima, Medard Beyanga and Emmanuel Magembe for their highly valuable assistance in the BMC microbiology laboratory in Mwanza, Tanzania.
Funding
This study was supported by funds from Catholic University of Health and Allied Sciences to NM.
Availability of data and materials
All data have been included in this manuscript.
Authors’ contributions
NM, HC and SEM designed the study. NM collected data. NM and SEM analyzed and interpreted the data. NM wrote the first draft of the manuscript. NM, HC and SEM revised the manuscript and approved the final version to be submitted.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The Joint BMC/Catholic University of Health and Allied Sciences Scientific and Ethics Review committee approved publication of these data.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Moremi, N., Claus, H. & Mshana, S.E. Antimicrobial resistance pattern: a report of microbiological cultures at a tertiary hospital in Tanzania. BMC Infect Dis 16, 756 (2016). https://doi.org/10.1186/s12879-016-2082-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-016-2082-1