Abstract
Background
Sarcopenia is a multifactorial pathophysiologic condition of skeletal muscle mass and muscle strength associated with aging. However, biomarkers for predicting the occurrence of sarcopenia are rarely discussed in recent studies. The aim of the study was to elucidate the relationship between sarcopenia and several pertinent biomarkers.
Methods
Using the Gene Expression Omnibus (GEO) profiles of the National Center for Biotechnology Information, the associations between mRNA expression of biomarkers and sarcopenia were explored, including high temperature requirement serine protease A1 (HtrA1), procollagen type III N-terminal peptide (P3NP), apelin, and heat shock proteins 70 (Hsp72). We enrolled 408 community-dwelling adults aged 65 years and older with sarcopenia and nonsarcopenia based on the algorithm proposed by the Asian Working Group for Sarcopenia (AWGS). Muscle strength is identified by hand grip strength using an analogue isometric dynamometer. Muscle mass is estimated by skeletal mass index (SMI) using a bioelectrical impedance analysis. Physical performance is measured by gait speed using 6 m walking distance. The associations between these biomarkers and sarcopenia were determined using receiver operating characteristic (ROC) curve analysis and multivariate regression models.
Results
From the GEO profiles, the sarcopenia gene set variation analysis score was correlated significantly with the mRNA expression of APLNR (p < 0.001) and HSPA2 (p < 0.001). In our study, apelin was significantly associated with decreased hand grip strength with β values of − 0.137 (95%CI: − 0.229, − 0.046) in men. P3NP and HtrA1 were significantly associated with increased SMI with β values of 0.081 (95%CI: 0.010, 0.153) and 0.005 (95%CI: 0.001, 0.009) in men, respectively. Apelin and HtrA1 were inversely associated with the presence of sarcopenia with an OR of 0.543 (95%CI: 0.397–0.743) and 0.003 (95%CI: 0.001–0.890) after full adjustment. The cutoff point of HtrA1 was associated with the presence of sarcopenia with an OR of 0.254 (95%CI: 0.083–0.778) in men. The cutoff point of apelin was negatively associated with the presence of sarcopenia with an OR of 0.254 (95%CI: 0.083–0.778).
Conclusion
Our study highlights that P3NP, HtrA, and apelin are useful for diagnosis of sarcopenia in the clinical setting.
Similar content being viewed by others
Introduction
The term “sarcopenia” was first proposed in 1989 to describe the age-related decline of muscle mass that affects mobility, nutritional status, and independence [1]. The operational definition for sarcopenia proposed by European Working Group on Sarcopenia in Older People (EWGSOP) in 2010 has become the most widely used in the world which both muscle quantity and quality were cardinal requirements [2]. In 2019, Asian Working Group for Sarcopenia (AWGS) proposed an updated diagnostic algorithm based on Asian data, which sarcopenia is defined as low skeletal muscle mass with low muscle strength or low physical performance [3]. Sarcopenia is suggested to be a progressive and degenerative loss of skeletal muscle mass and muscle strength that occurs with aging that is associated with increased adverse outcomes [4]. Since its original description, the concept of sarcopenia has become more complicated and now involves multifactorial pathogenesis. The pathophysiology mechanisms of sarcopenia includes endocrine dysfunctions, neuromuscular junction, growth factors, inflammatory conditions, and muscle protein turnover [5, 6]. Based on these pathways, several biomarkers were proposed to be used for detecting and identifying the development of sarcopenia [7, 8].
However, the diagnosis of sarcopenia depends only on clinical, functional, and imaging parameters valuated after disease onset. It is essential to apply molecular biomarkers in the early diagnosis and prognosis of sarcopenia [9]. A previous study have reported several biomarkers such as high temperature requirement serine protease A1 (HtrA1), procollagen III N-terminal peptide (P3NP), apelin, and heat shock protein 70 (Hsp72) are possibly involved in different components of sarcopenia [10]. The aim of this study is to identify the role of potential biomarkers in the early diagnosis of sarcopenia in a Taiwanese older adult population.
To estimate the sample size of the study, we originally admitt a degree of error in the estimate of 5% and a confidence interval of 95% and have 80% power. Based on the study proposed by Asian Working Group for Sarcopenia (AWGS), the prevalence of sarcopenia in Asian population is about 12.9–15.9 %[11]. It was calculated as the minimum sample size necessary to include at least 173–206 participants to obtain an acceptable estimate. Therefore, our sample size exceeds 200 cases that allow us to make a statistically reliable sample of the prevalence of sarcopenia.
Method
Study design and recruitment of participants
In this cross-sectional study, the population was obtained during routine health checkup at Tri-Service General Hospital (TSGH), Taiwan, during 2015–2018. According to the flow chart shown in Fig. 1, 436 people aged 65 years or more were included in the study. A series of examination was performed as follows: clinical measures of sarcopenia included muscle strength, skeletal muscle mass, and gait speed were collected. Baseline laboratory data and biomarkers, including P3NP, HtrA1, apelin, and Hsp72 were collected from the participants. These participants were asked whether they had smoking history and medical history of stroke, malignancy, and chronic disease such as chronic kidney disease, chronic obstructive pulmonary disease, arthritis, liver disease, dyslipidemia, hypertension, diabetes mellitus, and coronary artery disease. Exclusion criteria included those with history of stroke (n = 10), malignancy (n = 3), or missing data included sarcopenia profiles, laboratory data, biomarkers, and past history of chronic disease. Finally, 43 participants with sarcopenia and 365 participants without sarcopenia were included in the subsequent analysis. All procedures were approved by the Institutional Review Board of TSGH (No. 2–103–05-024) in accordance with the revised Helsinki Declaration. We have obtained patient permission before enrollment by asking them to complete a wriiten informed consent.
Gene expression analysis from the gene expression omnibus (GEO) profiles
Gene expression of APLNR (apelin), HTRA1 (HtrA1), COL3A1 (P3NP) and HSPA2 (Hsp72) was obtained from the NCBI GEO database (accession ID: GSE18732) [12]. This dataset contains various physiological measurements and whole gene expression data of skeletal muscle (salverus lateralis) from 118 participants. Clinical information was selected to include demographic characteristics, medication, laboratory data, and fat mass and lean mass in the arms, legs, and trunk. To assess the extent of Sarcopenia by gene expression, we used the Sarcopenia gene set downloaded from the Enrichr database [13], which includes 27 genes that have been linked to sarcopenia in various literatures. To obtain individual Sarcopenia scores, we used the Gene Set Variation Analysis (GSVA) scoring strategy with default parameter settings for the analysis [14]. The analysis was performed in R using GSVA package (Version 1.38.2) according to the reference manual, and the enrichment score (ES) of the Sarcopenia gene set was calculated for each subject. A high GSVA score represents an overall higher expression of the relevant genes in the sample, therefore reflects the degree of activation of the sarcopenia gene set in the sample.
Diagnosis of sarcopenia
According to the algorithm proposed by the AWGS in 201 9[3], possible sarcopenia is determined when low muscle strength with or without reduced physical performance occurs. A diagnosis of sarcopenia is defined when reduced skeletal muscle mass appears with low muscle strength or low physical performance. When all criteria are displayed, sarcopenia is considered severe. In our study, muscle strength is identified by hand grip strength using an analogue isometric dynamometer (North Coast Hydraulic Hand Dynamometer, North Coast Medical Inc., Morgan Hill, CA). Muscle mass is estimated by skeletal mass index (SMI) using a bioelectrical impedance analysis (BIA) (InBody720, Biospace, Inc., Cerritos, CA, USA). SMI is calculated by dividing the skeletal muscle mass (kg) by the square of the height (m2). Physical performance is measured by gait speed using six meters walk. The cutoff points of hand grip strength are 28.0 kg for men and 18.0 kg for women. The cutoff points of SMI measuring by BIA are 7.0 kg/m2 for men and 5.7 kg/m2 for women. The cutoff point of gait speed are 1.0 m/s for both [2].
Biomarkers
We examined these multidimensional biomarkers to determine the underlying pathophysiological mechanisms, including P3NP, HtrA1, apelin, and Hsp72. According to the manufacturer’s instructions, plasma P3NP (Wuhan Fine Biotech Co., Ltd., Wuhan. China), HtrA1 (Cloud-Clone Corp., Houston, TX, USA), apelin (Phoenix Pharmaceuticals, Belmont, CA, USA, and Hsp72 (Enzo Life Science Inc., Farmingdale, NY, USA) were performed using a competitive enzyme-linked immunoassay (ELISA). The lower detection limit was 156 pg/mL for P3NP, 31.2 pg/mL for HtrA1, 70 pg/mL for apelin and 200 pg/mL for Hsp72. During the sample analysis period, the variation of these assays ranged from 5 to 10%.
Study variables
Educational level, past history of cigarette smoking, and chronic disease included chronic kidney disease, chronic obstructive pulmonary disease, arthritis, liver disease, dyslipidemia, hypertension, diabetes mellitus, and coronary artery disease was obtained from the self-reported questionnaires. Laboratory data included creatinine (Cr), total cholesterol (TC), and alanine aminotransferase (ALT) were collected from blood samples using standard procedures.
Statistical analysis
For all statistical analyses, we used the Statistical Package for the Social Sciences, version 22.0 (SPSS Inc., Chicago, IL, USA), for Windows. The normal distribution of data and variables were checked by Kolmogorov–Smirnov normality test. We stratify all participants in to male and female group due to significant findings of the interaction testing. In order to compare nonparametric variables in the test groups, we used Pearson chi-square test for qualitative variables and the Mann-Whitney U test for two independent groups to estimate the distributions. Spearman’s R correlation test was used to evaluate the relationships between the analyzed variables. The threshold for statistical significance was defined as p-value lower than 0.05. Correlations among sarcopenia GSVA scoring and arm, leg, and trunk fat mass and lean mass were determined using Pearson’s correlation coefficient. We analyzed associations between biomarkers and sarcopenia parameters using a linear regression model. Natural Log transformation was used to normalize the distributions of the biomarkers. To adjust these relationships, we used the variables, including age, education, number of chronic disease, and history of cigarette smoking. In addition, explained variance (Nagelkerke R2) was used to select the optimal model for the biomarkers. We used a receiving operating characteristic (ROC) analysis to determine the cutoff points of biomarkers as well as the area under the ROC (AUROC). Logistic regression model was used to determine the relationships between the cutoff points of the biomarkers and the odds ratio (OR) of the occurrence of sarcopenia.
Result
Association of the sarcopenia GSVA score with the mRNA expression of APLNR, HTRA1, COL3A1, and HSPA2
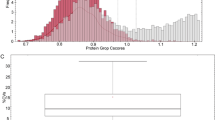
To determine which biomarkers were significantly correlated with the GSVA score of sarcopenia, we evaluated the relationships between several biomarkers and sarcopenia GSVA scoring via the DisGeNET databases (Fig. 2). Figure 2 shows that the sarcopenia GSVA score was significantly correlated with the mRNA expression of APLNR (p < 0.001) and HSPA2 (p < 0.001), but not with HTRA1 and COL3A1 mRNA expression. The lean mass of the arm, leg, and limb were not significantly negatively correlated with the mRNA expression of APLNR, HTRA1, COL3A1, and HSPA2.
Study population characteristics
Table 1 displays the baseline population and clinical variables of participants with and without sarcopenia. The mean ages of sarcopenia and control group were 76.92 ± 8.68 and 74.01 ± 37.63, respectively. Sarcopenia group had significantly higher proportion of smoking history than non-sarcopenia group. Participants without sarcopenia had significantly greater hand grip strength, SMI, and gait speed than those with sarcopenia. In terms of the biomarkers included in the study, sarcopenia group had significantly lower HtrA1 and apelin levels than control group, but no significant differences were found in other biomarkers.
Association between biomarkers and sarcopenia in gender difference
Table 2 analyzes the associations between P3NP, HtrA1, apelin, and Hsp72 and sarcopenia components, including grip strength, SMI, and gait speed, in both men and women. After full adjustment, apelin was significantly associated with decreased hand grip strength with β values of − 0.137 (95%CI: − 0.229, − 0.046) in men. P3NP and HtrA1 were significantly associated with increased SMI with β values of 0.081 (95%CI: 0.010, 0.153) and 0.005 (95%CI: 0.001, 0.009) in men, respectively. However, no significant difference was detected in the relationship between biomarkers and gait speed.
Association between biomarkers and the presence of sarcopenia
In Table 3, apelin was associated with dcreased presence of sarcopenia with an OR of 0.543 (95%CI: 0.397–0.743) after full adjustment. HtrA1 was associated with decreased presence of sarcopenia with an OR of 0.003 (95%CI: 0.001–0.890) after full adjustment. Nevertheless, no significant association was noted in other biomarkers.
Optimal cutoff points in P3NP, HtrA1, apelin, and Hsp72
Based on the results mentioned above, we further calculated the optimal cutoff points of P3NP, HtrA1, apelin, and Hsp72 using ROC curve analysis to identify the ability of these biomarkers to detect the presence of sarcopenia (Table 4). The AUROC was 0.578 (95%CI: 0.482–0.675; sensitivity: 48.8%, specificity: 66.5%), 0.593 (95%CI: 0.506–0.679; sensitivity: 38.0%, specificity: 89.5%), 0.709 (95%CI: 0.651–0.767; sensitivity: 47.3%, specificity: 95.8%), and 0.606 (95%CI: 0.524–0.688; sensitivity: 75.6%, specificity: 49.0%) in P3NP, HtrA1, apelin, and Hsp72, respectively. The cutoff point was determined by Youdan index that P3NP was 0.045 in men and 0.035 in women; HtrA1 was 0.067 in men and 0.035 in women; apelin was 0.911 in men and 0.863 in women; Hsp72 was 0.135 in men and 0.150 in women.
Association between cutoff points of biomarkers and the presence of sarcopenia in gender difference
In Table 5, HtrA1 was associated with the presence of sarcopenia with an OR of 0.254 (95%CI: 0.083–0.778) in men after full adjustment. Apelin was significantly associated with the presence of sarcopenia with an OR of 0.254 (95%CI: 0.083–0.778). In addition, apelin was negatively associated with the presence of sarcopenia with an OR of 0.283 (95%CI: 0.123–0.655) without adjustment in men, however, the p-value was slightly elevated after full adjustment that no significant difference was noted in the association with the presence of sarcopenia.
Discussion
Diverse pathophysiological mechanisms contribute to the process of sarcopenia. It is imperative to identify individuals at risk of sarcopenia and prevent the progression to disability and functional dependence, or even medical institutionalization of the older adult population. In the present study, we investigated the relationships between biomarkers and sarcopenia among Taiwanese older adults. We found that HtrA1 was associated with decreased presence of sarcopenia in older men and apelin is significantly associated with reduced occurrence of sarcopenia, in women. Our findings suggest that molecular biomarkers may represent a beneficial effect on aging and an essential tool for early diagnosis of sarcopenia in older adult population.
Apelin is a novel adipokine that is expressed in different tissues and secreted by.
both human and mouse adipocytes [15,16,17]. It is believed that apelin can be used to reduce insulin resistance with the aim of improving altered glucose metabolism [18]. Vinel et al. demonstrated that apelin synthesis in skeletal muscle is reduced and plasma apelin levels decrease in aged mouse [19]. Conversely, after supplementation with apelin injection per day, improved skeletal muscle capacities and myofiber hypertrophy were noted. Several researchers have considered apelin as an adipokine because of its upregulation in adiposity mass with obesity and improvement of cardiometabolic diseases [20,21,22]. This mediator has a beneficial effect on energy metabolism by increasing insulin sensitivity and glucose uptake [23]. Emerging research has indicated Apelin as a myokine from skeletal muscle that might have an influence on muscle physiology and function [24]. Apelin is suggested to induce the promotion of protein synthesis and inhibition of age-related proteolysis in aged myotubes [19]. On the other hand, apelin can trigger mechanisms of myogenesis and angiogenesis coupling that leads to an improvement in regenerative processes in the muscle cells of aged mice [25]. Apelin has been found as an independent predictor of bone mineral density in postmenopausal women [26]. It has an inhibitory effect on gonadotropin and prolactin secretion in females [27]. We speculated that the relationship between apelin and sarcopenia might be the interaction after menopause.
HtrA1, a protease of the family [28], is regarded as an important role in the inflammation via transforming growth factor-β (TGF-β) inhibition [29]. HtrA1 is involved in the pathophysiological mechanisms of these diseases, such as osteoarthritis, dementia, and age-related macular degeneration [30, 31]. Lorenzi et al. first described the association between HtrA1 and frailty in older population [32]. Loss of HtrA1 was reported to impair muscle cell maturation via TGF-β pathway [33]. Emerging study proposed that HtrA protease deficiency induced denervation-independent skeletal muscle degeneration with sarcopenia [34]. In our study, we found that HtrA1 was associated with the presence of sarcopenia, particularly in men. However, rare studies discussed the connection of HtrA1 and gender difference. The potential role of this biomarker warrants further longitudinal studies to explore in the association with sarcopenia.
Hormones, including testosterone, growth hormone, and dehydroepiandrosterone are potential biomarkers of sarcopenia [35,36,37]. Age-related reductions in protein synthesis and muscle function have been linked to age-related declines in sex hormones and growth factors [38]. Testosterone can increase plasma P3NP levels in a dose-dependent manner and stimulate the expression of several skeletal muscle transcripts and proteins [39]. Changes in levels of P3NP have been regarded as an early marker of muscle quality and function. In males, high levels of P3NP have been noted when the normal increases in skeletal mass in response to endurance-type exercise [40]. In a clinical trial of 106 elderly men, increases in P3NP induced by growth hormone treatment were found to be related to greater gains in total and appendicular skeletal mass [41]. The exercise intervention is believed to increase circulating P3NP and is positively correlated with changes in muscle mass [42]. Collectively, our findings are consistent with the results of these studies, however, future studies should examine other mechanisms as well, especially the role of apelin and P3NP in skeletal muscle function.
This study had some limitations that should be acknowledged. The major limitation is that we can’t identify the causal relationship between biomarkers and sarcopenia because of the study design. It is essential to conduct longitudinal studies to examine whether molecular biomarkers applying for early diagnosis and to evaluate clinical intervention in sarcopenia. Second, all participants included in this study were non-institutionalized and relatively healthy. It might lead to the underestimation of the prevalence of sarcopenia. Next, some potential confounding factors influencing sarcopenia, including physical activity, dementia status, nutritional status, number of medications, and body mass index were not included in the analysis, because these variables were not recorded in the health checkup. Last, we included only Taiwanese participants in this study. Thus, the generalization of the results to other population may be precluded.
Conclusion
In the present study, we found that HtrA1 and apelin were significantly associated with sarcopenia and provide a potential approach in molecular diagnosis of in older adult population. Our findings would help us more recognize the pathophysiologic mechanisms of this disorder. Sarcopenia is a multifactorial pathogenesis condition and is considered to be highly prevalent and carry a high risk for adverse health outcomes. It is important to determine a clear definition and diagnostic criteria of sarcopenia to guide both clinical practice and research design.
Availability of data and materials
The datasets generated and analyses performed during the current study are not publicly available due to the consent requirement of participants, but sex and age decade-stratified descriptive data are available from the corresponding author on reasonable request.
Change history
20 April 2022
A Correction to this paper has been published: https://doi.org/10.1186/s12877-022-02882-2
Abbreviations
- EWGSOP:
-
European Working Group on Sarcopenia in Older People
- AWGS:
-
Asian Working Group for Sarcopenia
- TSGH:
-
Tri-Service General Hospital
- P3NP:
-
Procollagen type III N-terminal peptide
- HtrA1:
-
High temperature requirement serine protease A1
- Hsp72:
-
Heat shock proteins 70
- GEO:
-
Gene Expression Omnibus
- SMI:
-
Skeletal mass index
- Cr:
-
Creatinine
- TC:
-
Total cholesterol
- ALT:
-
Alanine aminotransferase
- ROC:
-
Receiving operating characteristic
- CI:
-
Confidence interval
- OR:
-
Odds ratio
- IRB:
-
Institutional Review Board
- GSVA:
-
Gene Set Variation Analysis
- ELISA:
-
Enzyme-linked immunoassay
- TGF-β:
-
Transforming growth factor-β
References
Rosenberg IH. Summary comments. Am J Clin Nutr. 1989;50(5):1231–3.
Cruz-Jentoft AJ, Baeyens JP, Bauer JM, Boirie Y, Cederholm T, Landi F, Martin FC, Michel JP, Rolland Y, Schneider SM, et al. Sarcopenia: European consensus on definition and diagnosis: report of the European working group on sarcopenia in older people. Age Ageing. 2010;39(4):412–23.
Chen LK, Woo J, Assantachai P, Auyeung TW, Chou MY, Iijima K, Jang HC, Kang L, Kim M, Kim S, et al. Asian Working Group for Sarcopenia: 2019 Consensus Update on Sarcopenia Diagnosis and Treatment. J Am Med Dir Assoc. 2020;21(3):300–307.e302.
Cruz-Jentoft AJ, Sayer AA. Sarcopenia. Lancet. 2019;393(10191):2636–46.
Curcio F, Ferro G, Basile C, Liguori I, Parrella P, Pirozzi F, Della-Morte D, Gargiulo G, Testa G, Tocchetti CG, et al. Biomarkers in sarcopenia: a multifactorial approach. Exp Gerontol. 2016;85:1–8.
Malafarina V, Uriz-Otano F, Iniesta R, Gil-Guerrero L. Sarcopenia in the elderly: diagnosis, physiopathology and treatment. Maturitas. 2012;71(2):109–14.
Coto Montes A, Boga JA, Bermejo Millo C, Rubio González A, Potes Ochoa Y, Vega Naredo I, Martínez Reig M, Romero Rizos L, Sánchez Jurado PM, Solano JJ, et al. Potential early biomarkers of sarcopenia among independent older adults. Maturitas. 2017;104:117–22.
Kwak JY, Hwang H, Kim S-K, Choi JY, Lee S-M, Bang H, Kwon E-S, Lee K-P, Chung SG, Kwon K-S. Prediction of sarcopenia using a combination of multiple serum biomarkers. Sci Rep. 2018;8(1):8574.
Scharf G, Heineke J. Finding good biomarkers for sarcopenia. J Cachexia Sarcopenia Muscle. 2012;3(3):145–8.
Calvani R, Picca A, Marini F, Biancolillo A, Cesari M, Pesce V, Lezza AMS, Bossola M, Leeuwenburgh C, Bernabei R, et al. The “BIOmarkers associated with sarcopenia and PHysical frailty in EldeRly pErsons” (BIOSPHERE) study: rationale, design and methods. Eur J Intern Med. 2018;56:19–25.
Mayhew AJ, Amog K, Phillips S, Parise G, McNicholas PD, de Souza RJ, Thabane L, Raina P. The prevalence of sarcopenia in community-dwelling older adults, an exploration of differences between studies and within definitions: a systematic review and meta-analyses. Age Ageing. 2019;48(1):48–56.
Gallagher IJ, Scheele C, Keller P, Nielsen AR, Remenyi J, Fischer CP, Roder K, Babraj J, Wahlestedt C, Hutvagner G, et al. Integration of microRNA changes in vivo identifies novel molecular features of muscle insulin resistance in type 2 diabetes. Genome Med. 2010;2(2):9.
Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1):W90–7.
Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7.
Tatemoto K, Hosoya M, Habata Y, Fujii R, Kakegawa T, Zou MX, Kawamata Y, Fukusumi S, Hinuma S, Kitada C, et al. Isolation and characterization of a novel endogenous peptide ligand for the human APJ receptor. Biochem Biophys Res Commun. 1998;251(2):471–6.
Castan-Laurell I, Vitkova M, Daviaud D, Dray C, Kovacikova M, Kovacova Z, Hejnova J, Stich V, Valet P. Effect of hypocaloric diet-induced weight loss in obese women on plasma apelin and adipose tissue expression of apelin and APJ. Eur J Endocrinol. 2008;158(6):905–10.
Boucher J, Masri B, Daviaud D, Gesta S, Guigne C, Mazzucotelli A, Castan-Laurell I, Tack I, Knibiehler B, Carpene C, et al. Apelin, a newly identified adipokine up-regulated by insulin and obesity. Endocrinology. 2005;146(4):1764–71.
Wysocka MB, Pietraszek-Gremplewicz K, Nowak D. The role of Apelin in cardiovascular diseases, obesity and cancer. Front Physiol. 2018;9:557.
Vinel C, Lukjanenko L, Batut A, Deleruyelle S, Pradere JP, Le Gonidec S, Dortignac A, Geoffre N, Pereira O, Karaz S, et al. The exerkine apelin reverses age-associated sarcopenia. Nat Med. 2018;24(9):1360–71.
Yu X-H, Tang Z-B, Liu L-J, Qian H, Tang S-L, Zhang D-W, Tian G-P, Tang C-K. Apelin and its receptor APJ in cardiovascular diseases. Clin Chim Acta. 2014;428:1–8.
Dray C, Debard C, Jager J, Disse E, Daviaud D, Martin P, Attane C, Wanecq E, Guigne C, Bost F, et al. Apelin and APJ regulation in adipose tissue and skeletal muscle of type 2 diabetic mice and humans. Am J Physiol Endocrinol Metab. 2010;298(6):E1161–9.
Dray C, Knauf C, Daviaud D, Waget A, Boucher J, Buleon M, Cani PD, Attane C, Guigne C, Carpene C, et al. Apelin stimulates glucose utilization in normal and obese insulin-resistant mice. Cell Metab. 2008;8(5):437–45.
Bertrand C, Valet P, Castan-Laurell I. Apelin and energy metabolism. Front Physiol. 2015;6:115.
Besse-Patin A, Montastier E, Vinel C, Castan-Laurell I, Louche K, Dray C, Daviaud D, Mir L, Marques MA, Thalamas C, et al. Effect of endurance training on skeletal muscle myokine expression in obese men: identification of apelin as a novel myokine. Int J Obes. 2014;38(5):707–13.
Latroche C, Weiss-Gayet M, Muller L, Gitiaux C, Leblanc P, Liot S, Ben-Larbi S, Abou-Khalil R, Verger N, Bardot P, et al. Coupling between Myogenesis and angiogenesis during skeletal muscle regeneration is stimulated by restorative macrophages. Stem Cell Rep. 2017;9(6):2018–33.
Zhang H, Xie H, Zhao Q, Xie GQ, Wu XP, Liao EY, Luo XH. Relationships between serum adiponectin, apelin, leptin, resistin, visfatin levels and bone mineral density, and bone biochemical markers in post-menopausal Chinese women. J Endocrinol Investig. 2010;33(10):707–11.
Kurowska P, Barbe A, Różycka M, Chmielińska J, Dupont J, Rak A. Apelin in reproductive physiology and pathology of different species: a critical review. Int J Endocrinol. 2018;2018:9170480.
Tiaden AN, Richards PJ. The emerging roles of HTRA1 in musculoskeletal disease. Am J Pathol. 2013;182(5):1482–8.
Oka C, Tsujimoto R, Kajikawa M, Koshiba-Takeuchi K, Ina J, Yano M, Tsuchiya A, Ueta Y, Soma A, Kanda H, et al. HtrA1 serine protease inhibits signaling mediated by Tgfbeta family proteins. Development. 2004;131(5):1041–53.
Yang Z, Camp NJ, Sun H, Tong Z, Gibbs D, Cameron DJ, Chen H, Zhao Y, Pearson E, Li X, et al. A variant of the HTRA1 gene increases susceptibility to age-related macular degeneration. Science. 2006;314(5801):992–3.
Grau S, Baldi A, Bussani R, Tian X, Stefanescu R, Przybylski M, Richards P, Jones SA, Shridhar V, Clausen T, et al. Implications of the serine protease HtrA1 in amyloid precursor protein processing. Proc Natl Acad Sci U S A. 2005;102(17):6021–6.
Lorenzi M, Lorenzi T, Marzetti E, Landi F, Vetrano DL, Settanni S, Antocicco M, Bonassi S, Valdiglesias V, Bernabei R, et al. Association of frailty with the serine protease HtrA1 in older adults. Exp Gerontol. 2016;81:8–12.
Klose R, Prinz A, Tetzlaff F, Weis E-M, Moll I, Rodriguez-Vita J, Oka C, Korff T, Fischer A. Loss of the serine protease HTRA1 impairs smooth muscle cells maturation. Sci Rep. 2019;9(1):18224.
Zhou H, Yuan D, Gao W, Tian J, Sun H, Yu S, Wang J, Sun L. Loss of high-temperature requirement protein A2 protease activity induces mitonuclear imbalance via differential regulation of mitochondrial biogenesis in sarcopenia. IUBMB Life. 2020;72(8):1659–79.
Morley JE, Baumgartner RN, Roubenoff R, Mayer J, Nair KS. Sarcopenia. J Lab Clin Med. 2001;137(4):231–43.
Perrini S, Laviola L, Carreira MC, Cignarelli A, Natalicchio A, Giorgino F. The GH/IGF1 axis and signaling pathways in the muscle and bone: mechanisms underlying age-related skeletal muscle wasting and osteoporosis. J Endocrinol. 2010;205(3):201–10.
Valenti G, Denti L, Maggio M, Ceda G, Volpato S, Bandinelli S, Ceresini G, Cappola A, Guralnik JM, Ferrucci L. Effect of DHEAS on skeletal muscle over the life span: the InCHIANTI study. J Gerontol A Biol Sci Med Sci. 2004;59(5):466–72.
Ogawa K. The biomarkers of sarcopenia in elderly people. J Phys Fit Sports Med. 2013;2(2):209–14.
Chen F, Lam R, Shaywitz D, Hendrickson RC, Opiteck GJ, Wishengrad D, Liaw A, Song Q, Stewart AJ, Cummings CE, et al. Evaluation of early biomarkers of muscle anabolic response to testosterone. J Cachexia Sarcopenia Muscle. 2011;2(1):45–56.
Wallace JD, Cuneo RC, Lundberg PA, Rosen T, Jorgensen JO, Longobardi S, Keay N, Sacca L, Christiansen JS, Bengtsson BA, et al. Responses of markers of bone and collagen turnover to exercise, growth hormone (GH) administration, and GH withdrawal in trained adult males. J Clin Endocrinol Metab. 2000;85(1):124–33.
Bhasin S, He EJ, Kawakubo M, Schroeder ET, Yarasheski K, Opiteck GJ, Reicin A, Chen F, Lam R, Tsou JA, et al. N-terminal propeptide of type III procollagen as a biomarker of anabolic response to recombinant human GH and testosterone. J Clin Endocrinol Metab. 2009;94(11):4224–33.
Fragala MS, Jajtner AR, Beyer KS, Townsend JR, Emerson NS, Scanlon TC, Oliveira LP, Hoffman JR, Stout JR. Biomarkers of muscle quality: N-terminal propeptide of type III procollagen and C-terminal agrin fragment responses to resistance exercise training in older adults. J Cachexia Sarcopenia Muscle. 2014;5(2):139–48.
Acknowledgements
None.
Funding
This study was supported by the Tri-Service General Hospital (TSGH-C108–081) and the Ministry of Science and Technology (MOST 108–2314-B-016 -032), Taiwan, ROC.
Author information
Authors and Affiliations
Contributions
Conceptualization: YYC and WLC; Methodology: YYC, YLC, TWK, TCP, HFY, and WLC; Formal analysis and investigation: YYC, YLC, TWK, TCP, HFY, and WLC; Writing- original draft preparation: YYC; Writing- review and editing: WLC; Supervision: YYC, YLC, TWK, TCP, HFY, and WLC. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All procedures were conducted in accordance with the guidelines of TSGH. We have obtained patient permission before enrollment by asking them to complete a wriiten informed consent, and approval for the study was granted by the Institutional Review Board of TSGH, Taiwan.
Consent for publication
Not applicable.
Competing interests
The authors declared that they had no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Chen, YY., Chiu, YL., Kao, TW. et al. Cross-sectional associations among P3NP, HtrA, Hsp70, Apelin and sarcopenia in Taiwanese population. BMC Geriatr 21, 192 (2021). https://doi.org/10.1186/s12877-021-02146-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12877-021-02146-5