Abstract
Background
Recently, radiomics has been widely used in colorectal cancer, but many variable factors affect the repeatability of radiomics research. This review aims to analyze the repeatability of radiomics studies in colorectal cancer and to evaluate the current status of radiomics in the field of colorectal cancer.
Methods
The included studies in this review by searching from the PubMed and Embase databases. Then each study in our review was evaluated using the Radiomics Quality Score (RQS). We analyzed the factors that may affect the repeatability in the radiomics workflow and discussed the repeatability of the included studies.
Results
A total of 188 studies was included in this review, of which only two (2/188, 1.06%) studies controlled the influence of individual factors. In addition, the median score of RQS was 11 (out of 36), range-1 to 27.
Conclusions
The RQS score was moderately low, and most studies did not consider the repeatability of radiomics features, especially in terms of Intra-individual, scanners, and scanning parameters. To improve the generalization of the radiomics model, it is necessary to further control the variable factors of repeatability.
Similar content being viewed by others
Background
Colorectal cancer (CRC) is one of the most common clinical malignant tumors [1]. Medical imaging tools have become crucial in CRC for staging and treatment evaluation [2]. However, traditional radiology is mainly dependent on the subjective qualitative interpretations of the doctor [2], which often leads to suboptimal positive and negative predictive values [2, 3]. In recent years, with the rapid development of image analysis methods and pattern recognition tools, there is a growing shift away from qualitative to quantitative analysis of medical images [2].
As a quantitative analysis tool, radiomics extracts features from medical images through high-throughput computing and applies them to personalized clinical decisions to improve the accuracy of diagnosis and prognosis [4]. In recent years, radiomics showed a unique advantage for staging, differential diagnosis, and prognosis [5]. Although an increasing amount of radiomics research has been published, the comparability and repeatability of radiomics models remain a great challenge due to the lack of standardization in the field of radiomics [6, 7]. Assessing the repeatability of radiomics is necessary to achieve the clinical implementation of radiomics results and to ensure a high predictive capability of the radiomics model for a variety of populations and institutions [8]. In addition, several factors that affect the repeatability have been identified in the complicated workflow of radiomics, such as scanner [9,10,11], acquisition parameters [11,12,13,14,15,16], pretreatment method [17, 18], segmentation method [19,20,21,22], inter/intra-observer variability [16, 17, 19], feature selection method [23], modeling method [23].
Therefore, we conducted a systematic review to survey the repeatability of radiomics research in CRC. Furthermore, we gave some suggestions to increase radiomics repeatability for future research.
Methods
Review strategy
We conducted a systematic review according to the Preferred Reporting items for Systematic review and Meta-Analysis (PRISMA) checklist [24]. But the review was not registered before. The systematic search was conducted by two reviewers via PubMed and Embase databases until Jul 4, 2022. The full search strategies from Additional Text 1.
Study selection
Population
We included primary research assessing the role of radiomics for diagnostic or prognostic with CRC patients. However, studies consisting of animal subjects and other types of articles than original articles (reviews, case reports, brief communications, technical reports, letters to editors, comments, and conference proceedings) were excluded.
Intervention
To be included, studies had to use the radiomics analysis of the preoperative or postoperative medical images in CRC patients for stratification of the CRC, prediction of response to therapy, or prognosis.
Outcome
In this review, the primary outcome of interest was the repeatability in the whole process of the radiomics research (including intra-individual, imaging acquisition, segmentation, feature selection, modeling, and evaluation). Studies with insufficient information for assessing the methodological quality were excluded.
Study extraction and quality assessment
The following data from each eligible study was systematically recorded: author, year, purpose, type, sample size, imaging modality, acquisition parameters, reconstruction parameters, pretreatment method, feature selection method, modeling method, segmentation method, number of features, verification method, performance index, and clinical utility.
In addition, the methodological quality of the eligible studies was assessed by the Radiomics Quality Score (RQS) [4]. The RQS was a unique quality assessment tool in radiomics [25], which score was composed of 16 parts with a total score of 36. A higher score represents better quality of the article. There were great differences in the methods used in the eligible studies, so the meta-analysis did not conduct.
Risk of bias in individual studies
The common bias analysis tools were not applicable here for the following reasons. First, the systematic review aims to assess the repeatability of radiomics research rather than the clinical purpose and outcomes. Second, there is no strictly causal association between repeatability and outcomes (diagnostic or prognosis performance). So the TRIPOD (Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis) and ROBINS (Risk Of Bias in Non-randomized Studies) were not applicable. Finally, the purposes of the eligible studies were highly heterogeneous, including staging, diagnosis, prognosis, and evaluating treatment. Thus, QUADAS-2(Quality Assessment of Diagnostic Accuracy Studies), which assesses the risk of bias in diagnostic studies, and QUIPS (Quality in Prognosis Studies), which assesses the risk of bias in prognostic studies, were not applicable.
Quality assessment was conducted using the RQS. Furthermore, the risk of bias in the eligible studies was assessed by two reviewers from the following specific aspects:
-
1)
Sufficiency of method description and disclosure (imaging acquisition, stability of the segmentation, details of the selected features, methods for selecting features and modeling, and sufficiency of model performance description).
-
2)
Description of the code used to compute features, establish models, verify models, and statistical analysis.
-
3)
The methods to improve study reproducibility (phantom study and test–retest study) and validating the model in the external validation cohort.
Results
Study selection
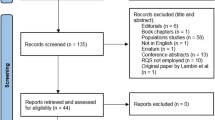
A total of 624 articles were retrieved from the comprehensive literature search (PubMed and Embase), of which were reduced to 358 based on screening of title and abstract. Of these studies, 188 articles were included after the full-text articles were assessed for eligibility. Include and exclude flowchart is shown in Fig. 1.
Statistics of the studies
The current situation of radiomics in patients with colorectal cancer was analyzed in the following aspects, and the included articles were given an overall overview. The key information of included studies is summarized in Additional Table 1.
Figure 2A shows the published number based on radiomics in patients with colorectal cancer in recent years. The number of articles continued to increase from 2016 to 2020 and decreased from 2020 to 2022. The publication number has an overall increasing trend.
The included studies were designed to assess the prognosis (101, 53.72%), response (74, 39.36%), staging (9, 4.79%), and diagnosis (4, 2.13%) of patients with colorectal cancer (Fig. 2B). Predictive performance was moderate to good in all radiomics models, but performance varied widely between models (the area under the curve values ranging from 0.56 to 0.98).
The imaging modalities of the studies include CT, MRI, PET, PET-CT, US, and multi-modality (Fig. 2C). It is worth noting that CT and MRI account for 35.11% (66/188) and 51.60% (97/188) of all studies.
Most radiomics of colorectal cancer were based on retrospective data sets (177, 94.15%), and only a few were prospective studies. Multicenter studies might fully reflect the overall situation and evaluate the generalization ability of the model. Of all the studies, only 9.04% (17/188) of articles had data from multiple medical institutions and 12.77% (24/188) of articles were dual-center studies.
Quality of the studies
RQS score
The included articles were scored by RQS, and the specific scores were shown in Additional Table 2. Its score ranges from-1 to 27 (-3.03% to 75.00%), the median was 11 (30.56%), of which 93.09% (175) studies scored less than 50%.
The total score of each item in the RQS is different, so it is difficult to compare the scores of each item. Therefore, the score ratio (actual score /total score*100%) was used to compare the scores of each item (Fig. 3).
Even though feature reduction was performed in all studies, 34.57% (65/188) of all studies still had the risk of overfitting, following the “one in ten” rule of thumb (at least ten patients for each feature in the model) [26]. Although well-documented image protocols for the studies were provided in 173 articles, only P Lovinfosse, et al. [27] showed [8] their study’s images were acquired and reconstructed according to the public image protocols. The cost-effectiveness of the clinical application has not been considered in the included studies, which is one of the most important components for the clinical application of the models. Only 0.53% (1/188) of the studies detect inter-scanner differences by phantom (Phantom study), and only 1.06% (2/188) analyzed feature robustness to temporal variabilities (Multiple time points).
Risk of bias
The potential risk of bias was present in the included studies to varying degrees and different aspects. The risk of bias in the studies was assessed in the terms of method details provided, code details provided, and repeatability (Additional Table 3).
Repeatability
The repeatability of radiomics features was directly related to the accuracy of model [11]. The repeatability might be affected by many factors in the radiomics process, Such as scanner [9,10,11], acquisition parameters [11,12,13,14,15,16], pretreatment method [17, 18], segmentation method [19,20,21,22], inter/intra-observer variability [16, 17, 19], feature selection method [23], modeling method [23]. The factors and solutions in the radiomics workflow are shown in Fig. 4.
Intra-individual repeatability
Radiomic features may be influenced by organ motion or expansion or shrinkage of the target volume caused by physiological factors such as respiration, bowel peristalsis, cardiac and cardiac activity [4]. The robustness of the radiomics features affected by various physiological factors of the individual was called intra-individual repeatability. The intra-individual repeatability was neglected in the majority of included studies, while the robustness of the features was analyzed by test–retest in only two (2/188, 1.06%) studies [28, 29]. X Ma, et al. [28] set the basis of an intraclass correlation coefficient (ICC) of 0.6 for test–retest analysis, but the population or phantom, time interval, and the ICC form in the test–retest analysis were not described. J Wang, et al. [29] performed a test–retest analysis on 40 patients with stage II rectal cancer, which scanned twice using the same scanner and imaging protocol before treatment, and calculated the Spearman’s correction coefficients for each feature. Although the test–retest analyses were taken, the methodology among the two studies varied considerably.
Acquisition parameters
The radiomics features were derived from the same scanner and imaging protocol in the 89 (89/188,47.34%) studies, and the impact on radiomics features due to scanner differences might be reduced. Notable was that K Nie, et al. [30] underwent quality assurance checks monthly and maintained bimonthly to ensure the image quality. And the standardized uptake value (SUV) measurement difference between the two scanners was reduced to less than 10 percent through regular standardization and quality assurance in the study of J Kang, et al. [31]. The effect of acquisition parameters on radiomics features might be further controlled by taking the above measures.
In addition, various image post-processing approaches were conducted to reduce the variation of the radiomics features. The image intensity discretization and normalization were investigated to reduce the noise and inconsistencies in the 60 (60/188, 31.91%) studies. In total, 42 (42/188, 22.34%) studies conducted the resampling, which was performed by linear interpolation, to mitigate the influence of the layer thickness. The size of voxels after resampling was not uniform between the included studies.
Segmentation
The radiomics features' robustness may be related to the segmentation of regions of interest (ROI) [10]. The segmentation method includes manual, semi-automatic, and automatic segmentation. Manual segmentation is usually regarded as the gold standard, but it was time-consuming [19] and suffered from great subjective differences among the observers [32, 33].
The advantages of semi-automatic segmentation in terms of segmentation time have been reported in the existing studies [19,20,21, 34, 35]. MM van Heeswijk, et al. [19] analyzed the segmentation methods and found that semi-automatic segmentation has similar accuracy to manual segmentation while reducing the time by 4 min (manual segmentation time 60–1118 s). Semi-automatic segmentations were used in 22 (11.70%) studies, while automatic segmentations were used in 11 (5.85%) studies. The segmentation methods were not mentioned in the twelve (6.38%) studies, manual segmentations were adopted in the remaining 143 (76.06%).
The radiomics features might be affected by segmentation variation (inter-observer variation and intra-observer variation), while multi-person/multi-method segmentation may reduce segmentation variation [4]. The multi-person/multi-method segmentation was not performed in 37 (37/188, 19.68%) studies. Segmentation variation was not measured quantitatively in 44 studies but was measured with different parameters in the remaining 111 studies. Among the included studies, intraclass correlation coefficient (ICC), Dice similarity Coefficient and/or Jaccard similarity coefficient, Bland–Altman plots, and Spearman correlation coefficient was used as evaluation parameters in 94 (94/111, 84.68%), 9 (9/111,8.11%), 1 (1/111, 0.90%), and 1 (1/111, 0.90%) articles, respectively. Notable was that P Lovinfosse, et al. [27] used automatic segmentation which repeatability was verified, and 4 articles [26, 36,37,38] defined a segmentation z-score that captures the robust features in the segmentation. In a word, most studies (84.68%) used ICC to evaluate segmentation variability, while I Fotina, et al. [39] preferred to use the Jaccard similarity coefficient, conformal number, or generalized conformability index to evaluate segmentation variability.
The ROI definition among studies varied considerably. The tumor region was defined as the ROI in most studies (174/188, 92.55%), while the peri-tumor region was also taken into account in 6 (6/188, 3.19%) studies. Three (3/188, 1.60%) studies used the entire parenchyma as the ROI. In addition, the tumor region and the lymph node region were defined as the ROIs in 3 (3/188, 1.60%) studies, and the ROIs were traced manually along the largest lateral pelvic lymph node in the study of R Nakanishi, et al. [40].
Feature selection
A large number of features were extracted in the radiomics studies, leading to the dimension disaster and the model overfitting so greatly reducing the generalization ability of the model [41, 42]. To reduce the false positive rate, A Chalkidou, et al. [42] proposed the following measures: (1) repeatability of features (2) cross-correlation analysis (3) inclusion of clinically important features (4) at least 10–15 patients with each feature (5) external verification.
The main purposes of feature selection were (1) to select repeatable features among the institutions, (2) to remove redundant features (highly related features between features), and (3) to select features with predictive potential. Various methods and combinations of feature selection were applied to reduce the number of features [43]. In total, only one feature selection method was used in 77 (77/188, 40.96%) articles, while two, three, four, five, and six feature selection methods were used in combination in 62 (62/188, 32.98%), 36 (36/188, 19.15%), 11 (11/188, 5.85%), 1(1/188, 0.53%), and 1(1/188, 0.53%) articles, respectively. L Boldrini, et al. [44] extracted only two features to predict the clinical complete response after neoadjuvant radio-chemotherapy, so no feature selection measures were taken. The Least Absolute Shrinkage and Selection operator (LASSO) (106/188, 56.38%) was the most commonly used, followed by correlation analysis (52/188, 27.66%). The feature selection scheme should be adjusted according to the number of features and samples [41].
The sample size of all studies ranged from 15 to 918, with a median of 149, and 66.49% (125/188) of the studies had a sample size of 0–200. To assess the adequacy of the sample size in the study, MA Babyak [26] suggested that at least 10–15 patients were needed for each feature. Based on this standard, 65 (65/188, 34.57%) of the included studies did not meet the above conditions except 4 (4/188, 2.13%) studies [44,45,46,47] which did not establish a model, and 14 (14/188, 7.45%) studies did not indicate the number of features (Fig. 5).
Modelling methodology
Five (5/188, 2.66%) studies [27, 44,45,46,47] analyzed the predictive performance of individual radiomics features, while no predictive models were constructed. M Hotta, et al. [46] reported the gray-level co-occurrence matrix entropy was the relevant feature for overall survival and progression-free survival. The delta radiomics features (L_least and glnu) were the most predictive feature ratios in clinical complete response prediction in the studies of L Boldrini, et al. [44]. Subsequently, D Cusumano, et al. [45] validated the prognostic potential of these delta radiomics features on an external validation cohort, while the accuracy of L_least is significantly higher than glnu. AA Negreros-Osuna, et al. [47] found that the spatial scaling factor was the potential biomarker for determining BRAF mutation status and predicting the 5-year overall survival.
Radiomics models were constructed in 183 (183/188, 97.34%) studies, while prediction models were constructed by combining clinical factors with radiomics features in 100 (100/184, 54.35%) studies. Z Liu, et al. [48] constructed a radiomics-clinical model which significantly improved the classification accuracy compared to the clinical model, based on the integrated discrimination improvement values. And the radiomics model outperformed the qualitative analysis by radiologists in the study of H Tibermacine, et al. [36].
A variety of models including machine learning models and statistical models were used in the radiomics studies, such as logistic regression (LR), random forest (RF), supporter vector machine (SVM), K nearest neighbor (KNN), neural network (NN), Bayesian network (BN). The LR (96/183, 52.46%), SVM (20/183, 10.93%) and RF (20/183, 10.93%) were the frequently used models in the included studies. Noteworthy, SP Shayesteh, et al. [49] observed the ensemble model of machine learning classifiers (SVM.NN.BN.KNN) showed the best predictive performance. And Z Zhang, et al. [50] performed a retrospective study on 189 patients with locally advanced rectal cancer to assess the performance and stability of classification methods (SVM, KNN and RF), the result showed that the RF outperformed KNN and SVM in terms of AUCs.
Evaluation
According to the principle of confirmatory analysis, the generalization ability of the model could be evaluated in the independent verification set [51]. However, only 144 (144/188, 76.60%) articles used the independent dataset validation, the internal validation was performed by applying resampling methods in the remaining studies.
The performance of the models was assessed in the terms of discrimination, calibration, and clinical utility. The discrimination statistics (C-statistic, ROC curve and AUC) of the models were reported in 173 (173/188, 92.02%) studies, and resampling analysis (bootstrapping and cross-validation) were applied in 78 (78/173, 45.09%) studies. The calibration statistics (calibration curve or Brier score) of the models were mentioned in only75 (75/188, 39.89%) articles s, of which 12 (12/75, 16.00%) used resampling analysis. In addition, the clinical utility was evaluated by the decision curve in 76 (76/188, 40.43%). The differences in evaluation metrics between studies lead to difficulties in comparing performance between models.
How to increase repeatability
Standardization protocol
D Mackin, et al. [9] performed a phantom analysis to compare the radiomics features extracted from four CT scanners (GE, Philips, Siemens, and Toshiba) and found that radiomics features might be affected by the scanners. The result was subsequently confirmed in the study of R Berenguer, et al. [11], who used two phantoms (the pelvic phantom and the phantom of different materials) to detect the features of intra-CT analysis (differences between different CT acquisition parameters) and inter-CT analysis(differences between five different scanners). R Berenguer, et al. [11] found that 71/177 were reproducible, and reported that the influence of scanners could be reduced by standardizing the acquisition parameters. RJ Gillies, et al. [37] have the same point of view.
The image Biomarker Standardization Initiative (IBSI) [52] standardized the definition, naming, and software of radiomics features. The Quantitative Imaging Network (QIN) [38] project initiated by NIC (National Cancer Institute) has promoted the standardization of imaging methods and imaging protocols. In addition, the Quantitative Imaging Biomarkers Alliance (QIBA) [53] organization sponsored by, the Radiological Society of North America (RSNA) has developed a standardized quantitative imaging document "Profiles" to promote clinical trials and practices of radiomics.
Test–retest reliability
JE van Timmeren, et al. [54] scanned forty patients with rectal cancer twice with the same scanning scheme at 15-min intervals and found that some radiomics features were not repeatable at different times for the same individual. But a set of highly reproducible radiomics features could be extracted by the test–retest based on phantom or patients [55, 56]. Moreover, JE van Timmeren, et al. [54] indicated that appropriate test–retest would be applied in the terms of the effects of hardware, acquisition, reconstruction, tumor segmentation, and feature extraction.
Post-processing
With the continuous emergence of new features, the efficiency of test–retest research becomes lower [18]. The variability of features might be reduced by the following post-processing methods.
Resampling and normalization: L He, et al. [15] demonstrated that acquisition parameters (slice thickness, convolution kernel, and enhancement) had affected the diagnostic performance of radiomics. Similarly, L Lu, et al. [14] demonstrated variation of radiomics features due to the slice thicknesses and reconstruction methods. Features associated with tumor size, border morphology, low-order density statistics, and coarse texture were more sensitive to variations in acquisition parameters. Subsequently, a more rigorous experiment [13] showed that 63/213 features were affected by voxels, but 42 features were significantly improved, and 21 features changed greatly after resampling. Therefore, some studies [12, 13, 57]showed that resampling could effectively improve the feature variation caused by voxel differences. However, resampling alone is not enough [13], normalization was used to reduce the influence of different gray ranges or the effects of low frequency and intensity inhomogeneity in some studies. Introducing noise, blurring the image, and causing the loss of image details are the disadvantages of normalization [58].
ComBat: Previously, genomics has been affected by batch effects, that is, systematic technical biases introduced by samples in different batches of processing and measurement that are not related to biological status [59]. WE Johnson, et al. [60] developed and validated a method to deal with the "batch effect"-ComBat. In radiomics, the impact of different scanners or scanning schemes is similar to the batches. Studies [61,62,63] showed that ComBat could reduce the feature differences caused by different scanners or scanning schemes, and retain the feature differences formed by biological variation. Although ComBat is practical, convenient and fast, it will be affected by the distribution of validated data sets [18], and it cannot be directly applied to imaging data [64]. So Y Li, et al. [18] developed a normalization method based on deep learning, which may effectively avoid the above problems.
Discussion
This review analyzed the repeatability of radiomics on patients with colorectal cancer by discussing the method details in 188 studies and evaluated the quality of studies by RQS. Although the included studies demonstrated excellent predictive performance, the methodology varied considerably among studies in the terms of imaging parameters, feature selection and modeling, so the comparison between study results is difficult. In addition, the values of test–retest have been investigated for improving study reproducibility, but test–retest was rarely used in the included studies. In the review, many radiomics studies had poor quality by RQS.
Since 2016, the number of radiomics studies on patients with colorectal cancer continued to increase, but the low RQS score (-1 ~ 27) was shown in many studies, indicating that radiomics was an immature new technology in the field of colorectal cancer. This finding has been confirmed in the previous study [65]. C Xue, et al. [8] demonstrated the RQS is highly correlated with reliability, especially in the phantom study and imaging at multiple time points. P Lambin, et al. [4] explained that multiple time point was used to eliminate the unstable radiomics features which strongly related to organ movement or the expansion or shrinkage of the target volume, and reduce the influence of intra-individual variability [55, 56]. J Wang, et al. [29] selected the most stable radiomics features based on the test–retest on 40 patients with rectal cancer. Although the subjects of the test–retest could be patients or phantoms [66, 67], the unnecessary radiation damage might be increased in the patients’ test–retest. In addition, JE van Timmeren, et al. [54]proposed that extensive retest-test experiments can provide a stable set of radiomics features, and emphasized that retest-test studies should be adopted in each step, including intra-individual variability, scan acquisition and reconstruction, tumor segmentation and feature extraction. Open science data includes opening scanning information, opening segmentations of ROIs, opening source code and opening feature extraction methods (including formulas)[4]. Opening science data is beneficial for other researchers to reproduce the research results (independent researchers use the same technology and different data to repeat the research results and independently verify the results) and promote the application of the research model in clinical practice [3, 4, 6].
There are many variable factors and a wide variety of diseases in radiology, so the standardization of scanning parameters, acquisition parameters and reconstruction parameters could effectively reduce variability [3, 6]. In addition, the influence of scanning parameters might be reduced by using the same scanning scheme, unstable features may be reduced by retest tests [66], resampling might be used to control the influence of slicer thickness [13], and the "batch effect" could be reduced by combat method [61,62,63]. To balance the deviation of imaging features from four institutions, Z Liu, et al. [48] normalized the data, adopted combat methods to control the deviation of radiomics features, and weighted the radiomics features with inverse probability of treatment weighting (IPTW) to eliminate the covariant effect among the four cohorts. M Taghavi, et al. [68] conducted two experiments with the fake label to verify the validity of the radiomics and the results show that the model was not influenced by noise (first experiment) or the patient distribution across hospitals (second experiment).
For the method of segmentation, semi-automatic segmentation was not only more efficient but also useful to reduce the variability of manual segmentation [19,20,21, 34, 35]. Of all the studies, 143 (143/188, 76.06%) were segmented manually, and only 22 (22/188, 11.70%) were segmented semi-automatically. For inter-observer variability, only 111 studies took into account inter-observer variability and excluded instability. Even though the Jaccard index, Cn and CIgen may be better as evaluation parameters [39], 94 (94/111, 84.68%) studies were evaluated by ICC.
According to the Harrell criterion [69], the sample size should be more than 10 times the number of variables, and feature selection can reduce redundant features and reduce the risk of model over-fitting [41, 42]. Such as, random forest on 89 patients was used to measure the Gini importance of parameters, and finally 10 important parameters were included in the study of C Yang, et al. [70]. M Sollini, et al. [71] recommended that at least 50 patients would be included in the radiomics studies because the quality of the studies and the credibility of the results might be seriously affected by the sample size [72]. But, M Hennessy, et al. [73] constructed the formal model, which was the mathematical algorithms formulated by the experts. The formal model was more interpretable than machine learning models and does not require large amounts of data for validation[74]. The formal models have been applied and have shown excellent predictive performance in previous studies [74,75,76,77,78,79]. However, the formal methods were not widely used and the predictive performance of the models needed to be further validated in the field of cancer.
It is also important to avoid overly optimistic results in addition to repeatability. In many studies, the type I errors might be increased by the combination of optimal cut-off method and multiple hypothesis testing [42], while the multiple hypothesis test correction (Holm Bonferroni or Benjamin-Hochberg pair) was helpful to reduce class I errors [6].
According to the review of 188 studies, the lack of repeatability was the key problem in radiomics studies, and the standardization of radiomics processes helped comparing the existing studies [37]. It is difficult to form global standardization at present [37], but the repeatability of studies might be improved through open science data [4], retest-test research [66] and post-processing. The radiomics was used to predict the sequencing results by the DNA microarray in some studies [80,81,82,83,84,85,86]. Notable was that there were no studies on the combination of liquid biopsy and radiomics. Studies have shown that the combination of liquid biopsy and imaging could play an early warning role in patients prone to recurrence and metastasis [87]. In the future, it may be possible to add biomarkers from other disciplines to the radiomics model. T Cheng, et al. [88] proposed that a pattern consisting of three or more biomarkers could improve the accuracy and specificity of tumor prediction, diagnosis and prognosis, that is, pattern recognition [88]. Delta radiomics is to quote the time component in the study, that is, to use the image data of multiple time points for radiomics analysis. This method is expected to improve the diagnosis, prognosis prediction and treatment response evaluation [4]. Since most radiomics of colorectal cancer were based on retrospective data sets (177, 94.15%), more prospective studies with large samples and multiple centers are needed to promote the development of radiomics in the field of colorectal cancer. Cost-effectiveness analysis from an economic point of view is necessary for the clinical application of colorectal cancer imaging in the future [4].
The main limitations of this review are shown below. First, the meta-analysis was not conducted due to there being great differences in the methods of the included studies. Second, some related studies might not be retrieved, such as some grey literature. In this paper, only RQS is used to evaluate the quality of the article.
Conclusion
Although existing studies showed that radiomics is helpful to the personalized treatment of patients in the field of colorectal cancer, there are still many challenges that remain to be solved. According to the RQS score, the quality of included studies was moderately low. Moreover, the main reason for the low RQS score was the lack of repeatability, most studies did not eliminate the influence of scanners, imaging parameters, and other factors. Therefore, these studies of lower quality and lack of repeatability mean that the results are not universal. In the future, larger samples and multicenter prospective high-quality studies are needed, and researches should focus on building a more stable and repeatable model.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- AUC:
-
Area Under the Curve
- CRC:
-
Colorectal cancer
- CRT:
-
Chemoradiotherapy
- CT:
-
Computed Tomography
- IBSI:
-
Image Biomarker Standardization Initiative
- ICC:
-
Intraclass Correlation Coefficient
- LASSO:
-
Least Absolute Shrinkage and Selection operator
- MRI:
-
Magnetic Resonance Imaging
- PET-CT:
-
Fluorodeoxyglucose Positron Emission Tomography-Computed Tomography
- PRISMA:
-
Preferred Reporting items for Systematic review and Meta-Analysis
- QIBA:
-
Quantitative Imaging Biomarkers Alliance
- QIN:
-
Quantitative Imaging Network
- RQS:
-
Radiomics Quality Score
- RSNA:
-
Radiological Society of North America
- SUV:
-
Standardized Uptake Value
- UA:
-
Ultrasound
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F: Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209-249
Staal FCR, van der Reijd DJ, Taghavi M, Lambregts DMJ, Beets-Tan RGH, Maas M. Radiomics for the Prediction of Treatment Outcome and Survival in Patients With Colorectal Cancer: A Systematic Review. Clin Colorectal Cancer. 2021;20(1):52–71.
Limkin EJ, Sun R, Dercle L, Zacharaki EI, Robert C, Reuzé S, Schernberg A, Paragios N, Deutsch E, Ferté C. Promises and challenges for the implementation of computational medical imaging (radiomics) in oncology. Ann Oncol. 2017;28(6):1191–206.
Lambin P, Leijenaar RTH, Deist TM, Peerlings J, De Jong EEC, Van Timmeren J, Sanduleanu S, Larue RTHM, Even AJG, Jochems A, et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14(12):749–62.
Forghani R, Savadjiev P, Chatterjee A, Muthukrishnan N, Reinhold C, Forghani B. Radiomics and Artificial Intelligence for Biomarker and Prediction Model Development in Oncology. Comput Struct Biotechnol J. 2019;17:995–1008.
Yip SS, Aerts HJ. Applications and limitations of radiomics. Phys Med Biol. 2016;61(13):R150-166.
Miles K. Radiomics for personalised medicine: the long road ahead. Br J Cancer. 2020;122(7):929–30.
Xue C, Yuan J, Lo GG, Chang ATY, Poon DMC, Wong OL, Zhou Y, Chu WCW. Radiomics feature reliability assessed by intraclass correlation coefficient: a systematic review. Quant Imaging Med Surg. 2021;11(10):4431–60.
Mackin D, Fave X, Zhang L, Fried D, Yang J, Taylor B, Rodriguez-Rivera E, Dodge C, Jones AK, Court L. Measuring Computed Tomography Scanner Variability of Radiomics Features. Invest Radiol. 2015;50(11):757–65.
Kumar V, Gu Y, Basu S, Berglund A, Eschrich SA, Schabath MB, Forster K, Aerts HJ, Dekker A, Fenstermacher D, et al. Radiomics: the process and the challenges. Magn Reson Imaging. 2012;30(9):1234–48.
Berenguer R, Pastor-Juan MDR, Canales-Vázquez J, Castro-García M, Villas MV, Mansilla Legorburo F, Sabater S. Radiomics of CT Features May Be Nonreproducible and Redundant: Influence of CT Acquisition Parameters. Radiology. 2018;288(2):407–15.
Mackin D, Fave X, Zhang L, Yang J, Jones AK, Ng CS, Court L. Harmonizing the pixel size in retrospective computed tomography radiomics studies. PLoS ONE. 2017;12(9):e0178524.
Shafiq-Ul-Hassan M, Zhang GG, Latifi K, Ullah G, Hunt DC, Balagurunathan Y, Abdalah MA, Schabath MB, Goldgof DG, Mackin D, et al. Intrinsic dependencies of CT radiomic features on voxel size and number of gray levels. Med Phys. 2017;44(3):1050–62.
Lu L, Ehmke RC, Schwartz LH, Zhao B. Assessing Agreement between Radiomic Features Computed for Multiple CT Imaging Settings. PLoS ONE. 2016;11(12):e0166550.
He L, Huang Y, Ma Z, Liang C, Liang C, Liu Z. Effects of contrast-enhancement, reconstruction slice thickness and convolution kernel on the diagnostic performance of radiomics signature in solitary pulmonary nodule. Sci Rep. 2016;6:34921.
Kim H, Park CM, Lee M, Park SJ, Song YS, Lee JH, Hwang EJ, Goo JM. Impact of Reconstruction Algorithms on CT Radiomic Features of Pulmonary Tumors: Analysis of Intra- and Inter-Reader Variability and Inter-Reconstruction Algorithm Variability. PLoS ONE. 2016;11(10):e0164924.
Traverso A, Kazmierski M, Shi Z, Kalendralis P, Welch M, Nissen HD, Jaffray D, Dekker A, Wee L. Stability of radiomic features of apparent diffusion coefficient (ADC) maps for locally advanced rectal cancer in response to image pre-processing. Phys Med. 2019;61:44–51.
Li Y, Han G, Wu X, Li Z, Zhao K, Zhang Z, Liu Z, Liang C: Normalization of multicenter CT radiomics by a generative adversarial network method. Phys Med Biol. 2021;(5):66.
van Heeswijk MM, Lambregts DM, van Griethuysen JJ, Oei S, Rao SX, de Graaff CA, Vliegen RF, Beets GL, Papanikolaou N, Beets-Tan RG. Automated and Semiautomated Segmentation of Rectal Tumor Volumes on Diffusion-Weighted MRI: Can It Replace Manual Volumetry? Int J Radiat Oncol Biol Phys. 2016;94(4):824–31.
Parmar C, Rios Velazquez E, Leijenaar R, Jermoumi M, Carvalho S, Mak RH, Mitra S, Shankar BU, Kikinis R, Haibe-Kains B, et al. Robust Radiomics feature quantification using semiautomatic volumetric segmentation. PLoS ONE. 2014;9(7):e102107.
Day E, Betler J, Parda D, Reitz B, Kirichenko A, Mohammadi S, Miften M. A region growing method for tumor volume segmentation on PET images for rectal and anal cancer patients. Med Phys. 2009;36(10):4349–58.
Trebeschi S, van Griethuysen JJM, Lambregts DMJ, Lahaye MJ, Parmar C, Bakers FCH, Peters N, Beets-Tan RGH, Aerts H. Deep Learning for Fully-Automated Localization and Segmentation of Rectal Cancer on Multiparametric MR. Sci Rep. 2017;7(1):5301.
Parmar C, Grossmann P, Bussink J, Lambin P, Aerts H. Machine Learning methods for Quantitative Radiomic Biomarkers. Sci Rep. 2015;5:13087.
McInnes MDF, Moher D, Thombs BD, McGrath TA, Bossuyt PM. Group atP-D: Preferred Reporting Items for a Systematic Review and Meta-analysis of Diagnostic Test Accuracy Studies: The PRISMA-DTA Statement. JAMA. 2018;319(4):388–96.
Chetan MR, Gleeson FV. Radiomics in predicting treatment response in non-small-cell lung cancer: current status, challenges and future perspectives. Eur Radiol. 2021;31(2):1049–58.
Babyak MA. What you see may not be what you get: a brief, nontechnical introduction to overfitting in regression-type models. Psychosom Med. 2004;66(3):411–21.
Lovinfosse P, Polus M, Van Daele D, Martinive P, Daenen F, Hatt M, Visvikis D, Koopmansch B, Lambert F, Coimbra C, et al. FDG PET/CT radiomics for predicting the outcome of locally advanced rectal cancer. Eur J Nucl Med Mol Imaging. 2018;45(3):365–75.
Ma X, Shen F, Jia Y, Xia Y, Li Q, Lu J. MRI-based radiomics of rectal cancer: preoperative assessment of the pathological features. BMC Med Imaging. 2019;19(1):86.
Wang J, Shen L, Zhong H, Zhou Z, Hu P, Gan J, Luo R, Hu W, Zhang Z. Radiomics features on radiotherapy treatment planning CT can predict patient survival in locally advanced rectal cancer patients. Sci Rep. 2019;9(1):15346.
Nie K, Shi L, Chen Q, Hu X, Jabbour SK, Yue N, Niu T, Sun X. Rectal Cancer: Assessment of Neoadjuvant Chemoradiation Outcome based on Radiomics of Multiparametric MRI. Clin Cancer Res. 2016;22(21):5256–64.
Kang J, Lee JH, Lee HS, Cho ES, Park EJ, Baik SH, Lee KY, Park C, Yeu Y, Clemenceau JR, et al. Radiomics features of18f-fluorodeoxyglucose positron-emission tomography as a novel prognostic signature in colorectal cancer. Cancers. 2021;13(3):1–17.
Rios Velazquez E, Aerts HJ, Gu Y, Goldgof DB, De Ruysscher D, Dekker A, Korn R, Gillies RJ, Lambin P. A semiautomatic CT-based ensemble segmentation of lung tumors: comparison with oncologists’ delineations and with the surgical specimen. Radiother Oncol. 2012;105(2):167–73.
van Dam IE. van Sörnsen de Koste JR, Hanna GG, Muirhead R, Slotman BJ, Senan S: Improving target delineation on 4-dimensional CT scans in stage I NSCLC using a deformable registration tool. Radiother Oncol. 2010;96(1):67–72.
Heye T, Merkle EM, Reiner CS, Davenport MS, Horvath JJ, Feuerlein S, Breault SR, Gall P, Bashir MR, Dale BM, et al. Reproducibility of dynamic contrast-enhanced MR imaging. Part II. Comparison of intra- and interobserver variability with manual region of interest placement versus semiautomatic lesion segmentation and histogram analysis. Radiology. 2013;266(3):812–21.
Egger J, Kapur T, Fedorov A, Pieper S, Miller JV, Veeraraghavan H, Freisleben B, Golby AJ, Nimsky C, Kikinis R. GBM volumetry using the 3D Slicer medical image computing platform. Sci Rep. 2013;3:1364.
Tibermacine H, Rouanet P, Sbarra M, Forghani R, Reinhold C, Nougaret S. Radiomics modelling in rectal cancer to predict disease-free survival: evaluation of different approaches. Br J Surg. 2021;108(10):1243–50.
Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures. They Are Data Radiology. 2016;278(2):563–77.
Clarke LP, Nordstrom RJ, Zhang H, Tandon P, Zhang Y, Redmond G, Farahani K, Kelloff G, Henderson L, Shankar L, et al. The Quantitative Imaging Network: NCI’s Historical Perspective and Planned Goals. Transl Oncol. 2014;7(1):1–4.
Fotina I, Lütgendorf-Caucig C, Stock M, Pötter R, Georg D. Critical discussion of evaluation parameters for inter-observer variability in target definition for radiation therapy. Strahlenther Onkol. 2012;188(2):160–7.
Nakanishi R, Akiyoshi T, Toda S, Murakami Y, Taguchi S, Oba K, Hanaoka Y, Nagasaki T, Yamaguchi T, Konishi T, et al. Radiomics Approach Outperforms Diameter Criteria for Predicting Pathological Lateral Lymph Node Metastasis After Neoadjuvant (Chemo)Radiotherapy in Advanced Low Rectal Cancer. Ann Surg Oncol. 2020;27(11):4273–83.
Avanzo M, Stancanello J, El Naqa I. Beyond imaging: The promise of radiomics. Phys Med. 2017;38:122–39.
Chalkidou A, O’Doherty MJ, Marsden PK. False Discovery Rates in PET and CT Studies with Texture Features: A Systematic Review. PLoS ONE. 2015;10(5):e0124165.
Guyon IM, Andr, Elisseeff. An introduction to variable and feature selection [J]. J Mach Learn Res. 2003.
Boldrini L, Cusumano D, Chiloiro G, Casà C, Masciocchi C, Lenkowicz J, Cellini F, Dinapoli N, Azario L, Teodoli S, et al. Delta radiomics for rectal cancer response prediction with hybrid 0.35 T magnetic resonance-guided radiotherapy (MRgRT): a hypothesis-generating study for an innovative personalized medicine approach. Radiol Med. 2019;124(2):145–53.
Cusumano D, Boldrini L, Yadav P, Yu G, Musurunu B, Chiloiro G, Piras A, Lenkowicz J, Placidi L, Romano A, et al. Delta radiomics for rectal cancer response prediction using low field magnetic resonance guided radiotherapy: an external validation. Physica Med. 2021;84:186–91.
Hotta M, Minamimoto R, Gohda Y, Miwa K, Otani K, Kiyomatsu T, Yano H. Prognostic value of (18)F-FDG PET/CT with texture analysis in patients with rectal cancer treated by surgery. Ann Nucl Med. 2021;35(7):843–52.
Negreros-Osuna AA, Parakh A, Corcoran RB, Pourvaziri A, Kambadakone A, Ryan DP, Sahani DV. Radiomics Texture Features in Advanced Colorectal Cancer: Correlation with BRAF Mutation and 5-year Overall Survival. Radiol Imaging Cancer. 2020;2(5):e190084.
Liu Z, Meng X, Zhang H, Li Z, Liu J, Sun K, Meng Y, Dai W, Xie P, Ding Y, et al. Predicting distant metastasis and chemotherapy benefit in locally advanced rectal cancer. Nat Commun. 2020;11(1):4308.
Shayesteh SP, Alikhassi A, Fard Esfahani A, Miraie M, Geramifar P, Bitarafan-Rajabi A, Haddad P. Neo-adjuvant chemoradiotherapy response prediction using MRI based ensemble learning method in rectal cancer patients. Phys Med. 2019;62:111–9.
Zhang Z, Jiang X, Zhang R, Yu T, Liu S, Luo Y. Radiomics signature as a new biomarker for preoperative prediction of neoadjuvant chemoradiotherapy response in locally advanced rectal cancer. Diagn Interv Radiol (Ankara, Turkey). 2021;27(3):308–14.
Taylor JM, Ankerst DP, Andridge RR. Validation of biomarker-based risk prediction models. Clin Cancer Res. 2008;14(19):5977–83.
Zwanenburg A, Vallières M, Abdalah MA, Aerts H, Andrearczyk V, Apte A, Ashrafinia S, Bakas S, Beukinga RJ, Boellaard R, et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiol. 2020;295(2):328–38.
Buckler AJ, Bresolin L, Dunnick NR, Sullivan DC. A collaborative enterprise for multi-stakeholder participation in the advancement of quantitative imaging. Radiol. 2011;258(3):906–14.
van Timmeren JE, Leijenaar RTH, van Elmpt W, Wang J, Zhang Z, Dekker A, Lambin P. Test-Retest Data for Radiomics Feature Stability Analysis: Generalizable or Study-Specific? Tomogr. 2016;2(4):361–5.
Balagurunathan Y, Kumar V, Gu Y, Kim J, Wang H, Liu Y, Goldgof DB, Hall LO, Korn R, Zhao B, et al. Test-retest reproducibility analysis of lung CT image features. J Digit Imaging. 2014;27(6):805–23.
Zhao B, Tan Y, Tsai WY, Qi J, Xie C, Lu L, Schwartz LH. Reproducibility of radiomics for deciphering tumor phenotype with imaging. Sci Rep. 2016;6:23428.
Shafiq-Ul-Hassan M, Latifi K, Zhang G, Ullah G, Gillies R, Moros E. Voxel size and gray level normalization of CT radiomic features in lung cancer. Sci Rep. 2018;8(1):10545.
Zwanenburg A, Leger S, Agolli L, Pilz K, Troost EGC, Richter C, Löck S. Assessing robustness of radiomic features by image perturbation. Sci Rep. 2019;9(1):614.
Lazar C, Meganck S, Taminau J, Steenhoff D, Coletta A, Molter C, Weiss-Solís DY, Duque R, Bersini H, Nowé A. Batch effect removal methods for microarray gene expression data integration: a survey. Brief Bioinform. 2013;14(4):469–90.
Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostat. 2007;8(1):118–27.
Orlhac F, Boughdad S, Philippe C, Stalla-Bourdillon H, Nioche C, Champion L, Soussan M, Frouin F, Frouin V, Buvat I. A Postreconstruction Harmonization Method for Multicenter Radiomic Studies in PET. J Nucl Med. 2018;59(8):1321–8.
Orlhac F, Frouin F, Nioche C, Ayache N, Buvat I. Validation of A Method to Compensate Multicenter Effects Affecting CT Radiomics. Radiol. 2019;291(1):53–9.
Fortin JP, Cullen N, Sheline YI, Taylor WD, Aselcioglu I, Cook PA, Adams P, Cooper C, Fava M, McGrath PJ, et al. Harmonization of cortical thickness measurements across scanners and sites. Neuroimage. 2018;167:104–20.
Ibrahim A, Primakov S, Beuque M, Woodruff HC, Halilaj I, Wu G, Refaee T, Granzier R, Widaatalla Y, Hustinx R, et al. Radiomics for precision medicine: Current challenges, future prospects, and the proposal of a new framework. Methods. 2021;188:20–9.
Wesdorp NJ, Hellingman T, Jansma EP, van Waesberghe JTM, Boellaard R, Punt CJA, Huiskens J, Kazemier G. Advanced analytics and artificial intelligence in gastrointestinal cancer: a systematic review of radiomics predicting response to treatment. Eur J Nucl Med Mol Imaging. 2021;48(6):1785–94.
Ligero M, Jordi-Ollero O, Bernatowicz K, Garcia-Ruiz A, Delgado-Muñoz E, Leiva D, Mast R, Suarez C, Sala-Llonch R, Calvo N, et al. Minimizing acquisition-related radiomics variability by image resampling and batch effect correction to allow for large-scale data analysis. Eur Radiol. 2021;31(3):1460–70.
Peerlings J, Woodruff HC, Winfield JM, Ibrahim A, Van Beers BE, Heerschap A, Jackson A, Wildberger JE, Mottaghy FM, DeSouza NM, et al. Stability of radiomics features in apparent diffusion coefficient maps from a multi-centre test-retest trial. Sci Rep. 2019;9(1):4800.
Taghavi M, Trebeschi S, Simões R, Meek DB, Beckers RCJ, Lambregts DMJ, Verhoef C, Houwers JB, van der Heide UA, Beets-Tan RGH, et al. Machine learning-based analysis of CT radiomics model for prediction of colorectal metachronous liver metastases. Abdom Radiol (NY). 2021;46(1):249–56.
Harrell F E . Regression modeling strategies: with applications to linear models, logistic regression, and survival analysis[M]. Springer, 2010.
Yang C, Jiang ZK, Liu LH, Zeng MS. Pre-treatment ADC image-based random forest classifier for identifying resistant rectal adenocarcinoma to neoadjuvant chemoradiotherapy. Int J Colorectal Dis. 2020;35(1):101–7.
Sollini M, Antunovic L, Chiti A, Kirienko M. Towards clinical application of image mining: a systematic review on artificial intelligence and radiomics. Eur J Nucl Med Mol Imaging. 2019;46(13):2656–72.
Moreira JM, Santiago I, Santinha J, Figueiredo N, Marias K, Figueiredo M, Vanneschi L, Papanikolaou N. Challenges and Promises of Radiomics for Rectal Cancer. Curr Colorectal Cancer Rep. 2019;15(6):175–80.
Hennessy M, Milner R. Algebraic laws for nondeterminism and concurrency. J ACM. 1985;32(1):137–61.
Brunese L, Mercaldo F, Reginelli A, Santone A. Prostate Gleason Score Detection and Cancer Treatment Through Real-Time Formal Verification. IEEE Access. 2019;7:186236–46.
Rocca A, Brunese MC, Santone A, Avella P, Bianco P, Scacchi A, Scaglione M, Bellifemine F, Danzi R, Varriano G et al: Early Diagnosis of Liver Metastases from Colorectal Cancer through CT Radiomics and Formal Methods: A Pilot Study. J Clin Med. 2021;11(1):31.
Santone A, Brunese MC, Donnarumma F, Guerriero P, Mercaldo F, Reginelli A, Miele V, Giovagnoni A, Brunese L. Radiomic features for prostate cancer grade detection through formal verification. Radiol Med. 2021;126(5):688–97.
Santone A, Belfiore MP, Mercaldo F, Varriano G, Brunese L: On the Adoption of Radiomics and Formal Methods for COVID-19 Coronavirus Diagnosis. Diagnostics (Basel). 2021;11(2):293.
Brunese L, Mercaldo F, Reginelli A, Santone A. Formal methods for prostate cancer Gleason score and treatment prediction using radiomic biomarkers. Magn Reson Imaging. 2020;66:165–75.
Brunese L, Mercaldo F, Reginelli A, Santone A: Radiomics for Gleason Score Detection through Deep Learning. Sensors (Basel). 2020;20(18):5411.
Wu X, Li Y, Chen X, Huang Y, He L, Zhao K, Huang X, Zhang W, Huang Y, Li Y, et al. Deep Learning Features Improve the Performance of a Radiomics Signature for Predicting KRAS Status in Patients with Colorectal Cancer. Acad Radiol. 2020;27(11):e254–62.
Yang L, Dong D, Fang M, Zhu Y, Zang Y, Liu Z, Zhang H, Ying J, Zhao X, Tian J. Can CT-based radiomics signature predict KRAS/NRAS/BRAF mutations in colorectal cancer? Eur Radiol. 2018;28(5):2058–67.
Li Y, Eresen A, Shangguan J, Yang J, Benson AB 3rd, Yaghmai V, Zhang Z. Preoperative prediction of perineural invasion and KRAS mutation in colon cancer using machine learning. J Cancer Res Clin Oncol. 2020;146(12):3165–74.
Oh JE, Kim MJ, Lee J, Hur BY, Kim B, Kim DY, Baek JY, Chang HJ, Park SC, Oh JH, et al. Magnetic Resonance-Based Texture Analysis Differentiating KRAS Mutation Status in Rectal Cancer. Cancer Res Treat. 2020;52(1):51–9.
Cui Y, Liu H, Ren J, Du X, Xin L, Li D, Yang X, Wang D. Development and validation of a MRI-based radiomics signature for prediction of KRAS mutation in rectal cancer. Eur Radiol. 2020;30(4):1948–58.
Horvat N, Veeraraghavan H, Pelossof RA, Fernandes MC, Arora A, Khan M, Marco M, Cheng CT, Gonen M, Golia Pernicka JS, et al. Radiogenomics of rectal adenocarcinoma in the era of precision medicine: A pilot study of associations between qualitative and quantitative MRI imaging features and genetic mutations. Eur J Radiol. 2019;113:174–81.
Shi R, Chen W, Yang B, Qu J, Cheng Y, Zhu Z, Gao Y, Wang Q, Liu Y, Li Z, et al. Prediction of KRAS, NRAS and BRAF status in colorectal cancer patients with liver metastasis using a deep artificial neural network based on radiomics and semantic features. Am J Cancer Res. 2020;10(12):4513–26.
Neri E, Del Re M, Paiar F, Erba P, Cocuzza P, Regge D, Danesi R. Radiomics and liquid biopsy in oncology: the holons of systems medicine. Insights Imaging. 2018;9(6):915–24.
Cheng T, Zhan X. Pattern recognition for predictive, preventive, and personalized medicine in cancer. Epma j. 2017;8(1):51–60.
Acknowledgements
Not applicable.
Funding
This work was supported by the Natural Science Foundation of Science and Technology Department of Sichuan Province (23NSFSC1943).
Author information
Authors and Affiliations
Contributions
YL and XQW contributed equally to the work. YL and XQW contributed to study design and wrote the study protocol. XF and YL performed the literature search. YL and XQW performed data collection and data interpretation and drafted the manuscript. XF, YL and GLF reviewed and critically revised the manuscript. YD were responsible for study supervision. All authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent to publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 2: Table 1.
Study characteristics of 188 papers. Table 2. RQS of studies. Table 3. Risk of bias in individualstudies.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Liu, Y., Wei, X., Feng, X. et al. Repeatability of radiomics studies in colorectal cancer: a systematic review. BMC Gastroenterol 23, 125 (2023). https://doi.org/10.1186/s12876-023-02743-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12876-023-02743-1