Abstract
Background
Approximately 5.0–24.2% of colorectal cancers (CRCs) have inactivating mutations in SMAD4, making it one of the frequently mutated genes in CRC. We thus carried out a comprehensive system review and meta-analysis investigating the prognostic significance and clinicopathological features of SMAD4 gene mutation in CRC patients.
Methods
A detailed literature search was conducted in PubMed, Web of Science and Embase databases to study the relationship between SMAD4 mutations and the demographic and clinicopathological characteristics in CRC patients. The hazard ratios (HRs) with 95% confidence intervals (CI) were used to evaluate the effect of SMAD4 mutations on overall survival (OS) and progression-free survival (PFS)/recurrence-free survival (RFS).
Results
Ten studies enrolling 4394 patients were eligible for inclusion. Data on OS were available from 5 studies and data on PFS/RFS were available from 3 studies. Comparing SMAD4-mutated CRC patients with SMAD4 wild-type CRC patients, the summary HR for OS was 1.46 (95% CI 1.28–1.67, P = 0.001), the summary HR for PFS/RFS was 1.59 (95% CI 1.14–2.22, P = 0.006). In terms of clinicopathology parameters, 9 studies have data that can be extracted, SMAD4 mutations were associated with tumor location (odds ratio [OR] = 1.15, colon/rectum, 95% CI 1.01–1.31, P = 0.042), TNM stage (OR = 1.28, stage IV/I–III, 95% CI 1.03–1.58, P = 0.025), lymph node metastasis (OR = 1.42, N1 + N2/N0, 95% CI 1.20–1.67, P < 0.001), mucinous differentiation (OR = 2.23, 95% CI 1.85–2.70, P < 0.001) and rat sarcoma viral oncogene homolog (RAS) mutation status (OR = 2.13, 95% CI 1.37–3.34, P = 0.001). No connection was found with age, gender, tumor grade, microsatellite instability status and b-viral oncogene homolog B1 mutation status. Besides, publication bias was not observed in any study.
Conclusions
This meta-analysis suggests that SMAD4 mutation was associated with OS, PFS/RFS, and clinicopathological parameters, including tumor site, disease stage, RAS status, lymph node metastasis and mucinous differentiation. Our meta-analysis indicated that SMAD4 mutations could predict the poor prognosis and aggressive clinicopathological characteristics of CRC. More large-sample cohort studies are needed to confirm this conclusion. Since SMAD4 mutations are closely related to RAS mutations, their relationship warrants further investigation.
Similar content being viewed by others
Background
Colorectal cancer (CRC) is the third most common cancer and the second most common cause of cancer-related death over the world [1]. Despite advances in early diagnosis and treatment, lymphatic metastasis and distant metastasis are still the main causes of death in newly diagnosed CRC patients, and the overall survival (OS) rate of advanced CRC is still unsatisfactory.
The Cancer Genome Atlas database revealed that the mutation frequency of SMAD4 is 10%, which is one of the most common mutated genes in CRC [2]. SMAD4 is an established tumor suppressor gene located in chromosome band 18q21, and one of the most commonly destroyed gene in cancer among SMAD family genes [3]. This gene encodes a member of the Smad family of signal transduction proteins, that is phosphorylated and activated by transmembrane serine-threonine receptor kinases in response to transforming growth factor beta (TGF-β) signal transduction. The product of this gene forms homomeric complexes and heteromeric complexes with other activated Smad proteins in the context of activating by TGF-β receptors, then accumulate in the nucleus and regulate the transcription of target genes [4]. Mutations or deletions in the SMAD4 gene have been shown to result in pancreatic cancer [5], juvenile polyposis syndrome [6], and hereditary hemorrhagic telangiectasia [7]. In the past 2 decades, many studies had shown that SMAD4 mutation can not cause tumorigenesis by itself, but it can promote tumor progression caused by other genes [8]. The role of SMAD4 in CRC is similar to that in pancreatic cancer. The prevalence of SMAD4 mutations have recently been reported in 5.0–24.2% of several retrospective studies of sporadic CRC from 1999 to 2020 [9,10,11,12,13,14]. However, whether pathogenic mutation of SMAD4 reduces the OS in all CRC patients remains unclear. Therefore, we conducted a meta-analysis to assess the association of SMAD4 mutations with OS and PFS/RFS, as well as the relationship between SMAD4 mutations and clinicopathological characteristics of early and advanced CRC.
Methods
Search strategy
We conducted this study based on the preferred reporting items for Systematic Reviews and Meta-Analyses 2009 guidelines and registered with the International Prospective Register of Systematic Reviews, PROSPERO (identification code CRD42021244570). Systematic review of several databases was conducted in December 2020 with no lower limit set for date of publication. Search for related articles published in English or Chinese in the following electronic databases: PubMed, Web of Science, and Embase. The keywords “SMAD4” or “DPC4” and “colorectal cancer” or “colon cancer” or “rectum cancer” were used for relative articles searching.
Study selection and inclusion criteria
All articles are limited to human studies published in English or Chinese that based on the following selection criteria: (1) Researches involved the prognostic of SMAD4 mutations in CRC patients, and provided sufficient information to obtain the Hazard ratios (HRs) and 95% confidence interval (CI) of OS or progression-free survival (PFS)/recurrence-free survival (RFS) directly or indirectly from the Kaplan–Meier curve. (2) Studies using surgical resection specimen of tumor to detect SMAD4 mutation in CRC. (3) The odds ratio (OR) associated with clinicopathologic features is given directly or can be obtained from computable data. (4) The study does not include CRC patients who received preoperative chemotherapy or radiotherapy. (5) Duplicate report results are unified by the latest or largest version.
Data extraction and quality assessment
The two authors (F.T. and L.T) extracted all data sets from the selected studies independently, if there are any objections, we resolved through consensus or consultation with the corresponding author. The following information was collected from each study: first author, year of publication, country, time of diagnosis, sample size, CRC cases with SMAD4 gene mutations, sequencing methods of SMAD4 gene, mean follow-up periods and participants’ characteristics, including median age, gender, lymph node metastasis status, rat sarcoma viral oncogene homolog (RAS), b-viral oncogene homolog B1 (BRAF), microsatellite instability (MSI) status and mucinous differentiation as well as tumor stage. HRs and 95% CI of OS and PFS/RFS were extracted directly from papers, if not, we choose to extract from Kaplan–Meier Curve via Engauge Digitizer Version 4.1 (http://markummitchell.github.io/engauge-digitizer/). We used the Newcastle–Ottawa Scale to assess the methods and report quality of the included studies, and ranked them by score (8–9 points for high quality; 5–7 points for medium quality; less than 5 points for low quality) [15].
Statistical analyses
HRs and ORs with their 95% CI were calculated. P value less than 0.05 was considered statistically significant. The Q statistic and I2 tests was used to estimate Heterogeneity among studies. The I2 statistic was ranged from 0 to 1. A random effect model was used for I2 > 0.5, which represented strong heterogeneity. Otherwise, fixed‐effect model would be applied. The analysis was performed to evaluate the impact of SMAD4 gene mutation on the prognosis of CRC. In addition, we evaluated the correlation between SMAD4 mutation status and different tumor grades, tumor differentiation, lymph node metastasis, and MSI/BRAF/RAS status. Sensitivity analysis was used to check data stability. The Egger’s test and Begg’s test were used for detection of publication bias, and P < 0.05 indicated significant bias. All analysis was performed with STATA 16.0 (Stata Corporation, College Station, TX, USA).
Results
Selection of studies
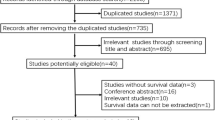
The flowchart of the study selection is shown in Fig. 1. There were 465 articles identified from PubMed, 686 articles from Web of Science, 895 articles from Embase database. A total of 2045 articles were initially identified by the search strategy, and 657 full-text articles were retrieved after screening. Each selected article is tracked forward and backward, in case they contain another research of interest that has not yet been identified. Second, 1280 unrelated titles and abstracts were excluded from the study, and 108 full-text articles were evaluated for applicability, 7 articles were found to have no available outcome indicators or clinicopathologic features, 63 articles relate to SMAD4 protein expression and survival data, 12 articles were non-human trials and 15 articles were reviews, letters or case reports. A total of 4394 patients were included in the final ten studies [9,10,11,12, 16,17,18,19,20,21]. The detailed features of these articles are listed in Tables 1 and 2. The quality evaluation table of all articles is attached in Table 3.
Relationship between SMAD4 mutations and CRC prognosis
A total of 5 articles provided OS related data. Due to the moderate heterogeneity (I2 = 41.6%, P heterogeneity = 0.144), we use the fixed-effect model to pool HR. Comparing SMAD4 mutant patients with SMAD4 wild-type patients in CRC, the summary HR for OS was 1.46 (95% CI 1.28–1.67, P = 0.001) (Fig. 2a).
A total of 3 articles provided PFS/RFS related data. Comparing SMAD4 mutant patients with SMAD4 wild-type patients in CRC, the summary HR for PFS/RFS was 1.59 (95% CI 1.14–2.22, P = 0.006) (Fig. 2b) and there was moderate heterogeneity between the studies (I2 = 48.2%, P heterogeneity = 0.145), so we use the fixed-effect model to pool HR.
Relationship between SMAD4 mutations and clinicopathologic features of CRC
A total of 9 studies have data that can be extracted from clinicopathologic results, the specific characteristics of which are detailed in Table 2. The clinicopathologic OR values of the final merger are presented in Table 4. In terms of clinicopathology parameters, SMAD4 mutations were associated with tumor location (OR = 1.15, for colon versus rectum, 95% CI 1.01–1.31, P = 0.042), pathological TNM stage (OR = 1.28, for stage IV vs I–III, 95% CI 1.03–1.58, P = 0.025), lymph node metastasis (OR = 1.42, for N1 + N2 vs N0, 95% CI 1.20–1.67, P < 0.001), mucinous differentiation (OR = 2.23, 95% CI 1.85–2.70, P < 0.001) and RAS mutations (OR = 2.13, 95% CI 1.37–3.34, P = 0.001). However, SMAD4 gene mutation has no connection with other clinicopathology parameters, including patient age, gender, tumor grade, MSI status and BRAF status.
Sensitivity analysis and publication bias
Our analysis of publication bias using correlation test revealed that there is no obvious publication bias for OS (P = 0.277 for Begg’s test and 0.221 for Egger's test) (Fig. 3a) and PFS/RFS (P = 0.235 for Begg's test and 1.000 for Egger's test) (Fig. 3b). In addition, the sensitivity analysis confirmed that the results were reliable for OS (Fig. 4a) and PFS/RFS (Fig. 4b).
Discussion
The role of Smad4 mutations of the prognosis and clinicopathological parameters in CRC has been investigated in several studies, but the results are inconsistent. In addition, no meta-analysis has been conducted to evaluate the impact of SMAD4 gene on the prognosis of CRC. Therefore, we conducted a meta-analysis and suggested that SMAD4 pathogenic mutations were associated with poor prognosis in CRC. Compared with the SMAD4 wild-type controls, SMAD4 mutations are associated with worse OS (pooled HR = 1.46, 95% CI 1.28–1.67, P < 0.001) and worse PFS/RFS (HR = 1.59, 95% CI 1.14–2.22, P = 0.006). In order to further investigate the role of SMAD4 gene in CRC, we also analyzed the relationship between SMAD4 status with clinical pathological parameters of CRC, the results show that patients with SMAD4 mutations have higher pathological TNM stages (stage IV/I–III; pooled OR = 1.28; 95% CI 1.03–1.58), that is, distant metastasis is more likely to occur in patients with SMAD4 mutations. And SMAD4 mutant patients were more likely to feature mucinous differentiation (pooled OR = 2.23; 95%CI 1.85–2.70, P = 0.000), tumors are more likely to occur in the colon (pooled OR = 1.15; 95% CI 1.01–1.31; P = 0.042), more prone to lymph node metastasis (N1 + N2/N0; pooled OR = 1.42; 95% CI 1.20–1.67; P = 0.000), and to harbor concurrent RAS mutations (pooled OR = 2.13; 95% CI 1.37–3.34; P = 0.001). Importantly, all of these parameters generally indicate a poor prognosis. Combined OR suggested that SMAD4 gene mutation has nothing to do with age, gender, tumor grade, MSI or BRAF status. The effect of SMAD4 gene on MSI or BRAF status remains to be elucidated. Another meta-analysis [14] showed that SMAD4-mutated patients were at a higher risk of distant metastasis (combined OR 2.04, 95% CI 1.41–2.95), which is consistent with our results.
Over the past 2 decades, many studies have shown that SMAD4 mutation does not cause tumorigenesis by itself, but it can promote tumor progression caused by other genes [8]. Ohtaki et al [22]. reported that the frequency of SMAD4 mutations were significantly higher in tumors with liver metastasis than in those without such metastasis. Inamoto et al. [23] reported that SMAD4-deficient colorectal tumor cells secreted more CCL9 and CCL15, these two chemokines recruit CCR1 + myeloid cells through CCL9-CCR1 and CCL15-CCR1 axis, resulting in metastasis. Vauthey et al. [24] concluded that patients with SMAD4 mutations are less likely to undergo repeated hepatectomy due to recurrent disease after the initial tumor resection. Alhopuro et al. [25] showed that SMAD4 is a predictive biomarker for 5-fluorouracil (5-Fu) based chemotherapy in CRC patients. Zhang et al. [26] discovered a novel mechanism mediated by SMAD4 to trigger 5-Fu chemosensitivity through cell cycle arrest by inhibiting the PI3K/Akt/CDC2/survivin cascade. Mei et al. [27] suggested that SMAD4 mutations could be potential biomarkers for poor prognosis of cetuximab-based therapy, which needs to be further validated in a larger patient cohort. Lin et al. [28] found that silencing SMAD4 reduces the sensitivity of CRC cells to cetuximab by promoting epithelial-mesenchymal transition (EMT), while the high expression of Smad4 may be clinically beneficial to cetuximab-based therapy. Mizuno et al. [11] found that SMAD4 mutation was significantly associated with poor OS following hepatic resection, which was independent of RAS mutation status. These findings indicate that SMAD4 pathogenic variants play a key role in tumor progression and the efficacy of target therapy in CRC patients.
In the current analysis, researchers found that SMAD4 gene alteration was significantly associated with loss of SMAD4 expression in CRC, and loss of SMAD4 disrupts canonical TGF-β signaling [29], because it is a signaling transcription factor. In addition, it is reported that the loss of SMAD4 function is independently associated with the reduction of RFS and OS in CRC patients, especially patients with advanced disease [30]. In contrast, CRC patients with high Smad4 expression had a much longer median OS than those with low Smad4 expression [31]. Germline mutations of TGF-β family signaling pathway genes significantly increase the risk of having colonic neoplasia [32]. The canonical TGF-β/Smad4 signaling pathway acts as a tumor suppressor in early stages, which is characterized by its anti-proliferative activity, ability to induce apoptosis and promote genome stability, while TGF-β acts as a metastasis promoter to stimulate the development of advanced tumors [33].
EMT is a well-coordinated process in which epithelial cells lose cell connectivity and polarity and transform into mesenchymal cells with migration and invasion capabilities. Studies have suggested that EMT is a key step in tumor progression and metastasis, and the TGF-β1 signaling plays a key role in EMT [34]. Functional study results indicate that TGF-β-induced Smad4-dependent EMT followed by apoptosis in CRC cells [35, 36]. Siraj et al. [37] identified TGF-β-induced EMT was insufficient to obtain invasive potential, while the activated RAS would alter the reaction, imparting tumorigenic and invasive potential. Therefore, the synergistic effect between Ras-Raf-MAPK and TGF-β/Smad cascades is a necessary condition for the acquisition of aggressive phenotype in cancer.
At present, RAS has been recognized as tumor driver gene, predictive biomarker and therapeutic target in CRC. The expression of RAS up-regulates the expression of phosphotyrosine kinase receptors ERBB1 (EGFR) and ERBB2 (HER2) and induces an aggressive phenotype. Smad4-dependent signal transduction negatively regulates the expression of these receptors and inhibits Ras-induced upregulation of EGFR and ERBB2, thus exerting an antiproliferative effect. The loss of oncogenic RAS and SMAD4 signals synergistically upregulate the abnormal expression of EGFR and ERBB2, leading to the development of neoplasm and the metastasis and spread of the primary tumor [38, 39]. TGF-β can quickly activated RAS and ERK pathway [40], in contrast, the ERK pathway inhibits the TGF-β/Smad4 pathway by phosphorylating Smad2 and Smad3 at serine or threonine residues in the linker region, so epithelial cells with oncogenic RAS mutations usually exhibit loss of TGF-β antiproliferative response [8]. Patients with RAS wild-type tumors and retained SMAD4 wild-type had longer OS than patients with both mutations [41]. However, SMAD4 mutations were significantly associated with poorer OS regardless of RAS mutation status or other clinicopathological factors. The precise cooperative mechanisms of SMAD4 with other genes of influence also requires further examination.
Given the relative frequency of SMAD4 mutations in CRC patients, routine SMAD4 testing may be appropriate. For individualized treatment of CRC, SMAD4, as a driver mutation, will become a novel target for precision medical treatment of CRC, and further research should be done for guiding clinical decision-making.
No heterogeneity or publication bias was found in this meta-analysis, and sensitivity analysis shows that our results are reliable. However, this analysis has several limitations. First, our meta-analysis included studies of qualified articles published in English or Chinese, and did not include relevant articles written in other languages or unpublished papers, which is likely to result in selection bias. Second, the use of specific therapies and tumor stage differed among the included articles. Third, the HR calculated from the data or extracted from the survival curve may not be as reliable as the HR calculated directly using the analysis of variance. Therefore, the results should be carefully interpreted. However, as far as we know, this is the first meta-analysis to demonstrate SMAD4 mutation by evaluating the pathological features and prognostication in CRC.
Conclusion
In conclusion, we found that SMAD4 mutation was associated with poor prognosis in CRC, but has nothing to do with MSI status, BRAF status or tumor grade. Further studies are needed to evaluate these findings and the clinical significance of SMAD4 status in CRC.
Availability of data and materials
All data generated or analyzed during this study are included in this article.
Abbreviations
- CRC:
-
Colorectal cancer
- OS:
-
Overall survival
- TGF-β:
-
Transforming growth factor beta
- HRs:
-
Hazard ratios
- ORs:
-
Odds ratios
- CI:
-
Confidence interval
- RAS:
-
Rat sarcoma viral oncogene homolog
- BRAF:
-
B-viral oncogene homolog B1
- MSI:
-
Microsatellite instability
- PFS:
-
Progression-free survival
- RFS:
-
Recurrence-free survival
- 5-Fu::
-
5-Fluorouracil
- EMT:
-
Epithelial-mesenchymal transition
References
Marmol I, Sanchez-de-Diego C, Pradilla Dieste A, Cerrada E, Rodriguez Yoldi MJ. Colorectal carcinoma: a general overview and future perspectives in colorectal cancer. Int J Mol Sci. 2017;18(1).https://www.ncbi.nlm.nih.gov/pubmed/28106826.
Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–7.
Itatani Y, Kawada K, Sakai Y. Transforming growth factor-beta signaling pathway in colorectal cancer and its tumor microenvironment. Int J Mol Sci. 2019;20(23). https://www.ncbi.nlm.nih.gov/pubmed/31756952.
Calonge MJ, Massague J. Smad4/DPC4 silencing and hyperactive Ras jointly disrupt transforming growth factor-beta antiproliferative responses in colon cancer cells. J Biol Chem. 1999;274(47):33637–43.
McCarthy AJ, Chetty R. Smad4/DPC4. J Clin Pathol. 2018;71(8):661–4.
Brosens LAA. Juvenile polyposis syndrome. World J Gastroenterol. 2011;17(44):4839.
Shovlin CL, Simeoni I, Downes K, Frazer ZC, Megy K, Bernabeu-Herrero ME, Shurr A, Brimley J, Patel D, Kell L, et al. Mutational and phenotypic characterization of hereditary hemorrhagic telangiectasia. Blood. 2020;136(17):1907–18.
Zhao M, Mishra L, Deng CX. The role of TGF-beta/SMAD4 signaling in cancer. Int J Biol Sci. 2018;14(2):111–23.
Fleming NI, Jorissen RN, Mouradov D, Christie M, Sakthianandeswaren A, Palmieri M, Day F, Li S, Tsui C, Lipton L, et al. SMAD2, SMAD3 and SMAD4 mutations in colorectal cancer. Cancer Res. 2013;73(2):725–35.
Sarshekeh AM, Advani S, Overman MJ, Manyam G, Kee BK, Fogelman DR, Dasari A, Raghav K, Vilar E, Manuel S, et al. Association of SMAD4 mutation with patient demographics, tumor characteristics, and clinical outcomes in colorectal cancer. PLoS ONE. 2017. https://doi.org/10.1371/journal.pone.0173345.
Mizuno T, Cloyd JM, Vicente D, Omichi K, Chun YS, Kopetz SE, Maru D, Conrad C, Tzeng CWD, Wei SH, et al. SMAD4 gene mutation predicts poor prognosis in patients undergoing resection for colorectal liver metastases. Eur J Surg Oncol. 2018;44(5):684–92. https://doi.org/10.1016/j.ejso.2018.02.247.
Miyaki M, Iijima T, Konishi M, Sakai K, Ishii A, Yasuno M, Hishima T, Koike M, Shitara N, Iwama T, et al. Higher frequency of Smad4 gene mutation in human colorectal cancer with distant metastasis. Oncogene. 1999;18(20):3098–103.
Kawaguchi Y, Kopetz S, Newhook TE, De Bellis M, Chun YS, Tzeng CD, Aloia TA, Vauthey JN. Mutation status of RAS, TP53, and SMAD4 is superior to mutation status of RAS alone for predicting prognosis after resection of colorectal liver metastases. Clin Cancer Res Off J Am Assoc Cancer Res. 2019;25(19):5843–51.
Huang D, Sun W, Zhou Y, Li P, Chen F, Chen H, Xia D, Xu E, Lai M, Wu Y, et al. Mutations of key driver genes in colorectal cancer progression and metastasis. Cancer Metastasis Rev. 2018;37(1):173–87.
Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25(9):603–5.
Oyanagi H, Shimada Y, Nagahashi M, Ichikawa H, Tajima Y, Abe K, Nakano M, Kameyama H, Takii Y, Kawasaki T, et al. SMAD4 alteration associates with invasive-front pathological markers and poor prognosis in colorectal cancer. Histopathology. 2019;74(6):873–82.
Liao X, Hao Y, Zhang X, Ward S, Houldsworth J, Polydorides AD, Harpaz N. Clinicopathological characterization of SMAD4-mutated intestinal adenocarcinomas: a case-control study. PLoS ONE. 2019;14(2):e0212142.
Stahler A, Stintzing S, von Einem JC, Westphalen CB, Heinrich K, Kramer N, Michl M, Modest DP, von Weikersthal LF, Decker T, et al. Single-nucleotide variants, tumour mutational burden and microsatellite instability in patients with metastatic colorectal cancer: next-generation sequencing results of the FIRE-3 trial. Eur J Cancer (Oxford, England: 1990). 2020;137:250–9.
Jia X, Shanmugam C, Paluri RK, Jhala NC, Behring MP, Katkoori VR, Sugandha SP, Bae S, Samuel T, Manne U. Prognostic value of loss of heterozygosity and sub-cellular localization of SMAD4 varies with tumor stage in colorectal cancer. Oncotarget. 2017;8(12):20198–212.
Ando T, Sugai T, Habano W, Jiao YF, Suzuki K. Analysis of SMAD4/DPC4 gene alterations in multiploid colorectal carcinomas. J Gastroenterol. 2005;40(7):708–15.
Khan M, Loree JM, Advani SM, Ning J, Li W, Pereira AAL, Lam M, Raghav K, Morris VK, Broaddus R, et al. Prognostic implications of mucinous differentiation in metastatic colorectal carcinoma can be explained by distinct molecular and clinicopathologic characteristics. Clin Colorectal Cancer. 2018;17(4):e699–709.
Ohtaki N, Yamaguchi A, Goi T, Fukaya T, Takeuchi K, Katayama K, Hirose K, Urano T. Somatic alterations of the DPC4 and Madr2 genes in colorectal cancers and relationship to metastasis. Int J Oncol. 2001;18(2):265–70.
Inamoto S, Itatani Y, Yamamoto T, Minamiguchi S, Hirai H, Iwamoto M, Hasegawa S, Taketo MM, Sakai Y, Kawada K. Loss of SMAD4 promotes colorectal cancer progression by accumulation of myeloid-derived suppressor cells through the CCL15-CCR1 chemokine axis. Clin Cancer Res Off J Am Assoc Cancer Res. 2016;22(2):492–501.
Vauthey JN, Kawaguchi Y. Innovation and future perspectives in the treatment of colorectal liver metastases. J Gastrointest Surg Off J Soc Surg Aliment Tract. 2020;24(2):492–6.
Alhopuro P, Alazzouzi H, Sammalkorpi H, Davalos V, Salovaara R, Hemminki A, Jarvinen H, Mecklin JP, Schwartz S Jr, Aaltonen LA, et al. SMAD4 levels and response to 5-fluorouracil in colorectal cancer. Clin Cancer Res Off J Am Assoc Cancer Res. 2005;11(17):6311–6.
Zhang B, Leng C, Wu C, Zhang Z, Dou L, Luo X, Zhang B, Chen X. Smad4 sensitizes colorectal cancer to 5-fluorouracil through cell cycle arrest by inhibiting the PI3K/Akt/CDC2/survivin cascade. Oncol Rep. 2016;35(3):1807–15.
Mei Z, Shao YW, Lin P, Cai X, Wang B, Ding Y, Ma X, Wu X, Xia Y, Zhu D, et al. SMAD4 and NF1 mutations as potential biomarkers for poor prognosis to cetuximab-based therapy in Chinese metastatic colorectal cancer patients. BMC Cancer. 2018;18(1):479.
Lin Z, Zhang L, Zhou J, Zheng J. Silencing Smad4 attenuates sensitivity of colorectal cancer cells to cetuximab by promoting epithelial-mesenchymal transition. Mol Med Rep. 2019;20:3735–45.
Tiwari A, Saraf S, Verma A, Panda PK, Jain SK. Novel targeting approaches and signaling pathways of colorectal cancer: an insight. World J Gastroenterol. 2018;24(39):4428–35.
Wasserman I, Lee LH, Ogino S, Marco MR, Wu C, Chen X, Datta J, Sadot E, Szeglin B, Guillem JG, et al. SMAD4 loss in colorectal cancer patients correlates with recurrence, loss of immune infiltrate, and chemoresistance. Clin Cancer Res Off J Am Assoc Cancer Res. 2019;25(6):1948–56.
Isaksson-Mettavainio M, Palmqvist R, Dahlin AM, Van Guelpen B, Rutegard J, Oberg A, Henriksson ML. High SMAD4 levels appear in microsatellite instability and hypermethylated colon cancers, and indicate a better prognosis. Int J Cancer. 2012;131(4):779–88.
Bellam N, Pasche B. Tgf-beta signaling alterations and colon cancer. Cancer Treat Res. 2010;155:85–103.
Javelaud D, Mauviel A. Crosstalk mechanisms between the mitogen-activated protein kinase pathways and Smad signaling downstream of TGF-beta: implications for carcinogenesis. Oncogene. 2005;24(37):5742–50.
Dai G, Sun B, Gong T, Pan Z, Meng Q, Ju W. Ginsenoside Rb2 inhibits epithelial-mesenchymal transition of colorectal cancer cells by suppressing TGF-beta/Smad signaling. Phytomed Int J Phytother Phytopharmacol. 2019;56:126–35.
Farooqi AA, de la Roche M, Djamgoz MBA, Siddik ZH. Overview of the oncogenic signaling pathways in colorectal cancer: mechanistic insights. Semin Cancer Biol. 2019;58:65–79.
Siraj AK, Pratheeshkumar P, Divya SP, Parvathareddy SK, Bu R, Masoodi T, Kong Y, Thangavel S, Al-Sanea N, Ashari LH, et al. TGFbeta-induced SMAD4-dependent apoptosis proceeded by EMT in CRC. Mol Cancer Ther. 2019;18(7):1312–22.
Safina AF, Varga AE, Bianchi A, Zheng Q, Kunnev D, Liang P, Bakin AV. Ras alters epithelial-mesenchymal transition in response to TGFbeta by reducing actin fibers and cell-matrix adhesion. Cell Cycle. 2009;8(2):284–98.
Zhao S, Wang Y, Cao L, Ouellette MM, Freeman JW. Expression of oncogenic K-ras and loss of Smad4 cooperate to induce the expression of EGFR and to promote invasion of immortalized human pancreas ductal cells. Int J Cancer. 2010;127(9):2076–87.
Grusch M, Petz M, Metzner T, Ozturk D, Schneller D, Mikulits W. The crosstalk of RAS with the TGF-beta family during carcinoma progression and its implications for targeted cancer therapy. Curr Cancer Drug Targets. 2010;10(8):849–57.
Yue J, Mulder KM. Requirement of Ras/MAPK pathway activation by transforming growth factor beta for transforming growth factor beta 1 production in a Smad-dependent pathway. J Biol Chem. 2000;275(40):30765–73.
Masetti M, Acquaviva G, Visani M, Tallini G, Fornelli A, Ragazzi M, Vasuri F, Grifoni D, Di Giacomo S, Fiorino S, et al. Long-term survivors of pancreatic adenocarcinoma show low rates of genetic alterations in KRAS, TP53 and SMAD4. Cancer Biomark. 2018;21(2):323–34.
Acknowledgements
Not applicable.
Funding
The authors declare that they did not receive funding for this research from any source.
Author information
Authors and Affiliations
Contributions
All authors provided intellectual input into the study design and methodology. TF, TL, and YW screened texts, performed data extraction and risks of bias assessment. TF and TL drafted the manuscript. HW, SL, LX and JL carried out the statistical analysis of studies. CW and YT provided comments and edited the manuscript to become the final version for submission. All authors approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Fang, T., Liang, T., Wang, Y. et al. Prognostic role and clinicopathological features of SMAD4 gene mutation in colorectal cancer: a systematic review and meta-analysis. BMC Gastroenterol 21, 297 (2021). https://doi.org/10.1186/s12876-021-01864-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12876-021-01864-9