Abstract
Background
Acute myocardial infarction (AMI) is a leading cause of death worldwide. Mitochondrial dysfunction is a key determinant of cell death post-AMI. Preventing mitochondrial dysfunction is thus a key therapeutic strategy. This study aimed to explore key genes and target compounds related to mitochondrial dysfunction in AMI patients and their association with major adverse cardiovascular events (MACE).
Methods
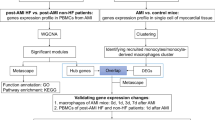
Differentially expressed genes in AMI were identified from the Gene Expression Omnibus (GEO) datasets (GSE166780 and GSE24519), and mitochondria-related genes were obtained from MitoCarta3.0 database. By intersection of the two gene groups, mitochondria-related genes in AMI were identified. Next, the identified genes related to mitochondria were subject to Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) functional enrichment analyses. Protein-protein interaction (PPI) network was constructed, and key genes were screened. Then, targeted drug screening and molecular docking were performed. Blood samples from AMI patients and healthy volunteers were analyzed for the key genes expressions using quantitative real time polymerase chain reaction (qRT-PCR). Later, receiver operating characteristic (ROC) curves assessed the diagnostic value of key genes, and univariate and multivariate COX analyses identified risk factors and protective factors for MACE in AMI patients.
Results
After screening and identification, 138 mitochondria-related genes were identified, mainly enriched in the processes and pathways of cellular respiration, redox, mitochondrial metabolism, apoptosis, amino acid and fatty acid metabolism. According to the PPI network, 5 key mitochondria-related genes in AMI were obtained: translational activator of cytochrome c oxidase I (TACO1), cytochrome c oxidase subunit Va (COX5A), PTEN-induced putative kinase 1 (PINK1), SURF1, and NDUFA11. Molecular docking showed that Cholic Acid, N-Formylmethionine interacted with COX5A, nicotinamide adenine dinucleotide + hydrogen (NADH) and NDUFA11. Subsequent basic experiments revealed that COX5A and NDUFA11 expressions were significantly lower in the blood of patients with AMI than those in the corresponding healthy volunteers; also, AMI patients with MACE had lower COX5A and NDUFA11 expressions in the blood than those without MACE (P < 0.01). ROC analysis also showed high diagnostic value for COX5A and NDUFA11 [area under the curve (AUC) > 0.85]. In terms of COX results, COX5A, NDUFA11 and left ventricular ejection fraction (LVEF) were protective factors for MACE in AMI, while C-reactive protein (CRP) was a risk factor.
Conclusion
COX5A and NDUFA11, key mitochondria-related genes in AMI, may be used as biomarkers to diagnose AMI and predict MACE.
Similar content being viewed by others
Introduction
Myocardial infarction is defined as a clinical or pathological event characterized by myocardial injury due to myocardial ischemia. According to data from the American Heart Association in 2023, the overall prevalence of myocardial infarction in adults over 20 years of age can be up to 3.2% [1]. More terribly, the hospital mortality rate of myocardial infarction is 7%, and more than 7 million people worldwide die of it each year [2, 3]. Acute myocardial infarction (AMI) has become a global public health problem and a significant challenge for cardiovascular physicians due to its high prevalence, mortality, and heavy socioeconomic burden. Although advanced treatment for myocardial infarction can improve short-term survival, the incidence of heart failure and recurrence, as well as long-term mortality rates, remain high [4]. Currently, independent predictors for assessing the risk of adverse cardiovascular events in patients with myocardial infarction include age, systolic blood pressure, history of heart failure, initial cardiac markers, and ST segment deviation. However, there are few investigations on the novel biomarkers for the prognosis of myocardial infarction [5].
Mitochondria are the center of cellular energy metabolism. The functional integrity of mitochondria is one of the necessary survival conditions for high-energy-metabolism cells such cardiomyocytes. Myocardial infarction is characterized by a reduction in coronary blood flow, and the resulting cardiac ischemia leads to mitochondrial dysfunction, activation of ischemic cascade reactions and cell death [6]. Mitochondrial proteins have been shown to be associated with AMI. To be specific, mitochondrial DNA (mtDNA) participates in cardiac remodeling through inducing myocardial inflammation after myocardial infarction [7]. In addition, myocardial infarction is closely related to abnormalities in the production pathway of mitochondrial energy. During myocardial ischemia and hypoxia, mitochondrial adenosine triphosphate (ATP) production decreases, and a large amount of lactic acid is produced; then, mitochondria are damaged by intracellular acidosis. While during reperfusion, the extremely rapid increase in local blood oxygen concentration and the persistence of mitochondrial dysfunction together contribute to excessive production of reactive oxygen species (ROS) in the mitochondria. Excessive ROS may disrupt the dynamic balance of redox and help the release of apoptotic factors into the cytoplasm. Hence, it is possible to initiate apoptosis when there is an excess of ROS [8]. Mitochondria can also protect the heart after myocardial infarction by eliminating damaged mitochondria through autophagy [9]. Ale-Agha et al. also stated that the mitochondrial telomerase reverse transcriptase (TERT) improved complex I subunit composition, thereby preventing myocardial ischemia/reperfusion injury through enhancing endothelial cell migration and myofibroblast differentiation [10]. It has been discovered that cardiac dysfunction and cardiomyocyte death are intimately related to mitochondrial damage. Therefore, in-depth study of the role of mitochondria-related genes in the development of AMI is of great significance in elucidating the pathogenesis of AMI and developing new therapeutic approaches for it.
In this paper, we investigated the key mitochondria-related genes and target compounds for mitochondrial dysfunction in AMI patients; also, their association with the diagnosis of AMI was explored through the analysis of the Gene Expression Omnibus (GEO) database. Then, the markers with prognostic value in myocardial infarction were identified, offering a novel perspective on how to safeguard the mitochondrial activity of cardiomyocytes.
Materials and methods
Data acquisition
Myocardial infarction-related expression profile datasets were searched in the GEO database with the keyword “myocardial infarction”. All searched datasets were screened according to the following inclusion criteria: (1) the disease was AMI; (2) the source was human; (3) the datasets contained healthy controls. Finally, the datasets GSE166780 (8 AMI samples and 8 control samples) and GSE24519 (34 AMI samples and 4 control samples) were obtained [11, 12]. Finally, 1136 human mitochondria-related genes were obtained from the MitoCarta 3.0 database (https://www.broadinstitute.org/mitocarta/mitocarta30-inventory-mammalian-mitochondrial-proteins-and-pathways) [13].
Data preprocessing and differentially expressed gene screening
The gene expression profiles of GSE166780 and GSE24519 datasets were downloaded from GEO database and converted into TPM format. The probe IDs of GSE166780 and GSE24519 were converted into gene symbols according to the probe annotation information of GPL20795 and GPL2895 platforms, respectively. The median value was calculated as the expression value of the gene if more than one probe corresponded to the same gene symbol. The probes matching multiple genes were deleted. Normalization, logarithmization, background correction and screening of differentially expressed genes (DEGs) were performed using the R package “limma”. DEG screening criteria were shown as follows: |log fold change (FC)| > 1 and adjusted p < 0.05. The intersection between the two sets of DEGs and mitochondrial-related genes were obtained separately using the online tool, Evenn (http://www.ehbio.com/test/venn/#/).
Gene functional enrichment analysis
To explore the pathophysiological changes in patients with AMI, the screened genes were subject to functional enrichment analysis using the R package “clusterProfiler”. Gene Oncology (GO) analysis included three dimensions, namely, biological processes (BP), cellular component (CC), and molecular function (MF), which described the functional annotation entries of different differential genes enriched. In Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis, hypergeometric test was used to reveal the genetic functions of genes based on gene enrichment pathways.
Protein-protein interaction network analysis
The STRING database was adopted to analyze protein-protein interaction (PPI) [14], and subsequently, the PPI networks were constructed. The interaction score > 0.4 was considered as a critical criterion, and active interaction sources, including text mining, experiments, databases, co-expression, neighboring genes, gene fusion and co-occurrence, were set as data mining parameters. Next, the PPI interactions obtained from STRING analysis were imported into Cytoscape for visualization. Subsequently, the top ten genes of Degree, Maximum Neighborhood Component (MNC), Edge Percolated Component (EPC), Closeness, and Radiality in the network were computationally acquired using the cytohubba plugin [15], and their expression regulations were annotated. As complementary indicators, betwweennes and bottleneck were used to identify key genes in the network.
Targeted drug screening and molecular docking
The DrugBank (https://go.drugbank.com/) database [16] was utilized to find the association between key genes and drugs. The two dimensional structures of the compounds were downloaded from Pubchem (https://pubchem.ncbi.nlm.nih.gov/) and Protein Data Bank (PDB, https://www.rcsb.org/) databases [17, 18]. Then, two dimensional structures downloaded were converted into three dimensional structures by ChemBioDraw 14.0 software. The protein crystals and compounds were pre-treated by original ligand elimination, dehydration and hydrogenation using AutoDock Tools 1.5.6 and Discovery Studio 2016. Molecular docking between active ingredients and proteins was carried out by AutoDockVina 1.1.2 software with the binding sites of protein self-contained ligands as active pockets.
Study subjects
This study enrolled AMI patients (AMI group, n = 114) who were diagnosed and treated in the Affiliated Hospital of Nantong University from January 2022 to January 2023 and healthy volunteers (Healthy group, n = 45) during the same period. Inclusion criteria: (1) patients were willing to participate in this study and were informed about it; (2) men or non-pregnant women were aged from 18 to 85 years old; and (3) the AMI patients enrolled were characterized with typical chest pain, elevated troponin I (TnI) levels, and ischemic-looking ST-T segment changes on the electrocardiogram (ECG). Exclusion criteria were listed as follows: (1) patients with a history of myocardial infarction, cardiac surgery, severe hepatic or renal failure; (2) patients with uncontrolled severe hypertension (systolic blood pressure > 180 mmHg and diastolic blood pressure > 100 mmHg after standard treatment); (3) patients with recent trauma or a history of major surgery within 30 days; (4) patients with severe breathing difficulties, such as bronchial asthma and chronic obstructive pulmonary disease; (5) patients with active malignancies and autoimmune diseases; (6) pregnant or lactating women; and (7) patients with poor compliance.
The following baseline information was collected from subjects in the AMI and Healthy groups: gender, age, body mass index (BMI), smoking history, drinking history, diabetes mellitus history, hypertension history, left ventricular ejection fraction (LVEF) (%), leukocytes (109/L), erythrocytes (1012/L), hemoglobin (g/L), B-type natriuretic peptide (BNP) (pg/ml), TnI (ng/ml), C-reactive protein (CRP) (mg/L). This study was approved by the Ethics Committee of Affiliated Hospital of Nantong University (B2018-154 (2)), and all patients signed the informed consent form.
Follow-up
Patients in the AMI group were followed up for 6 months with a follow-up endpoint of occurrence of major adverse cardiovascular events (MACE). MACE included heart failure, nonfatal re-infarction, recurrent angina, rehospitalization for cardiovascular-related illness, revascularization, and all-cause mortality. The 114 patients in the AMI group were categorized into MACE (n = 26) and non-MACE (n = 88) groups dependent on whether MACE occurred.
Quantitative real-time polymerase chain reaction
First off, 5mL of venous blood was extracted from patients in the AMI group before hospitalization and patients in the Healthy group during physical examination, respectively. The blood samples extracted were then stored in heparin anticoagulant tubes. Next, total RNA in peripheral blood was extracted by QIAGENRNeasyMini kit (74,106, QIAGEN, Duesseldorf, Germany), and then reversely transcribed into cDNA by One Step RT-PCR kit (TQ2601, Shanghai Yuanmu Biotechnology Co., Ltd., Shanghai, China). TB Green Premix Ex Taq II kit (RR820A, Takara, Tokyo, Japan) was used for quantitative real-time polymerase chain reaction (qRT-PCR). The messenger ribonucleic acid (mRNA) expression of key genes in peripheral blood was determined on the PCR instrument (7500 Fast Real-Time PCR System, ABI, USA). The reaction system was 20 µL in total, including 1 µL of complementary deoxyribonucleic acid (cDNA), 0.5 µL each of forward and reverse primers (Table 1), 10 µL of PCR buffer, and 8 µL of distilled water. The reaction conditions were displayed as follows: pre-denaturation at 95 °C for 3 min, denaturation at 95 °C for 15 s, annealing at 60 °C for 30 s, and extension at 70 °C for 30 s, with a total of 40 cycles. The relative expression of cytochrome c oxidase subunit Va (COX5A) and NDUFA11 was calculated by the 2−ΔΔCt method with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as the internal reference gene.
Statistical analysis
Data were processed using SPSS23.0, GraphPadPrism7.0 and MedCalc15.8. Measurement data conforming to normal distribution were expressed as mean ± standard deviation (SD), and those did not conform to normal distribution were expressed as median (range). Count data were expressed as n (%), and comparisons between two groups were made using independent samples t-test and chi-square test, respectively. Pearson correlation analysis was used to explore the expression correlation between key genes. The median of gene expression was considered as the threshold value, and the study subjects were categorized into high-expression and low-expression groups. The predictive value of the key genes was assessed using the receiver operating characteristic curve (ROC). The analysis of MACE was performed using univariate and multivariate Cox proportional-hazards models. Factors with P < 0.05 in univariate Cox regression analysis were included in multivariate Cox regression analysis for screening of the risk factors of MACE. P < 0.05 was considered a statistically significant difference.
Results
Identification of mitochondria-related differentially expressed genes in acute myocardial infarction
Gene expression data of GSE24519 and GSE166780 were acquired from the GEO database, and significant DEGs were identified through differential analysis with a criteria of |logFC |> 1 and P < 0.05. Briefly, 2,044 down-regulated genes and 304 up-regulated genes were screened for GSE166780 (Fig. 1A); 42 down-regulated genes and 439 up-regulated genes were screened for GSE24519 (Fig. 1B). Besides, 1136 human mitochondria-associated genes were obtained from the MitoCarta3.0 database. The two sets of DEGs were intersected with the 1136 mitochondria-associated genes, and 117 (8 up-regulated DEGs and 109 down-regulated DEGs) and 22 (18 up-regulated DEGs and 7 down-regulated DEGs) mitochondria-associated DEGs were obtained, respectively (Fig. 1C, D). Finally, 138 mitochondria-associated DEGs were obtained by combining the intersected genes and removing duplicated ones.
Screening for mitochondria-associated differentially expressed genes in acute myocardial infarction. A–B, Visualization of DEGs sourced from datasets GSE166780 (A) and GSE24519 (B), with heatmaps (top 200) on the left and volcano plots on the right (screening conditions of |logFC| > 1 and P < 0.05, blue indicating significantly down-regulated genes in AMI and red indicating significantly up-regulated genes in AMI); C–D, Venn diagrams showing the intersection of DEGs from the GSE166780 (C) and GSE24519 (D) datasets with mitochondria-associated genes. DEGs, differentially expressed genes; AMI, acute myocardial infarction
Functional enrichment analyses of differentially expressed genes
Subsequently, functional enrichment analyses were performed on the 138 mitochondria-related DEGs obtained. GO enrichment analysis results showed that 138 DEGs were enriched to BP including energy derivation by oxidation of organic compounds, cellular respiration, mitochondrial transmembrane transport, regulation of mitochondrion organization, organic acid catabolic process and nucleotide biosynthetic process (Fig. 2A); to CCs including mitochondrial matrix, mitochondrial inner membrane, mitochondrial outer membrane, organelle outer membrane, outer membrane, and integral component of the mitochondrial membrane (Fig. 2B); and to MFs including flavin adenine dinucleotide binding, oxidoreductase activity, acting on the CH-CH group of donors, acyl-CoA dehydrogenase activity, oxidoreductase activity, acting on the CH-CH group of donors, with a flavin as acceptor, oxidoreductase activity, acting on a sulfur group of donors, and protein-disulfide reductase activity (Fig. 2C). Additionally, KEGG enrichment analysis revealed the correlation of 138 DEGs with mitophagy-animal, biosynthesis of cofactors, valine, leucine and isoleucine degradation, fatty acid metabolism, thermogenesis and apoptosis-multiple species pathways (Fig. 2D).
Gene Ontology and Kyoto Encyclopedia of Genes and Genomes enrichment analyses of mitochondria-associated differentially expressed genes.A–D, Mitochondria-associated differentially expressed genes enriched for biological processes (BP, A, top 6), cellular components (CC, B, top 6), molecular functions (MF, C, top 6) and KEGG signaling pathways (D, top 6)
Construction of protein-protein interaction network and screening of key genes
The PPI (an interaction score > 0.4) of 138 DEGs were further analyzed. As shown in Fig. 3A, the PPI network consisted of 27 nodes and 79 edges. The top 10 hub genes of each algorithm [Matthews Correlation Coefficient (MCC), Density of MNC (DMNC), MNC, Degree and EPC] in the cytohubba plugin were obtained. After intersecting theses hub genes using the Venn diagram, translational activator of cytochrome c oxidase I (TACO1), COX5A, PTEN-induced putative kinase 1 (PINK1), SURF1 and NDUFA11 were screened out (Fig. 3B). Further, the correlation analysis of the hub genes in the two datasets were performed separately. Specifically, the GSE166780 dataset exhibited a strong and positive correlations between the hub genes, while the GSE24519 dataset displayed a significant and positive correlation between the expression of COX5A and SURF1 (Fig. 3C, D).
Construction of protein-protein interaction network for mitochondria-associated differentially expressed genes and screening of key genes. (A), PPI network of mitochondria-associated DEGs; (B), Intersection of the top 10 genes-key genes in the five algorithms of MCC, DMNC, MNC, Degree, and EPC in the cytohubba plugin; C–D, correlation analysis of mitochondria-related key DEGs in the datasets GSE24519 (C) and GSE166780 (D). PPI, protein-protein interaction; DEGs, differentially expressed genes; MCC, Matthews Correlation Coefficient; MNC, Maximum Neighborhood Component; DMNC, Density of MNC; EPC, Edge Percolated Component
Targeted drug screening and molecular docking
The DrugBank database was used to explore associations between key genes and drugs. Briefly speaking, Cholic Acid and N-Formylmethionine were found to interact with COX5A, nicotinamide adenine dinucleotide + hydrogen (NADH) and NDUFA11. The molecular docking results predicted a minimum binding energy of -6.3 kcal mol− 1 for Cholic Acid and COX5A, -3.9 kcal mol− 1 for N-Formylmethionine and COX5A, and − 8.3 kcal mol− 1 for NDUFA11 and NADH (Fig. 4A–C). However, no drugs targeting AMI that bind to TACO1, PINK1, and SURF1 have been identified.
Baseline information
A total of 114 AMI patients and 45 healthy controls were included in this study. Their baseline data were shown in Table 2. We could see that there was no significant difference in gender, BMI, smoking history, drinking history, diabetes mellitus history, hypertension history, erythrocytes, and hemoglobin between the subjects in the AMI group and the Healthy group (P > 0.05). Notably, compared with the Healthy group, the age, leukocyte count, BNP, TnI, and CRP levels of the subjects in the AMI group were much higher, while the LVEF was much lower (P < 0.05).
Validation of COX5A and NDUFA11 expression in peripheral blood of patients with acute myocardial infarction
Subsequently, we detected the expression of COX5A and NDUFA11 in the collected clinical blood samples. The results showed that the expression levels of COX5A and NDUFA11 in the peripheral blood of patients in the AMI group were much lower than those in the Healthy group (P < 0.01, Fig. 5A, B). Next, the patients with AMI were divided into MACE group and non-MACE group according to the presence of MACE. Briefly, patients in the MACE group exhibited an obvious drop in the expression levels of COX5A and NDUFA11 in peripheral blood compared with those in the non-MACE group (P < 0.01, Fig. 5C, D). In a nutshell, low expression levels of COX5A and NDUFA11 were associated with the occurrence of AMI or MACE in AMI patients.
Validation of COX5A and NDUFA11 expression in peripheral blood of patients with acute myocardial infarction. A–B, qRT-PCR to detect the relative mRNA expression levels of COX5A (A) and NDUFA11 (B) in the peripheral blood of subjects in the Healthy group and the AMI group; C–D, qRT-PCR to detect the relative mRNA expression levels of COX5A (C) and NDUFA11 (D) in the peripheral blood of subjects in the non-MACE group and the MACE group. **P < 0.01. AMI, acute myocardial infarction; qRT-PCR, quantitative real-time polymerase chain reaction; COX5A, cytochrome c oxidase subunit Va; mRNA, messenger ribonucleic acid; MACE, major adverse cardiovascular events
Diagnostic value of COX5A and NDUFA11 in acute myocardial infarction
ROC curves were used to analyze the value of COX5A and NDUFA11 in the diagnosis of AMI. As shown in Fig. 6, area under the curve (AUC) of COX5A, NDUFA11 and the combination of the two in predicting the occurrence of AMI was 0.8901, 0.8559 and 0.9370, respectively, and the AUC of the three was greater than 85%. Such results suggested that COX5A, NDUFA11 and their combination had good diagnostic value in AMI. It was worth noting that the diagnostic value of COX5A combined with NDUFA11 was better than that of COX5A and NDUFA11 genes alone.
Receiver operating characteristic curves for determining the diagnostic value of COX5A, NDUFA11 expression and their combination in acute myocardial infarction. The black line represented COX5A, the red line represented NDUFA11, and the blue line represented COX5A combined with NDUFA11. COX5A, cytochrome c oxidase subunit Va
COX proportional-hazards models for predicting the occurrence of major adverse cardiovascular events in patients with acute myocardial infarction
Finally, we assessed the factors influencing the occurrence of MACE in AMI patients by univariate and multivariate COX regression analyses (Table 3). The results of univariate analysis showed that BNP [hazard ratio (HR) = 1.001), TnI (HR = 1.044), and CRP (HR = 1.027) were risk factors for the occurrence of MACE in AMI patients, whereas COX5A (HR = 0.109), NDUFA11 (HR = 0.085), and LVEF (HR = 0.126) acted as protective factors for it.
With respect to multivariate analysis, COX5A (HR = 0.326, P = 0.04), NDUFA11 (HR = 0.221, P = 0.011), and LVEF (HR = 0.381, P = 0.033) were the protective factors for the occurrence of MACE, while CRP (HR = 1.018, P = 0.003) was a risk factor for it.
Discussion
As one of the common cardiovascular diseases, AMI is featured with rapid onset and high mortality rate and may be accompanied by MACE. The rapid development and wide application of microarray technology has opened up a new avenue for deciphering the etiology of AMI and identifying its main targets. Prolonged hypoxia triggers stress responses such as reduced ATP production and calcium overload in cardiomyocytes, as well as overproduction of ROS in mitochondria. These stress responses result in excessive mitochondrial damage and the release of pro-apoptotic factors like cytochrome C, thereby activating downstream apoptotic pathways [6].
In this study, the DEGs of myocardial infarction were screened by bioinformatics, followed by the intersection with mitochondria-related genes. The observation results revealed that the main cellular functions involved in AMI included cellular respiration, redox, apoptosis, amino acid and fatty acid metabolism, and that the key genes included TACO1, COX5A, PINK1, SURF1, and NDUFA11. Of them, COX5A, a constitutive subunit of cytochrome C oxidase in the mitochondrial respiratory chain, has been reported to have considerably low expression in the hearts and cardiomyocytes of rats with myocardial infarction induced by hypoxia. Also, COX5A acts as a downstream regulatory factor of microRNA-26a/b-5p to promote myocardial infarction-induced cell death [19]. PINK is a mitochondrial protein kinase directly tied to the maintenance of mitochondrial homeostasis. As reported in a recent research, PINK has a major role in stress-induced mitochondrial autophagy, which can protect cardiomyocytes from ischemic injury [20]. TACO1 is a mitochondrial protein that participates in the transcriptional activation of cytochrome c oxidase subunit I in mitochondria [21]. SURF1, a protein localized in the inner mitochondrial membrane, is implicated in the assembly and stability of cytochrome c oxidase [22]. NDUFA11, an accessory subunit of mitochondrial respiratory chain complex I, is essential for the generation of ATP in the mitochondria [23]. In fact, TACO1, SURF1, and NDUFA11 are all linked to mitochondrial function and their mutations can cause mitochondrial dysfunction [21, 23, 24]. Studies have shown that ischemia-reperfusion (I/R) injury impacts mitochondrial biosynthesis through various post-transcriptional mechanisms, including the regulation of mitochondrial RNA stability, translation, and the modification of mitochondrial proteins [25, 26]. Additionally, post-translational modifications such as phosphorylation, acetylation, and ubiquitination of mitochondrial proteins are crucial in responding to the stress induced by I/R injury, ensuring proper mitochondrial function and promoting cell survival [27, 28]. These specific regulatory mechanisms play a vital role in maintaining mitochondrial integrity and function, thus contributing to cardiomyocyte maturation and survival during and after I/R injury. Nevertheless, no studies have yet explore the direct connection between these genes and myocardial infarction. Therefore, more studies are required to clarify the connection.
In recent years, the application of targeted drugs in myocardial infarction has received widespread attention. Examples include interleukin (IL)-1β antagonists that target inflammatory responses, Monocyte chemoattractant protein (MCP)-1/ C-C motif ligand 2 (CCL2) signaling pathway antagonists, and antibodies to leukocyte surface molecules that target immune cells [29,30,31]. However, both therapeutic outcomes and clinical translation of these targeted medications have fallen short of expectations. The prevention or mitigation of cardiac I/R injury can be aided by improving mitochondrial dysfunction now. Mitochondrial dysfunction can be improved to attenuate myocardial infarction through multiple pathways and different mechanisms with the help of modern medicine, traditional Chinese medicine (TCM), and other therapies such as hyperbaric oxygen [32]. The most widely used western medicines for improving myocardial injury include mitochondrial permeability transition pore (mPTP) inhibitors such cyclosporin A and coenzyme Q10, modulators of mitochondrial dynamics such as Mdivi-1, and mitochondria-targeted antioxidants such as MitoQ and SkQ1 [33]. In terms of TCM, danshen injection and carthamus tinctorius linne are usually adopted for the improvement of myocardial I/R injury [34, 35]. Our results demonstrated that Cholic Acid, N-Formylmethionine and NADH all produce potential therapeutic effects on AMI through binding to COX5A and NDUFA11. Cholic Acid has a major role in the management of serum cholesterol levels, which are linked to cardiac health, as well as the digestion and absorption of fats. Yunlong Xia et al. put forward that endogenous bile acids may exert cardioprotective effects against acute ischemic cardiac injury by binding to mesenchymal stromal cells [36]. N-Formylmethionine aids in protein synthesis in bacteria. The role of N-Formylmethionine may be connected to the mechanism of bacterial development and infection in cases when certain bacterial infections result in heart inflammation or infective endocarditis. In addition, N-Formylmethionine has been shown to correlate with the left ventricular mass index and may be involved in left ventricular remodeling and the BP of subclinical heart failure [37]. NADH is crucial in energy metabolism in the heart. In conditions such as myocardial ischemia and myocardial infarction, energy metabolism may be severely disturbed, thereby decreasing NADH accumulation and energy production [38]. In our study, molecular docking results elucidated that COX5A and NDUFA11 were the main target genes for drugs mentioned above. Therefore, we examined the expression of COX5A and NDUFA11 in the peripheral blood of AMI patients and healthy volunteers. The examination outcomes indicated that the expression levels of COX5A and NDUFA11 were significantly reduced in the peripheral blood of the AMI group compared with those in the Healthy group; also, they were much lower in the peripheral blood of patients in the MACE group than in the non-MACE group. It can be concluded that COX5A and NDUFA11 may serve as biomarkers for the diagnosis of AMI and prediction of MACE. The ROC curves further disclosed that the two genes had certain diagnostic value for myocardial infarction and their combination had a higher diagnostic efficacy than each one alone. Additionally, the results of COX regression analyses indicated that COX5A, NDUFA11, LVEF, and CRP in peripheral blood may be independent influencing factors of MACE, suggesting the correlation of COX5A, NDUFA11, LVEF, and CRP with the occurrence of MACE after AMI.
While our study identifies key mitochondria-related genes (TACO1, COX5A, PINK1, SURF1, and NDUFA11) associated with AMI, several limitations must be acknowledged. Firstly, the limited sample size of the GEO database made it impossible to obtain more detailed clinical data and prognostic information. This limitation highlights the need for larger, more diverse cohorts to validate our findings across different populations. Secondly, we did not do prognosis analyses for adverse outcomes or assess the correlation between key gene expression and risk factors of AMI. Moreover, further functional experiments are needed to confirm that key genes and targeted drugs identified by bioinformatics analysis are involved in regulating BP such as energy metabolism and mitochondrial respiratory function after AMI. Finally, our current research is based on bioinformatics analysis and experimental validation at the mRNA level. Additional experiments, such as cell-based studies involving RNA interference or overexpression of target genes, are necessary to observe their impact on cell phenotype and validate their functional roles in AMI. Despite these limitations, our study contributes to the understanding of mitochondrial dysfunction in AMI and suggests potential biomarkers and therapeutic targets. Further research addressing these limitations will enhance the feasibility and clinical applicability of our findings, ultimately advancing medical development in the treatment of myocardial infarction.
Conclusion
In summary, the key mitochondria-related differential genes in AMI patients screened by bioinformatics are TACO1, COX5A, PINK1, SURF1 and NDUFA11, while the targeted drugs identified include Cholic Acid, N-Formylmethionine and NADH. These drugs can bind to COX5A and NDUFA11, respectively. Furthermore, COX5A and NDUFA11 are significantly down-regulated in the peripheral blood of AMI patients and AMI patients with MACE, and the down-regulation is closely related to the occurrence of MACE in AMI patients. More importantly, the two key genes have high diagnostic value in AMI. Taken together, COX5A and NDUFA11 may be potential biomarkers for the diagnosis of AMI and the prediction of MACE.
Data availability
The datasets used or analyzed during the current study are available from the corresponding author on reasonable request.
References
Tsao CW, et al. Heart Disease and Stroke Statistics-2023 update: a Report from the American Heart Association. Circulation. 2023;147(8):e93–621.
Reed GW, Rossi JE, Cannon CP. Acute Myocard Infarct Lancet. 2017;389(10065):197–210.
Kochar A et al. Long-term mortality of older patients with Acute myocardial infarction treated in US clinical practice. J Am Heart Assoc, 2018. 7(13).
Damluji AA, et al. Mechanical complications of Acute Myocardial Infarction: A Scientific Statement from the American Heart Association. Circulation. 2021;144(2):e16–35.
Fox KA, et al. Prediction of risk of death and myocardial infarction in the six months after presentation with acute coronary syndrome: prospective multinational observational study (GRACE). BMJ. 2006;333(7578):1091.
Ramachandra CJA, et al. Mitochondria in acute myocardial infarction and cardioprotection. EBioMedicine. 2020;57:102884.
Enzan N, et al. ZBP1 protects against mtDNA-Induced myocardial inflammation in failing hearts. Circ Res. 2023;132(9):1110–26.
Qi B, et al. Cardiac-specific overexpression of Ndufs1 ameliorates cardiac dysfunction after myocardial infarction by alleviating mitochondrial dysfunction and apoptosis. Exp Mol Med. 2022;54(7):946–60.
Zhang H, et al. The zinc transporter ZIP7 (Slc39a7) controls myocardial reperfusion injury by regulating mitophagy. Basic Res Cardiol. 2021;116(1):54.
Ale-Agha N, et al. Mitochondrial telomerase reverse transcriptase protects from myocardial Ischemia/Reperfusion Injury by improving Complex I composition and function. Circulation. 2021;144(23):1876–90.
Wang C, et al. Gene expression analysis suggests immunological changes of Peripheral Blood monocytes in the progression of patients with coronary artery disease. Front Genet. 2021;12:641117.
Su J, et al. Genes associated with inflammation and the cell cycle may serve as biomarkers for the diagnosis and prognosis of acute myocardial infarction in a Chinese population. Mol Med Rep. 2018;18(2):1311–22.
Rath S, et al. MitoCarta3.0: an updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic Acids Res. 2021;49(D1):D1541–7.
Szklarczyk D, et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49(D1):D605–12.
Chin CH, et al. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8(Suppl 4):S11.
Wishart DS, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46(D1):D1074–82.
Kim S, et al. PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res. 2021;49(D1):D1388–95.
Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28(1):235–42.
Jung SE, et al. MicroRNA-26a/b-5p promotes myocardial infarction-induced cell death by downregulating cytochrome c oxidase 5a. Exp Mol Med. 2021;53(9):1332–43.
Tu M, et al. RhoA signaling increases mitophagy and protects cardiomyocytes against ischemia by stabilizing PINK1 protein and recruiting Parkin to mitochondria. Cell Death Differ. 2022;29(12):2472–86.
Richman TR, et al. Loss of the RNA-binding protein TACO1 causes late-onset mitochondrial dysfunction in mice. Nat Commun. 2016;7:11884.
Khan TR, et al. Natural History of SURF1 Deficiency: a Retrospective Chart Review. Pediatr Neurol. 2023;140:40–6.
Knapp-Wilson A et al. Maintenance of complex I and its supercomplexes by NDUF-11 is essential for mitochondrial structure, function and health. J Cell Sci, 2021. 134(13).
Lee IC, Chiang KL. Clinical diagnosis and treatment of Leigh Syndrome based on SURF1: genotype and phenotype. Antioxid (Basel), 2021. 10(12).
Hayashida K, et al. Mitochondrial transplantation therapy for ischemia reperfusion injury: a systematic review of animal and human studies. J Transl Med. 2021;19(1):214.
Yang S, et al. Protective effects of salvianic acid A against multiple-organ ischemia-reperfusion injury: a review. Front Pharmacol. 2023;14:1297124.
Sluijter JPG, Xiao J. Post-translational modifications upon mitochondrial dysfunction in heart failure. Eur Heart J. 2023;44(44):4713–4.
Alves-Figueiredo H, et al. A systematic review of post-translational modifications in the mitochondrial permeability transition pore complex associated with cardiac diseases. Biochim Biophys Acta Mol Basis Dis. 2021;1867(1):165992.
Abbate A, et al. Interleukin-1 blockade with anakinra to prevent adverse cardiac remodeling after acute myocardial infarction (Virginia Commonwealth University Anakinra Remodeling Trial [VCU-ART] Pilot study). Am J Cardiol. 2010;105(10):1371–e13771.
Dhayni K, et al. Targeting CXCR1 and CXCR2 receptors in cardiovascular diseases. Pharmacol Ther. 2022;237:108257.
Kyaw T, et al. Cytotoxic and proinflammatory CD8 + T lymphocytes promote development of vulnerable atherosclerotic plaques in apoe-deficient mice. Circulation. 2013;127(9):1028–39.
Hu X, et al. The role of Oxygen Therapy in Normoxemic Acute Coronary Syndrome: a systematic review of Randomized controlled trials. J Cardiovasc Nurs. 2018;33(6):559–67.
Xin Y, et al. New insights into the role of Mitochondria Quality Control in Ischemic Heart Disease. Front Cardiovasc Med. 2021;8:774619.
Zhang Y, et al. A Chinese patent medicine Salvia miltiorrhiza depside salts for infusion combined with conventional treatment for patients with angina pectoris: a systematic review and meta-analysis of randomized controlled trials. Phytomedicine. 2017;25:100–17.
Bai J, et al. Study on the protective effects of danshen-honghua herb pair (DHHP) on myocardial ischaemia/reperfusion injury (MIRI) and potential mechanisms based on apoptosis and mitochondria. Pharm Biol. 2021;59(1):335–46.
Xia Y, et al. Mesenchymal stromal cells overexpressing farnesoid X receptor exert cardioprotective effects against Acute Ischemic Heart Injury by binding endogenous bile acids. Adv Sci (Weinh). 2022;9(24):e2200431.
Razavi AC, et al. Pseudouridine and N-formylmethionine associate with left ventricular mass index: Metabolome-wide association analysis of cardiac remodeling. J Mol Cell Cardiol. 2020;140:22–9.
Zhai X, et al. Exogenous supplemental NAD + protect myocardium against myocardial ischemic/reperfusion injury in swine model. Am J Transl Res. 2019;11(9):6066–74.
Acknowledgements
Not applicable.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
The conception and design of the research, drafting of the manuscript, and analysis and interpretation of data are done by Jun Qiu, Yiyang Gu. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was approved by the Ethics Committee of Affiliated Hospital of Nantong University (B2018-154 (2) ), and all patients signed the informed consent form.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it.The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Qiu, J., Gu, Y. Analysis of the prognostic value of mitochondria-related genes in patients with acute myocardial infarction. BMC Cardiovasc Disord 24, 408 (2024). https://doi.org/10.1186/s12872-024-04051-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12872-024-04051-2