Abstract
Background
As a novel circRNA, BTBD7_hsa_circ_0000563 has not been fully investigated in coronary artery disease (CAD). Our aim is to reveal the possible functional role and regulatory pathway of BTBD7_hsa_circ_0000563 in CAD via exploring genes combined with BTBD7_hsa_circ_0000563.
Methods
A total of 45 peripheral blood mononuclear cell (PBMC) samples of CAD patients were enrolled. The ChIRP-RNAseq assay was performed to directly explore genes bound to BTBD7_hsa_circ_0000563. The Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis were conducted to reveal possible functions of these genes. The interaction network was constructed by the STRING database and the Cytoscape software. The Cytoscape software were used again to identify clusters and hub genes of genes bound to BTBD7_hsa_circ_0000563. The target miRNAs of hub genes were predicted via online databases.
Results
In this study, a total of 221 mRNAs directly bound to BTBD7_hsa_circ_0000563 were identified in PBMCs of CAD patients via ChIRP-RNAseq. The functional enrichment analysis revealed that these mRNAs may participate in translation and necroptosis. Moreover, the interaction network showed that there may be a close relationship between these mRNAs. Eight clusters can be further subdivided from the interaction network. RPS3 and RPSA were identified as hub genes and hsa-miR-493-5p was predicted to be the target miRNA of RPS3.
Conclusions
BTBD7_hsa_circ_0000563 and mRNAs directly bound to it may influence the initiation and progression of CAD, among which RPS3 and RPSA may be hub genes. These findings may provide innovative ideas for further research on CAD.
Similar content being viewed by others
Background
Coronary artery disease (CAD) still poses a great threat to human health and quality of lives, although the current treatment strategies for CAD are quite mature [1, 2]. Since CAD is considered to be influenced by a combination of genetic and lifestyle factors, deeper genetic factors deserve to be explored [3].
In eukaryotes, circular RNAs (circRNAs) are covalently closed and endogenous biomolecules without 5′ and 3′ ends and poly-a-tail. The synthesis of circRNAs relies on reverse splicing, which includes connection between the donor site and the upstream receptor site, making circRNAs containing a single exon or multiple exons [4]. Currently, the functions of circRNAs that have been revealed mainly include “microRNAs (miRNAs) sponges”, regulation of transcription, combination with RNA binding proteins (RBPs), and encoding small peptides [4,5,6]. Moreover, circRNAs have been shown to not only serve as biomarkers to assist diagnosis in diseases such as cerebrovascular disease, various cancers, and liver disease [7,8,9], but also as therapeutic targets for these diseases [9,10,11]. Due to the characteristics of expression abundant and evolutionarily conserved, the impact of circRNAs on the pathogenesis of CAD is gradually being emphasized and research on the mechanism of circRNAs involvement in CAD is rapidly developing as well [12, 13]. However, it remains unclear whether circRNAs can interact with RNAs other than miRNAs to regulate gene expression.
In our previous study, we found that the expression level of BTBD7_hsa_circ_0000563 was significantly lower in human coronary artery samples with severe atherosclerotic stenosis than that with moderate atherosclerotic stenosis [14]. Simultaneously, we observed that the level of BTBD7_hsa_circ_0000563 was significantly lower in PBMCs of CAD patients compared to healthy controls [15]. However, it is crucial to investigate the function and regulatory pathway of BTBD7_hsa_circ_0000563 to explore the relationship between BTBD7_hsa_circ_0000563 and CAD.
In the present investigation, we profiled the expression of transcriptome-wide RNAs bound to BTBD7_hsa_circ_0000563 (except for miRNA) in peripheral blood mononuclear cells (PBMCs) of subjects with CAD by means of chromatin isolation by RNA purification followed by RNA sequencing (ChIRP-RNAseq). Moreover, bioinformatics analyses were conducted to predict functions of RNAs bound to BTBD7_hsa_circ_0000563.
Materials and methods
Study subjects
From May 2021 to October 2021, a total of 45 subjects with CAD were enrolled from the First Affiliated Hospital of Nanjing Medical University. Subjects with infectious diseases, malignant neoplasms, cerebrovascular disease, congenital heart disease, myocardiopathy, rheumatic valvular disease, and dysfunction with adrenal, liver, kidney or thyroid were excluded from the present study. All subjects underwent coronary artery angiography. According to the ACC/AHA classification, the diagnosis criteria of CAD as following: stenosis of major epicardial coronary artery ≥50% [16]. The baseline characteristics of the subjects were shown in Table 1.
Written informed consent from all subjects was obtained. All experimental protocols were approved by the ethics committee of the First Affiliated Hospital of Nanjing Medical University and were conducted in accordance with the Declaration of Helsinki.
PBMCs isolation
Ten ml of artery blood samples were drawn from the arterial sheaths in all subjects by artery puncture before coronary angiography. After being treated with Lymphocyte Separation Medium (TBD, Tianjin, China), PBMCs were isolated from the diluted whole blood to the middle white monolayer by density gradient centrifugation. Next, after repeated centrifugations, PBMCs were completely separated and precipitated. The detailed process is provided in Additional file 1. The isolated PBMC samples were preserved at − 80 °C until use.
Chromatin isolation by RNA purification (ChIRP) assays
The ChIRP assay was used to study the interaction between BTBD7_hsa_circ_0000563 and RNA on a genome-wide scale [17]. Four probes targeting the splice-junction sequence of BTBD7_hsa_circ_0000563 were designed and synthesized by Ribobio Co., Ltd. (Ribobio, China) (Probe_1 CATTTCTTGTGCAAGCATGT−/3bio/; Probe_2 TCATCCATTTCTTGTGCAAG−/3bio/; Probe_3 CCATTTCTTGTGCAAGCATG−/3bio/; Probe_4 ATCCATTTCTTGTGCAAGCA−/3bio/). The specificity and accuracy of the BTBD7_hsa_circ_0000563 probes were verified in our previous study [15].
Forty-five PBMC samples were mixed into one. RNA bound to BTBD7_hsa_circ_0000563 was harvested using ChIRP kit (Gzscbio, China) according to instructions of the manufacturer. First, after crosslinking with 1% formaldehyde at room temperature for 10 min, PBMCs were lysed using lysis buffer. Second, a total of 5 μl lysates were taken from the cleaned lysates as the input group for standby, and 500 μl lysates were taken from the cleaned lysates as the target group which were hybridized with 100 pmol biotinylated BTBD7_hsa_circ_0000563 specific probes at 37 °C for 4 h. Third, after 4-hour hybridization, streptavidin magnetic beads were hybridized with the mixture of lysates and probes at 37 °C for 30 min. Finally, the input group and eluted and purified products of the target group were applied to isolation of RNA. And the isolated RNA was used to construct the RNA sequencing library.
Library preparation and high-throughput sequencing
RNA quality control was conducted using NanoDrop 2000 (Thermo Fisher Scientific, USA). RNA libraries were constructed using NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (NEB, USA) according to the manufacturer’s instructions and were purified using AMPure beads (Beckman Coulter, USA). The quality and quantity of libraries were evaluated using Ailgent 4200 (Ailgent Technologies, USA). Sequencing was performed using illumina Novaseq 6000 (Illumina, USA) with 150 bp paired end reads. Then, raw data was obtained from sequencing. After removing adapter-containing and low-quality reads from the raw data, clean data was obtained. Quality control of the raw data and clean data were completed by FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and the summarized plots of quality control reports were created by MultiQC (v 1.11) [18].
Identification of differentially expressed genes
Clean reads were mapped to the human reference genome (UCSC hg38) via HISAT2 software (v 2.2.0) [19]. In order to quantify high-quality mapped reads, featureCounts tool of subread software (v 2.0.1) [20] was employed. Then, edgeR software (v 3.16) was used to normalize the original readcount and calculate the fold change, P-value, and false discovery rate (FDR) of RNAs between the target group and the negative control group. RNAs that met the criterion of |log2(FoldChange)| > 1 and FDR < 0.05 were defined as differentially expressed genes (DEGs).
Functional enrichment analysis
Gene Ontology (GO) functional enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses of upregulated DEGs and genes of clusters were performed using the clusterProfiler 4.0 package in R [21]. The ggplot2 package and the DOSE package [22] were employed to visualize enrichment results in R.
Interaction network and clusters construction and hub genes analysis
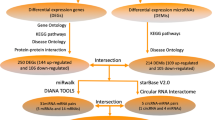
The protein–protein interaction (PPI) analysis of upregulated DEGs was performed using the STRING database (STRING v11.5) (https://string-db.org/). Simultaneously, the interaction data obtained from STRING was loaded into the Cytoscape software (v 3.9.1) to structure interaction networks. In the Cytoscape software, the cytoHubba tool [23] was used to calculate the candidate hub genes and the cytoCluster tool [24] was used to make cluster analysis. Thereafter, candidate target miRNAs of hub genes were predicted via the TargetScan database [25] (TargetScanHuman v8.0) (https://www.targetscan.org/vert_80/), the miRTargBase database [26] (miRTargBase v9.0) (https://mirtarbase.cuhk.edu.cn/~miRTarBase/miRTarBase_2022/php/index.php), and the StarBase database [27] (https://starbase.sysu.edu.cn/). In addition, Venn diagrams were generated by EVenn online (http://www.ehbio.com/test/venn/#/) [28] to figure out intersection of different gene sets.
Statistical analysis
Categorical variables are displayed as counts (percentages). Normally distributed continuous variables are presented as mean ± standard deviation, while non-normally distributed continuous variables are described as median (25th–75th interquartile range). All data analyses were performed using SPSS 21 software. Two-tailed P values < 0.05 were considered to be statistically significant.
Results
Quality control of the sequencing data
Through ChIRP-RNAseq assay, a total of 55,004,477 raw reads and 10,292,036 raw reads were obtain from the target group and the input group, respectively. After removing adapter-containing and low-quality reads, a total of 53,528,122 clean reads in the target group and 10,003,634 clean reads in the input group were left. The quality control results of raw data and clean data were visualized and presented in Fig. 1. The mean phred scores of each position of reads were almost higher than 30 in all groups (Fig. 1A and B). Moreover, Q30 percentage of input group was > 90% while Q30 percentage of the target group was < 90% (read1 = 61.94%, read2 = 87.16%) in raw data. Nevertheless, in clean data, Q30 percentages of all groups were > 93% (Fig. 1C and D), which indicated that the quality of clean data was high. Additionally, the average GC content of each sample was between 55 and 70% (Fig. 1E and F). Furthermore, properly mapped ratios of the target group and the input group reached 81.71 and 87.63%, respectively. To sum up, our results of ChIRP-RNAseq assay and relevant analysis were reliable.
The quality control of the sequencing data. A Mean quality score of each group in raw data. The Green area represents excellent quality. The yellow area represents medium quality. The red area represents poor quality. B Mean quality score of each group in clean data. C Per sequence quality score of each group in raw data. D Per sequence quality score of each group in clean data. E Per sequence GC content of each group in raw data. F Per sequence GC content of each group in clean data
Identification of mRNA bound to BTBD7_hsa_circ_0000563
After normalizing and comparing expression levels of mRNAs captured by probes between the target group and the input group, a total of 509 differentially expressed mRNAs were found, among which 221 mRNAs were upregulated and 288 mRNAs were downregulated in the target group (Fig. 2). Compared with the input group, mRNAs upregulated in the target group were identified as mRNAs directly combined with BTBD7_hsa_circ_0000563 in PBMC of CAD patients through the ChIRP-RNAseq assay, because the input group, as the control group, was used to eliminate the background factors and the unavoidable systematical errors. Therefore, subsequent analyses would be based on mRNAs upregulated in the target group. In addition, to be intuitionistic, top 30 upregulated mRNAs were presented in Table 2.
GO enrichment analysis for mRNA bound to BTBD7_hsa_circ_0000563
The GO enrichment analysis was used to reveal possible biological roles of those mRNAs directly bound to BTBD7_hsa_circ_0000563 from three aspects: biological process (BP), cellular component (CC), and molecular function (MF). Top 20 terms of BP, CC, and MF were shown in Fig. 3. Especially, the terms with top 3 adjusted P value of each part and their details were listed as follows: BP (Fig. 3A): SRP-dependent cotranslational protein targeting to membrane (GO:0006614, p.adjust = 9.69 × 10−19), cotranslational protein targeting to membrane (GO:0006613, p.adjust = 9.69 × 10−19), protein targeting to ER (GO:0045047, p.adjust = 1.00 × 10−18); CC (Fig. 3B): cytosolic ribosome (GO:0022626, p.adjust = 3.02 × 10−22), cytosolic part (GO:0044445, p.adjust = 6.17 × 10−18), ribosomal subunit (GO:0044391, p.adjust = 1.15 × 10−17); MF (Fig. 3C): structural constituent of ribosome (GO:0003735, p.adjust = 1.40 × 10−10), cadherin binding (GO:0045296, p.adjust = 0.041), mRNA 3′-UTR binding (GO: 0003730, p.adjust = 0.152).
The functional enrichment analysis for mRNAs bound to BTBD7_hsa_circ_0000563. A The top 20 biological process terms of GO analysis for mRNAs bound to BTBD7_hsa_circ_0000563. B The top 20 cellular component terms of GO analysis for mRNAs bound to BTBD7_hsa_circ_0000563. C The top 20 molecular function terms of GO analysis for mRNAs bound to BTBD7_hsa_circ_0000563. D The top 20 pathways of KEGG analysis for mRNAs bound to BTBD7_hsa_circ_0000563. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes
KEGG enrichment analysis for mRNA bound to BTBD7_hsa_circ_0000563
In order to further evaluate the biological significance of these mRNAs directly bound to BTBD7_hsa_circ_0000563, the KEGG enrichment analysis was conducted (Fig. 3D). According to the adjusted P value, the top five pathways were ribosome (hsa03010, p.adjust = 1.84 × 10−14), coronavirus disease - COVID-19 (hsa05171, p.adjust = 3.33 × 10−12), necroptosis (hsa04217, p.adjust = 0.019), African trypanosomiasis (hsa05143, p.adjust = 0.073), and FoxO signaling pathway (hsa04068, p.adjust = 0.076).
Interaction network construction and cluster analysis for mRNA bound to BTBD7_hsa_circ_0000563
The String database was used to calculate the interaction degree between mRNAs bound to BTBD7_hsa_circ_0000563. Before calculating, the minimum required interaction score was set to 0.40 and the max number of first shell interactors to show was set to no more than 5 interactors. As a result, a mRNA interaction network composed of 158 nodes and 660 edges was constructed using the Cytoscape software (Fig. 4A). The P value of the interaction network was < 1.00 × 10−16. From this network, we can find out that there was one large cluster with close internal relations, among which RPS3 has the highest node degree.
The interaction network and clusters of mRNAs bound to BTBD7_hsa_circ_0000563. A The interaction network of mRNAs bound to BTBD7_hsa_circ_0000563. Each node indicates one gene and each edge indicates one interaction between genes. The larger the size of the node is, the greater the degree value of the gene in the interaction network is. The redder the node color is, the greater the log2FC value of the gene is. The thicker the edge is, the more reliable the data support of the relationship between genes is. B Eight clusters with a P value < 0.05 extracted by the cytoCluster tool. FC, fold change
In order to further calculate main clusters of this interaction network, the cytoCluster tool in the Cytoscape software was used in following analyses. Through merging highly overlapping mRNA nodes (the overlap threshold set to default value 0.8), the ClusterONE algorithm of the cytoCluster tool output a total of 13 clusters that contained no less than three mRNAs and whose density was larger than 0.05, among which 8 clusters had a P value < 0.05 (Fig. 4B). The nodes of these clusters were attributed using yFiles radial layout algorithm, which showed that the 8 clusters might respectively centered on EIF4A1 (Cluster 1), RHOA (Cluster 2), SIRT1 (Cluster 3), GRN (Cluster 4 and 7), DYNLL2 (Cluster 5), HIST2H2AC (Cluster 6), IL6 (Cluster 8).
Functional enrichment analysis for clusters
Considering that these clusters separated from the whole interaction network can refine the relationship between genes, GO and KEGG functional enrichment analyses were performed again for the 8 clusters. The results of GO analyses revealed that the 8 clusters were most likely to participate in translational initiation (Cluster 1), post-translational protein modification (Cluster 2), glucose homeostasis (Cluster 3), neutrophil degranulation (Cluster 4 and 7), regulation of gene expression, epigenetic (Cluster 5), microtubule-based movement (Cluster 6), regulation of lymphocyte proliferation (Cluster 8), and other biological processes, respectively (Fig. 5A). Furthermore, the outcomes of KEGG analyses indicated that the 8 clusters were most likely to take part in ribosome (Cluster 1), Pathogenic Escherichia coli infection (Cluster 2), FoxO signaling pathway (Cluster 3), lysosome (Cluster 4 and 7), necroptosis (Cluster 5), Amyotrophic lateral sclerosis (Cluster 6), JAK-STAT signaling pathway (Cluster 8), and other pathways, respectively (Fig. 5B).
Identification of hub genes and target miRNA prediction for hub genes
According to the global-based method, five algorithms were performed respectively to calculate the top 10 genes, namely the Maximal Clique Centrality (MCC) algorithm, the Degree algorithm, the Edge Percolated Component (EPC) algorithm, the Closeness algorithm, and the Radiality algorithm of the cytoHubba tool. After intersecting the above predicted genes and the top 30 genes bound to BTBD7_hsa_circ_0000563, RPS3 and RPSA were identified as hub genes (Fig. 6A). The detailed information of the two hub genes were presented in Table 3.
Venn diagrams for screening hub genes and target miRNA. A The Venn diagram of five top 10 candidate hub genes sets respectively calculated by the MCC, Degree, EPC, Closeness, and Radiality algorithms and top 30 mRNA bound to BTBD7_hsa_circ_0000563. The intersection contains RPS3 and RPSA. B The Venn diagram of three candidate target miRNA sets of RPS3 respectively obtained from the TargertScan, miRTargBase, and StarBase databases. The intersection contains hsa-miR-493-5p. C The Venn diagram of three candidate target miRNA sets of RPSA respectively obtained from the TargertScan, miRTargBase, and StarBase databases. There is no intersection between these candidate sets. MCC, Maximal Clique Centrality; EPC, Edge Percolated Component; RPS3, ribosomal protein S3; RPSA, ribosomal protein SA
In order to obtain highly reliable target miRNA of hub genes, intersections of results from the TargertScan database, the miRTargBase database, and the StarBase database were identified as target miRNA of hub genes. Ultimately, hsa-miR-493-5p was identified as target miRNA of RPS3 (Fig. 6B), while there was no predicted target miRNA of RPSA that met the above requirements (Fig. 6C).
Discussion
In this investigation, through ChIRP-RNAseq assay, we revealed the profile of RNA bound to BTBD7_hsa_circ_0000563 in PBMCs of CAD patients. A total of 221 mRNAs were identified to bind directly to BTBD7_hsa_circ_0000563. GO and KEGG functional enrichment analyses suggested that these mRNAs may influence CAD via involving in translation and necroptosis processes. We further constructed an interaction network of mRNAs directly bound to BTBD7_hsa_circ_0000563, which showed that there may be a close relationship between these mRNAs. Through more in-depth analyses of this interaction network, we separated eight clusters with a P value < 0.05 and calculated that RPS3 and RPSA may be hub genes. Functional enrichment analyses of eight clusters indicated that genes of these clusters may participate in translation, regulation of gene expression, regulation of immune cells, and other processes. After synthesizing the results of multiple databases, hsa-miR-493-5p was identified as target miRNA of RPS3, while RPSA had none.
BTBD7_hsa_circ_0000563 was first reported in the circRNAs profile of animals by Memczak et al. [29]. Subsequently, scientists also discovered BTBD7_hsa_circ_0000563 in normal human tissues [30,31,32]. BTBD7_hsa_circ_0000563, a circRNA with 1268 bases, is located on chromosome 14. The gene code of BTBD7_hsa_circ_0000563 starts at position 93,760,203 and ends at position 93,762,503 according to the Circbank database (http://www.circbank.cn/). Available studies have corroborated that BTBD7_hsa_circ_0000563 can inhibit adipogenesis [33] and inactivate the p38 mitogen-activated protein kinase (MAPK) signaling pathway [34] by acting as competitive endogenous RNAs (ceRNAs). Moreover, based on the Circbank database, BTBD7_hsa_circ_0000563 has Open Reading Frame (ORF) and internal ribosome entry site (IRES), indicating its enormous potential for encoding polypeptides. The diagnostic value and RBPs of BTBD7_hsa_circ_0000563 in CAD were verified in our previous studies [14, 15]. Recently, multiple studies in various areas introduce ChIRP-seq technology to explore genes directly bound to noncoding RNA (ncRNA) [17, 35,36,37]. However, in mechanism study of CAD, gene profiles directly bound to ncRNA are rarely explored, especially that ChIRP-RNAseq, a highly sensitive and specific technology to reveal RNA binding RNAs, is also rarely applied. Therefore, in this investigation, we introduced ChIRP-RNAseq to explore the profile of mRNA bound to BTBD7_hsa_circ_0000563.
By means of GO and KEGG enrichment analyses, we found that most enriched cellular component and molecular function of these mRNAs bound to BTBD7_hsa_circ_0000563 were both constituent of ribosome and these mRNAs may mainly involve in translation and necroptosis processes. Moreover, the most enriched biological process with most significant P value of these mRNAs was SRP-dependent cotranslational protein targeting to membrane.
The signal recognition particle (SRP), a predominant and universally conserved protein delivery machine, can combine with ribosome and signal peptide, suspend translation and transport the free ribosome – nascent polypeptide chain to the target membrane, which is defined as the cotranslational translocation process [38]. Correct protein function depends on proper localization of proteins, and it is the cotranslational translocation process that is able to ensure that the ongoing protein translation complex can be deliver to the correct cellular or subcellular compartment [39]. Protein is the basis for maintaining normal life activities. It is accepted that the disorder of protein translation will lead to a series of cardiovascular diseases such as cardiac hypertrophy and failure, ischemic heart diseases, atherosclerosis, and hypertension [40, 41]. Therefore, BTBD7_hsa_circ_0000563 is likely to affect the protein translation by regulating the SRP-dependent cotranslation process, and ultimately interfere with the occurrence and development of CAD. In addition, the necroptosis pathway was significant enriched in our results as well. Necroptosis is a form of programmed necrotic cell death, having a significant impact on diseases, of which receptor-interacting serine-threonine kinase 3 (RIPK3) and mixed lineage kinase domain-like (MLKL) act as key mediators [42]. Karunakaran et al. [43] have confirmed that low-density lipoprotein can bring about atherosclerosis through increase RIPK3 and MLKL transcription and phosphorylation, which are two critical steps in the execution of necroptosis [42]. On this basis, they find that the necroptosis inhibitor Nec-1 can reduce atherosclerotic lesion size and markers of plaque instability, including necrotic core formation. Furthermore, Hu et al. [44] find that plasma RIPK3 levels of patients with CAD were significantly higher than those of controls and plasma RIPK3 levels increased linearly with the severity of CAD. According to the evidence above, BTBD7_hsa_circ_0000563 might be able to influence the initiation and progression of CAD via regulating the necroptosis pathway.
After constructing the interaction network of mRNAs bound to BTBD7_hsa_circ_0000563, interaction clusters and hub genes were identified. Although the integral interaction network can reflect the overall connection, the subdivided clusters can present more details of mRNAs bound to BTBD7_hsa_circ_0000563. For example, the functional enrichment analysis of clusters showed that, in addition to translation and necroptosis, other processes that mRNA bound to BTBD7_hsa_circ_0000563 might take part in, such as post-translational protein modification [45, 46], glucose homeostasis [47, 48], neutrophil and lymphocyte [49, 50], and FoxO signaling pathway [51], can make an impact on CAD or atherosclerosis as well.
RPS3 (Ribosomal Protein S3) and RPSA (Ribosomal Protein SA), hub genes of mRNAs bound to BTBD7_hsa_circ_0000563, are both genes encoding ribosomal proteins [52, 53]. In addition to being involved in translation as a component of ribosomes, they also have an undeniable impact on DNA repair and apoptosis [54,55,56], which are important pathogenesis of atherosclerosis [57, 58]. Furthermore, researchers reported that RPS3 is an essential subunit of nuclear factor-kappaB (NF-κB) and influences rapid cellular activation responses or target genes transcription of NF-κB [59, 60]. Pertschy et al. found that RPS3 can further interact with the NF-κB inhibitor IκBα, regulating the activity of the NF-κB signaling pathway [61]. Interestingly, mounting researchers have confirmed that regulating NF-κB signaling pathway, an important signaling pathway involved in immune and inflammatory reactions, can reduce atherosclerosis and protect coronary artery [62,63,64]. This makes RPS3 of great research value in CAD. In addition, another researcher believed that RPS3 can enhance cell stress and lead to cell apoptosis by activating c-Jun N-terminal kinase (JNK) [65]. The JNK signaling pathway had also been confirmed to be involved in the foam cell formation in atherogenesis [66]. With regard to RPSA, the protein encoded by RPSA is also known as 37-kDa laminin receptor precursor/67-kDa laminin receptor [53], playing an important role in cell adhesion and signaling transduction [67, 68]. Li et al. corroborated that RPSA can act as a cell surface receptor, binding to pigment epithelium-derived factor (PEDF) to inhibits angiogenesis by inducing endothelial cells apoptosis and reducing cell migration [69]. Of note, PEDF is not only proved to prevent atherosclerosis by inhibiting thrombosis or inhibiting inflammatory reaction of vascular endothelial cells caused by oxidative stress in vivo and in vitro experiments [70, 71], but also proved to be significantly overexpressed in CAD patients in the population, which may act as a protective response and biomarker of CAD [72,73,74]. Recently, small molecule inhibitors have gradually become a new hot topic in the treatment strategy of chronic diseases. Polyphenol (−)-epigallocatechin-3-O-gallate (EGCG), one such small molecule, has been reported to suppress lipopolysaccharide-mediated inflammation in human vascular endothelial cells by binding to cell surface receptor RPSA [75, 76]. This reveals the vascular protective value of RPSA.
As predicted target miRNA of PRS3, hsa-miR-493-5p has been proven to be closely related to the pathological mechanisms of various cancers [77, 78] and osteoporosis [79]. Simultaneously, hsa-miR-493-5p shows its ability to combine with circRNA and lncRNA to influence diseases development [79,80,81]. Although hsa-miR-493-5p has not been fully investigated in CAD, its research prospects are limitless.
To sum up, the above evidence implies that BTBD7_hsa_circ_0000563 together with mRNA bound to it may have an impact on initiation and progression of CAD by participating in translation and necroptosis mainly. The present study might provide innovative ideas for further research on the occurrence and development of CAD.
The present investigation had several limitations. First, the small sample size in our study was not enough to minimize the experimental bias and contingency of the results. The mRNAs directly bound to BTBD7_hsa_circ_0000563 should be continuously verified in a large-scale and multicentre cohort to increase the credibility of the results. Second, in our present study, the exploration of mRNAs bound to BTBD7_hsa_circ_0000563 was still in the qualitative stage. In order to further clarify the application value of these mRNAs in the real world, it is necessary to verify the potential expression of these mRNAs, especially the hub genes, in the patients and healthy populations in the future. Third, the mechanisms in which these mRNAs involved were only based on the bioinformatic analysis. Therefore, for ultimately benefiting clinical transformation, rigorous in vivo and in vitro experiments of mRNAs bound to BTBD7_hsa_circ_0000563 should be formulated and implemented to reveal their deeper mechanism and function in CAD.
Conclusions
In conclusion, a total of 221 mRNAs directly bound to BTBD7_hsa_circ_0000563 were identified in PBMCs of CAD patients in the present investigation through ChIRP-RNAseq. The functional enrichment analysis suggested that these mRNAs might mainly located in ribosome and take part in translation and necroptosis pathways. Based on the interaction network of these mRNAs, RPS3 and RPSA were presumed as hub genes and eight subclusters were analysed. And hsa-miR-493-5p was considered to be the target miRNA of RPS3. These findings may shed light on the possible potential mechanism of BTBD7_hsa_circ_0000563 influencing CAD, and provide innovative research ideas for the mechanism and diagnosis and treatment strategy of CAD.
Availability of data and materials
The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE239635 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE239635).
Reviewer access KEY: udodoqaozxwnlyn.
Abbreviations
- CAD:
-
Coronary artery disease
- PBMC:
-
Peripheral blood mononuclear cell
- GO:
-
Gene ontology
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- miRNAs:
-
microRNAs
- RBPs:
-
RNA binding proteins
- ChIRP-RNAseq:
-
Chromatin isolation by RNA purification followed by RNA sequencing
- FDR:
-
False discovery rate
- DEGs:
-
Differentially expressed genes
- PPI:
-
Protein–protein interaction
- BP:
-
Biological process
- CC:
-
Cellular component
- MF:
-
Molecular function
- MCC:
-
Maximal clique centrality
- EPC:
-
Edge percolated component
- MAPK:
-
Mitogen-activated protein kinase
- ceRNAs:
-
Competitive endogenous RNAs
- ORF:
-
Open Reading frame
- IRES:
-
Internal ribosome entry site
- RIPK3:
-
Receptor-interacting serine-threonine kinase 3
- MLKL:
-
Mixed lineage kinase domain-like
- SRP:
-
Signal recognition particle
- RPS3:
-
Ribosomal protein S3
- RPSA:
-
Ribosomal protein SA
- JNK:
-
C-Jun N-terminal kinase
- PEDF:
-
Pigment epithelium-derived factor
- EGCG:
-
(−)-epigallocatechin-3-O-gallate
References
Bauersachs R, Zeymer U, Brière JB, Marre C, Bowrin K, Huelsebeck M. Burden of coronary artery disease and peripheral artery disease: a literature review. Cardiovasc Ther. 2019;2019:8295054.
Doenst T, Haverich A, Serruys P, Bonow RO, Kappetein P, Falk V, Velazquez E, Diegeler A, Sigusch H. PCI and CABG for treating stable coronary artery disease: JACC review topic of the week. J Am Coll Cardiol. 2019;73(8):964–76.
Khera AV, Emdin CA, Drake I, Natarajan P, Bick AG, Cook NR, Chasman DI, Baber U, Mehran R, Rader DJ, et al. Genetic risk, adherence to a healthy lifestyle, and coronary disease. N Engl J Med. 2016;375(24):2349–58.
Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–91.
Zang J, Lu D, Xu A. The interaction of circRNAs and RNA binding proteins: an important part of circRNA maintenance and function. J Neurosci Res. 2020;98(1):87–97.
Gao X, Tian X, Huang Y, Fang R, Wang G, Li D, Zhang J, Li T, Yuan R. Role of circular RNA in myocardial ischemia and ageing-related diseases. Cytokine Growth Factor Rev. 2022;65:1–11.
Lu D, Ho ES, Mai H, Zang J, Liu Y, Li Y, Yang B, Ding Y, Tsang CK, Xu A. Identification of blood circular RNAs as potential biomarkers for acute ischemic stroke. Front Neurosci. 2020;14:81.
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X, Huang S. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25(8):981–4.
Meng H, Niu R, Huang C, Li J. Circular RNA as a novel biomarker and therapeutic target for HCC. Cells. 2022;11(12).
Yang Z, Huang C, Wen X, Liu W, Huang X, Li Y, Zang J, Weng Z, Lu D, Tsang CK, et al. Circular RNA circ-FoxO3 attenuates blood-brain barrier damage by inducing autophagy during ischemia/reperfusion. Mol Ther : J Am Soc Gene Ther. 2022;30(3):1275–87.
Li J, Sun D, Pu W, Wang J, Peng Y. Circular RNAs in Cancer: biogenesis, function, and clinical significance. Trends Cancer. 2020;6(4):319–36.
Cao Q, Guo Z, Du S, Ling H, Song C. Circular RNAs in the pathogenesis of atherosclerosis. Life Sci. 2020;255:117837.
Tong X, Dang X, Liu D, Wang N, Li M, Han J, Zhao J, Wang Y, Huang M, Yang Y, et al. Exosome-derived circ_0001785 delays atherogenesis through the ceRNA network mechanism of miR-513a-5p/TGFBR3. J Nanobiotechnol. 2023;21(1):362.
Chen JX, Hua L, Zhao CH, Jia QW, Zhang J, Yuan JX, Zhang YJ, Jin JL, Gu MF, Mao ZY, et al. Quantitative proteomics reveals the regulatory networks of circular RNA BTBD7_hsa_circ_0000563 in human coronary artery. J Clin Lab Anal. 2020;34(11):e23495.
Zhou H, Gan X, He S, Wang Y, Zhang S, Chen J, Zhou Y, Hou C, Hua L, Zhang Q, et al. Identification of circular RNA BTBD7_hsa_circ_0000563 as a novel biomarker for coronary artery disease and the functional discovery of BTBD7_hsa_circ_0000563 based on peripheral blood mononuclear cells: a case control study. Clin Proteomics. 2022;19(1):37.
Ryan TJ, Faxon DP, Gunnar RM, Kennedy JW, King SB 3rd, Loop FD, Peterson KL, Reeves TJ, Williams DO, Winters WL Jr, et al. Guidelines for percutaneous transluminal coronary angioplasty. A report of the American College of Cardiology/American Heart Association task force on assessment of diagnostic and therapeutic cardiovascular procedures (subcommittee on percutaneous transluminal coronary angioplasty). Circulation. 1988;78(2):486–502.
Xiu B, Chi Y, Liu L, Chi W, Zhang Q, Chen J, Guo R, Si J, Li L, Xue J, et al. LINC02273 drives breast cancer metastasis by epigenetically increasing AGR2 transcription. Mol Cancer. 2019;18(1):187.
Ewels P, Magnusson M, Lundin S, Käller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics (Oxford, England). 2016;32(19):3047–8.
Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37(8):907–15.
Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics (Oxford, England). 2014;30(7):923–30.
Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, Feng T, Zhou L, Tang W, Zhan L, et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (Cambridge (Mass)). 2021;2(3):100141.
Yu G, Wang LG, Yan GR, He QY. DOSE: an R/Bioconductor package for disease ontology semantic and enrichment analysis. Bioinformatics (Oxford, England). 2015;31(4):608–9.
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8(Suppl 4):S11.
Li M, Li D, Tang Y, Wu F, Wang J. CytoCluster: a Cytoscape plugin for cluster analysis and visualization of biological networks. Int J Mol Sci. 2017;18(9).
McGeary SE, Lin KS, Shi CY, Pham TM, Bisaria N, Kelley GM, Bartel DP. The biochemical basis of microRNA targeting efficacy. Science (New York, NY). 2019;366(6472).
Huang HY, Lin YC, Cui S, Huang Y, Tang Y, Xu J, Bao J, Li Y, Wen J, Zuo H, et al. miRTarBase update 2022: an informative resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. 2022;50(D1):D222-d230.
Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–7.
Chen T, Zhang H, Liu Y, Liu YX, Huang L. EVenn: easy to create repeatable and editable Venn diagrams and Venn networks online. J Genet Genom = Yi Chuan Xue Bao. 2021;48(9):863–6.
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–8.
Xu T, Wu J, Han P, Zhao Z, Song X. Circular RNA expression profiles and features in human tissues: a study using RNA-seq data. BMC Genomics. 2017;18(Suppl 6):680.
Maass PG, Glažar P, Memczak S, Dittmar G, Hollfinger I, Schreyer L, Sauer AV, Toka O, Aiuti A, Luft FC, et al. A map of human circular RNAs in clinically relevant tissues. J Mol Med (Berlin, Germany). 2017;95(11):1179–89.
Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58(5):870–85.
Ma Z, Chen Y, Qiu J, Guo R, Cai K, Zheng Y, Zhang Y, Li X, Zan L, Li A. CircBTBD7 inhibits adipogenesis via the miR-183/SMAD4 axis. Int J Biol Macromol. 2023;253(Pt 2):126740.
Bian Q, Chen B, Weng B, Chu D, Tang X, Yan S, Yin Y, Ran M. circBTBD7 promotes immature porcine Sertoli cell growth through modulating miR-24-3p/MAPK7 Axis to inactivate p38 MAPK signaling pathway. Int J Mol Sci. 2021;22(17).
Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44(4):667–78.
Luo C, Xiong S, Huang Y, Deng M, Zhang J, Chen J, Yang R, Ke X. The novel non-coding transcriptional regulator Gm18840 drives cardiomyocyte apoptosis in myocardial infarction post ischemia/reperfusion. Front Cell Dev Biol. 2021;9:615950.
Singh AP, Luo H, Matur M, Eshelman MA, Hamamoto K, Sharma A, Lesperance J, Huang S. A coordinated function of lncRNA HOTTIP and miRNA-196b underpinning leukemogenesis by targeting FAS signaling. Oncogene. 2022;41(5):718–31.
Akopian D, Shen K, Zhang X, Shan SO. Signal recognition particle: an essential protein-targeting machine. Annu Rev Biochem. 2013;82:693–721.
Cross BC, Sinning I, Luirink J, High S. Delivering proteins for export from the cytosol. Nat Rev Mol Cell Biol. 2009;10(4):255–64.
Minamino T, Komuro I, Kitakaze M. Endoplasmic reticulum stress as a therapeutic target in cardiovascular disease. Circ Res. 2010;107(9):1071–82.
Simpson LJ, Reader JS, Tzima E. Mechanical regulation of protein translation in the cardiovascular system. Front Cell Dev Biol. 2020;8:34.
Galluzzi L, Kepp O, Chan FK, Kroemer G. Necroptosis: mechanisms and relevance to disease. Annu Rev Pathol. 2017;12:103–30.
Karunakaran D, Geoffrion M, Wei L, Gan W, Richards L, Shangari P, DeKemp EM, Beanlands RA, Perisic L, Maegdefessel L, et al. Targeting macrophage necroptosis for therapeutic and diagnostic interventions in atherosclerosis. Sci Adv. 2016;2(7):e1600224.
Hu XM, Chen X, Pang HY, Liu HH, Chen PP, Shi JL, Tang S, Wu ZH, Zhang SY. Plasma levels of receptor interacting protein kinase-3 correlated with coronary artery disease. Chin Med J. 2019;132(12):1400–5.
Li P, Ge J, Li H. Lysine acetyltransferases and lysine deacetylases as targets for cardiovascular disease. Nat Rev Cardiol. 2020;17(2):96–115.
Tang M, Kalim S. Avenues for post-translational protein modification prevention and therapy. Mol Asp Med. 2022;86:101083.
Schmidt AM, Hori O, Chen JX, Li JF, Crandall J, Zhang J, Cao R, Yan SD, Brett J, Stern D. Advanced glycation endproducts interacting with their endothelial receptor induce expression of vascular cell adhesion molecule-1 (VCAM-1) in cultured human endothelial cells and in mice. A potential mechanism for the accelerated vasculopathy of diabetes. J Clin Invest. 1995;96(3):1395–403.
Verdoia M, Schaffer A, Cassetti E, Barbieri L, Di Ruocco MV, Perrone-Filardi P, Marino P, De Luca G. Glycosylated hemoglobin and coronary artery disease in patients without diabetes mellitus. Am J Prev Med. 2014;47(1):9–16.
Döring Y, Soehnlein O, Weber C. Neutrophil extracellular traps in atherosclerosis and Atherothrombosis. Circ Res. 2017;120(4):736–43.
Li J, Ley K. Lymphocyte migration into atherosclerotic plaque. Arterioscler Thromb Vasc Biol. 2015;35(1):40–9.
Tucka J, Yu H, Gray K, Figg N, Maguire J, Lam B, Bennett M, Littlewood T. Akt1 regulates vascular smooth muscle cell apoptosis through FoxO3a and Apaf1 and protects against arterial remodeling and atherosclerosis. Arterioscler Thromb Vasc Biol. 2014;34(11):2421–8.
Zhang XT, Tan YM, Tan YH. Isolation of a cDNA encoding human 40S ribosomal protein s3. Nucleic Acids Res. 1990;18(22):6689.
Ardini E, Pesole G, Tagliabue E, Magnifico A, Castronovo V, Sobel ME, Colnaghi MI, Ménard S. The 67-kDa laminin receptor originated from a ribosomal protein that acquired a dual function during evolution. Mol Biol Evol. 1998;15(8):1017–25.
Park YJ, Kim SH, Kim TS, Lee SM, Cho BS, Seo CI, Kim HD, Kim J. Ribosomal protein S3 associates with the TFIIH complex and positively regulates nucleotide excision repair. Cell Mol Life Sci : CMLS. 2021;78(7):3591–606.
Jang CY, Lee JY, Kim J. RpS3, a DNA repair endonuclease and ribosomal protein, is involved in apoptosis. FEBS Lett. 2004;560(1–3):81–5.
DiGiacomo V, Meruelo D. Looking into laminin receptor: critical discussion regarding the non-integrin 37/67-kDa laminin receptor/RPSA protein. Biol Rev Camb Philos Soc. 2016;91(2):288–310.
Durik M, Kavousi M, van der Pluijm I, Isaacs A, Cheng C, Verdonk K, Loot AE, Oeseburg H, Bhaggoe UM, Leijten F, et al. Nucleotide excision DNA repair is associated with age-related vascular dysfunction. Circulation. 2012;126(4):468–78.
Duan H, Zhang Q, Liu J, Li R, Wang D, Peng W, Wu C. Suppression of apoptosis in vascular endothelial cell, the promising way for natural medicines to treat atherosclerosis. Pharmacol Res. 2021;168:105599.
Wier EM, Neighoff J, Sun X, Fu K, Wan F. Identification of an N-terminal truncation of the NF-κB p65 subunit that specifically modulates ribosomal protein S3-dependent NF-κB gene expression. J Biol Chem. 2012;287(51):43019–29.
Wan F, Anderson DE, Barnitz RA, Snow A, Bidere N, Zheng L, Hegde V, Lam LT, Staudt LM, Levens D, et al. Ribosomal protein S3: a KH domain subunit in NF-kappaB complexes that mediates selective gene regulation. Cell. 2007;131(5):927–39.
Stanborough T, Niederhauser J, Koch B, Bergler H, Pertschy B. Ribosomal protein S3 interacts with the NF-κB inhibitor IκBα. FEBS Lett. 2014;588(5):659–64.
Farina FM, Serio S, Hall IF, Zani S, Cassanmagnago GA, Climent M, Civilini E, Condorelli G, Quintavalle M, Elia L. The epigenetic enzyme DOT1L orchestrates vascular smooth muscle cell-monocyte crosstalk and protects against atherosclerosis via the NF-κB pathway. Eur Heart J. 2022;43(43):4562–76.
Karunakaran D, Nguyen MA, Geoffrion M, Vreeken D, Lister Z, Cheng HS, Otte N, Essebier P, Wyatt H, Kandiah JW, et al. RIPK1 expression associates with inflammation in early atherosclerosis in humans and can be therapeutically silenced to reduce NF-κB activation and Atherogenesis in mice. Circulation. 2021;143(2):163–77.
Zheng Y, Li Y, Ran X, Wang D, Zheng X, Zhang M, Yu B, Sun Y, Wu J. Mettl14 mediates the inflammatory response of macrophages in atherosclerosis through the NF-κB/IL-6 signaling pathway. Cell Mol Life Sci : CMLS. 2022;79(6):311.
Jang CY, Kim HD, Kim J. Ribosomal protein S3 interacts with TRADD to induce apoptosis through caspase dependent JNK activation. Biochem Biophys Res Commun. 2012;421(3):474–8.
Ricci R, Sumara G, Sumara I, Rozenberg I, Kurrer M, Akhmedov A, Hersberger M, Eriksson U, Eberli FR, Becher B, et al. Requirement of JNK2 for scavenger receptor A-mediated foam cell formation in atherogenesis. Science (New York, NY). 2004;306(5701):1558–61.
Vilas-Boas F, Bagulho A, Tenente R, Teixeira VH, Martins G, da Costa G, Jerónimo A, Cordeiro C, Machuqueiro M, Real C. Hydrogen peroxide regulates cell adhesion through the redox sensor RPSA. Free Radic Biol Med. 2016;90:145–57.
Kim JE, Park H, Lee JE, Kang TC. Blockade of 67-kDa laminin receptor facilitates AQP4 Down-regulation and BBB disruption via ERK1/2-and p38 MAPK-mediated PI3K/AKT activations. Cells. 2020;9(7).
Bernard A, Gao-Li J, Franco CA, Bouceba T, Huet A, Li Z. Laminin receptor involvement in the anti-angiogenic activity of pigment epithelium-derived factor. J Biol Chem. 2009;284(16):10480–90.
Takenaka K, Yamagishi S, Matsui T, Nakamura K, Jinnouchi Y, Yoshida Y, Ueda S, Katsuki Y, Katsuda Y, Imaizumi T. Pigment epithelium-derived factor (PEDF) administration inhibits occlusive thrombus formation in rats: a possible participation of reduced intraplatelet PEDF in thrombosis of acute coronary syndromes. Atherosclerosis. 2008;197(1):25–33.
Yamagishi S, Inagaki Y, Nakamura K, Abe R, Shimizu T, Yoshimura A, Imaizumi T. Pigment epithelium-derived factor inhibits TNF-alpha-induced interleukin-6 expression in endothelial cells by suppressing NADPH oxidase-mediated reactive oxygen species generation. J Mol Cell Cardiol. 2004;37(2):497–506.
Wang F, Ma X, Zhou M, Pan X, Ni J, Gao M, Lu Z, Hang J, Bao Y, Jia W. Serum pigment epithelium-derived factor levels are independently correlated with the presence of coronary artery disease. Cardiovasc Diabetol. 2013;12:56.
Kajikawa M, Maruhashi T, Iwamoto Y, Iwamoto A, Oda N, Kishimoto S, Matsui S, Aibara Y, Hidaka T, Kihara Y, et al. Circulating level of pigment epithelium-derived factor is associated with vascular function and structure: a cross-sectional study. Int J Cardiol. 2016;225:91–5.
Tahara N, Yamagishi S, Tahara A, Nitta Y, Kodama N, Mizoguchi M, Mohar D, Ishibashi M, Hayabuchi N, Imaizumi T. Serum level of pigment epithelium-derived factor is a marker of atherosclerosis in humans. Atherosclerosis. 2011;219(1):311–5.
Li J, Ye L, Wang X, Liu J, Wang Y, Zhou Y, Ho W. (−)-epigallocatechin gallate inhibits endotoxin-induced expression of inflammatory cytokines in human cerebral microvascular endothelial cells. J Neuroinflammation. 2012;9:161.
Zidane N, Ould-Abeih MB, Petit-Topin I, Bedouelle H. The folded and disordered domains of human ribosomal protein SA have both idiosyncratic and shared functions as membrane receptors. Biosci Rep. 2012;33(1):113–24.
Wang G, Fang X, Han M, Wang X, Huang Q. MicroRNA-493-5p promotes apoptosis and suppresses proliferation and invasion in liver cancer cells by targeting VAMP2. Int J Mol Med. 2018;41(3):1740–8.
Cui FC, Chen Y, Wu XY, Hu M, Qin WS. MicroRNA-493-5p suppresses colorectal cancer progression via the PI3K-Akt-FoxO3a signaling pathway. Eur Rev Med Pharmacol Sci. 2020;24(8):4212–23.
Xue J, Liu L, Liu H, Li Z. LncRNA SNHG14 activates autophagy via regulating miR-493-5p/Mef2c axis to alleviate osteoporosis progression. Commun Biol. 2023;6(1):1120.
Liu Z, Wang Y, Ding Y. Circular RNA circPRKDC promotes tumorigenesis of gastric cancer via modulating insulin receptor substrate 2 (IRS2) and mediating microRNA-493-5p. Bioengineered. 2021;12(1):7631–43.
Zhang D, Yang X, Luo Q, Fu D, Li H, Zhang P, Tie C. Circular RNA CSPP1 motivates renal cell carcinoma carcinogenesis and the Warburg effect by targeting RAC1 through microRNA-493-5p. Acta Biochim Pol. 2023;70(3):693–701.
Acknowledgements
This study received support from the National Natural Science Foundation of China (grant 81970302), the Science and Technique Planed Projects of Wuzhong District (grant 202103) and the Priority Academic Development Programme of the Jiangsu Higher Education Institutions.
Funding
This study received support from the National Natural Science Foundation of China (No. 81970302), the Science and Technique Planed Projects of Wuzhong District (No. 202103) and the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Author information
Authors and Affiliations
Contributions
As guarantors, Enzhi Jia, Jun Liu, and Zhengxian Tao conceived the study. Hanxiao Zhou wrote the manuscript. Hanxiao Zhou, Ning Guo and Qian Zhang enrolled participants and completed the experiments. Yahong Fu, Qiaowei Jia, and Xiongkang Gan performed the analysis and collected the data. Yanjun Wang, Shu He, and Chengcheng Li coordinated the study. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The methods were performed according to the approved guidelines, and all experimental protocols used were approved by the ethics committee of the First Affiliated Hospital of Nanjing Medical University. Written informed consent was obtained from all subjects or their families.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Guo, N., Zhou, H., Zhang, Q. et al. Exploration and bioinformatic prediction for profile of mRNA bound to circular RNA BTBD7_hsa_circ_0000563 in coronary artery disease. BMC Cardiovasc Disord 24, 71 (2024). https://doi.org/10.1186/s12872-024-03711-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12872-024-03711-7