Abstract
Background
Coronary atherosclerosis (CA) is the most common type of atherosclerosis. However, the inherent pathogenesis and mechanisms of CA are unclear, and the relationship with ferroptosis-related genes (FRGs) has not been reported. The purpose of this study was to use bioinformatics techniques to evaluate potential therapeutic targets for CA.Please provide the given name for author “Dingshun”.Please provide the given name for author “Dingshun”.
Methods
First, the GSE132651 dataset was acquired from the Gene Expression Omnibus database. Gene Ontology enrichment analysis, Kyoto Encyclopedia of Genes and Genomes enrichment analysis, and Protein–Protein interaction network were successively conducted. Next, overlapping genes between hub genes and CA genes were found. FRGs were found when comparing the CA group with the normal group. The correlation between overlapping genes and FRGs was further analyzed. At last, we performed Elisa to validate the expression of these genes in human blood specimens. Mice aortic tissues were used for western blot to detect the expression of proteins.
Results
Based on the GSE132651 dataset, 102 differentially expressed genes were identified. Five overlapping genes between hub genes and CA genes were found (CCNA2, RRM2, PBK, PCNA, CDK1). TFRC and GPX4 were found to be FRGs. TFRC was positively correlated with CCNA2, PBK, PCNA, CDK1, RRM2, with CDK1 being the strongest correlation. GPX4 was negatively correlated with these genes, among which CCNA2 was the strongest correlation. The ELISA results showed that CCNA2, CDK1, and TFRC expression were markedly increased in serum of the CA samples compared with controls, while GPX4 expression was markedly decreased in the CA samples. The western blot results show that GPX4 expression was lower in the model group, TFRC, CDK1, and CCNA2 protein expression were high in the model group.
Conclusions
Ferroptosis-related genes GPX4 and TFRC were closely correlated with the identified overlapping genes CCNA2 and CDK1, which may serve as targeted therapies for the treatment of CA.
Similar content being viewed by others
Introduction
According to the American Heart Association's Heart Disease and Stroke Statistics (2020 Edition), cardiovascular disease (CVD) is a growing public health concern worldwide, nearly 18 million people die from CVD each year, accounting for more than 30% of all deaths worldwide [1]. Atherosclerosis is the pathological basis of CVD, of which coronary atherosclerosis (CA) has the highest incidence. Controlling CA in a timely and effective manner is critical in the prevention and treatment of CVD. However, the precise pathogenic mechanism underlying CA is still unspecified. Acquiring a fundamental understanding of the molecular and pathological mechanisms may contribute to future therapeutic targets for CA.
The current pathogenesis of CA includes theories of the inflammatory response, oxidative stress, disturbance of lipid energy metabolism, and ferroptosis, among which ferroptosis is a current research hotspot. Ferroptosis is a relatively new type of death caused by a massive accumulation of reactive oxygen species because of lipid peroxidation [2, 3]. Distinguished from apoptosis, necrosis, autophagy, and scorching cell death, ferroptosis is morphologically, biochemically, and genetically distinct. Morphological manifestations include shrinking of mitochondria, swelling of cytoplasm and organelles, rupture of the plasma membrane, and formation of double-membrane vesicles. In molecular biology, ferroptosis is manifested by a significant decrease in glutathione peroxidase 4 (GPX4) levels, reduced glutamate-cystine reverse transport system (XC-) activity, and inhibition of expression [4]. Ferroptosis is a complicated system regulated by multiple mechanisms that are a significant therapeutic target in tumors, liver diseases, neurodegenerative diseases, endocrine, cardiovascular, and other diseases [5,6,7,8,9].
The Gene Expression Omnibus (GEO) database contains gene profiles primarily derived from DNA molecular techniques [10]. This study aimed to screen potential DEGs of CA, genes associated with ferroptosis, and their mechanisms of action, using bioinformatics methods. The GEO database was used to download raw data from microarray analyses performed on Blood Outgrowth Endothelial Cells (BOEC) of abnormal (ABNL) coronary endothelial function samples and normal Blood Outgrowth Endothelial Cells (BOEC) samples. The STRING and DAVID databases were used for functional enrichment analyses and the construction of the protein–protein interaction (PPI) network of DEGs, respectively. Then, overlapping genes between hub genes and CA genes were found. Finally, we explored the correlation between FRGs and overlapping genes, which contributes to furthering our understanding of CA pathophysiological mechanisms and treatment approaches.
Materials and methods
Gene microarray data acquisition
The GSE132651 gene dataset was selected from the GEO database (https//www.ncbi.nlm.nih.gov/geo/) for this analysis and contained gene expression data for 13 BOEC of abnormal coronary endothelial function samples and 6 normal BOEC. The data sets were normalized and log2ized uniformly. To normalize the data, the normalized quantiles function from the R software preprocess Core package had been used.
Screening for DEGs
DEGs of mRNA was obtained by using the Limma package of R software (version: 3.40.2). “P < 0.05 and log2 (fold change) > 1.5 or log2 (fold change) < − 1.5” was used as the threshold for screening mRNA differential expression. The R package ggplot2 was used to create the box line plots; expression heat maps were presented by the R package pheatmap [11].
Screening of CA genes
The GeneCards (https://www.genecards.org/) database was used to search for disease genes using the English term “coronary atherosclerosis” for CA.
GO functional analysis and KEGG pathway enrichment analysis of DEGs
The DAVID database (http://david.abcc.ncifcrf.gov) was used for GO function analysis and KEGG pathway enrichment of DEGs.
Construction of PPI network and screening hub genes
The DEGs were imported into the STRING database, and the species “homo sapiens” was selected with the lowest interaction score with maximum confidence of 0.15 and then obtained the PPI network map of the differential genes. The PPI network map was uploaded into Cytoscape 3.6.0 software, and the top 20 most significant hub genes were obtained using the MCC algorithm in the cytoHubba plug-in.
Overlapping genes between hub genes and CA genes
The hub gene and CA genes were imported into Venny2.1 (http://bioinfogp.cnb.csic.es/tools/venny/index.html) to obtain overlapping genes. These overlapping genes were thought to be key genes in the treatment of CA.
Identification of FRGs
FRGs were derived from a systematic analysis of the abnormalities and functions of ferroptosis in disease by Ze-Xian Liu et al. [12]. The box plot is implemented by the R software package ggplot2; The R package was implemented by R foundation for statistical computing (2020) version 4.0.3.
Correlation analysis of overlapping genes and FRGs
Screening for differences in FRGs in CA was performed, followed by correlation analysis between FRGs and overlapping genes. Box-line plots were constructed by the R package ggplot2; two-gene correlation plots were implemented by the R package ggstatsplot [13,14,15].
ELISA
The research had been performed following the Declaration of Helsinki. The ethics committee of the First Affiliated Hospital of Hainan Medical University gave its approval to this experiment. Patients with CA and normal people signed an informed consent form informing them that 2 ml of blood would be taken. A total of 32 blood specimens from the control group and 34 blood specimens from the CA group were finally collected. Serum specimens were collected in anticoagulation tubes and blood samples were centrifuged at 4 °C for 10 min at 3000 rpm. The ELISA kits (GPX4, TFRC, CDK1, and CCNA2) were used following the instructions provided by the manufacturer (MEIMIAN, Jiangsu Biological Industrial Co., Ltd., China). The catalog numbers of these four ELISA kits are MM-60328H1, MM-1504H1, MM-12973H1, MM-60616H1.
Animal experiment to verify the efficacy
Twenty male C57BL/6 mice, 8–10 weeks old, weighing 18–22 g, were purchased from Changsha Tianqin Biotechnology Co. Mice were housed at room temperature of 20–24 °C, relative humidity of 50–60%, and light and dark cycles (12 h/12 h). Common and high-fat diets (1% cholesterol, 5% lard, 94% standard diet) purchased from Guangdong Medical Laboratory Animal Center. After 1 week of acclimatization feeding, 10 of 20 mice were randomly selected as the normal group and given normal chow; 10 mice were given high-fat chow as the model group for 6 months. The experiments were approved by the Institutional Animal Care and Use Committee of Hainan Medical University. After the end of modeling, the mice were anesthetized by pentobarbital sodium (200 mg/kg), then the aorta was dissected (operated on ice). Small pieces of the aorta were cut out, the aorta were lysed by adding RIPA lysis solution, total protein was extracted, and the operation was performed according to the instructions of the BCA kit for the determination of protein concentration. Prepare the electrophoresis gel required for WB, perform sample addition, electrophoresis, electrotransfer membrane instrument transfer, 5% skim milk room temperature closure. The PDVF membrane was cropped prior to incubation of the primary antibody, and all subsequent developments are cropped images, then add GPX4, TFRC, CDK1, CCNA2 primary antibody (1:1000, Proteintech Group Inc., Wuhan Sanying, Wuhan, China) for incubation, shaking at 4 °C overnight, recovering primary antibody, washing the membrane 3 times with TBST, dressing secondary antibody(Biyuntian Co. Shanghai, China) at room temperature, washing the membrane again 3 times, developing a solution for luminescence development, and exposure imaging with gel imager. The strips were analyzed for grayscale using Image J software and processed for semi-quantitative analysis.
Statistical analysis
The data is expressed in the form of the mean ± SD. Prism 8.0 was used for statistical analysis. The unpaired t-test was used to make group comparisons. Wilcox test for comparison of two independent samples where the population does not follow a normal distribution. The correlations between quantitative variables that did not follow a normal distribution were examined by using Spearman's correlation analysis. A p-value < 0.05 was considered statistically significant.
Results
Overall study protocol
The general flow chart of this study is displayed in Fig. 1.
Analysis of differential expression
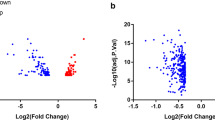
This study included 13 patients with ABNL and 6 normal subjects. Microarray data of the GSE132651 dataset were standardized (Fig. 2A). After setting the cutoff at FDR < 0.05, and |log2 (FC)|> 1.5, 102 DEGs were discovered. Finally, 48 up-regulated genes and 54 down-regulated genes were identified (Fig. 2B)(Additional file 1: Table S1). The heat map depicts the 48 up-regulated genes and 50 down-regulated genes with the greatest differential alteration (Fig. 2C).
DEGs between abnormal (ABNL) coronary endothelial function and normal groups. A Box line plot after data normalization, different colors represent different data sets, rows represent samples, and represent gene expression values in the samples. B Differential gene volcano plot: volcano plot using Fold change and corrected p-values. The red dots in the figure indicate genes that are differentially up-regulated, while the blue dots represent genes that are differentially down-regulated. C Differential gene expression heat map: different colors represent expression trends in different tissues. Showing the 48 most differentially altered up-regulated genes and 50 down-regulated genes
GO functional enrichment analysis and KEGG pathway analysis of DEGs
The DAVID database was used to perform the GO analysis. Enriched GO terms were classified into three categories: biological process (BP), cellular component (CC), and molecular function (MF). As shown in Table 1 and Fig. 3A, in BP analysis, DEGs mainly enriched in the “positive regulation of cell proliferation”, “signal transduction”. CC analysis indicated that the DEGs were mainly enriched in the “proteinaceous extracellular matrix” “extracellular space” (Table 2 and Fig. 3A). In terms of MF, DEGs were most enriched in “endopeptidase inhibitor activity”, “endopeptidase regulator activity” and “peptidase inhibitor activity” (Table 3 and Fig. 3A). After uploading the 102 DEGs to the DAVID database, KEGG analysis was performed to investigate the pathways of these 102 DEGs. KEGG analysis of DEGs revealed that they were primarily enriched in “MAPK signaling pathway”, “HLTV-1 infection”, “osteoclast differentiation” and “Cytokine-cytokine receptor interaction signaling pathway” (Table 4 and Fig. 3B).
DEG distribution in CA for GO and KEGG enrichment analysis. A GO enrichment analysis of DEGs. Green denotes biological processes (BP), orange denotes cellular components (CC), and purple denotes molecular function (MF). B KEGG enrichment analysis of DEGs. The different colors represent the significance of the differential enrichment results, and the size of the circle represents the number of enriched genes. Enrichment results with p < 0.05 were considered as enrichment to significant pathways
PPI network analysis of DEGs and obtained the overlapping genes
The PPI network was obtained by importing the differentially expressed genes into STRING, which consisted of 102 nodal proteins and 679 interactions (Fig. 4A). The PPI network was loaded into the Cytoscape 3.6.0 software, and the MCC algorithm in cytoHubba plug-in was used to identify the top 20 most significant hub genes, namely HJURP, OIP5, SHCBP1, CCNA2, CEP55, PTTG1, TRIP13, MELK, RRM2, PBK, PCNA, MAD2L1, CKS1B, HMGB2, NCAPG2, CDK1, RFC3, SPC25, DEPDC1, RAD51AP1 (Fig. 4B). The overlapping genes between hub genes and CA genes were then validated using a Venn diagram (Fig. 5). Finally, the five overlapping genes were obtained: CCNA2, RRM2, PBK, PCNA, CDK1.
Differential expression of FRGs in CA
A comparison of FRGs was performed in CA and normal groups, and significant differences were found for GPX4 and TFRC (Fig. 6A). Among them, GPX4 was highly expressed in the normal group and low expressed in the ABNL group. TFRC was low expressed in the normal group and highly expressed in the ABNL group (Fig. 6B and C).
Identification of differentially expressed ferroptosis-related genes. A Results of ferroptosis-associated genes analysis. Box line plot: expression distribution of ferroptosis-related genes in the normal group (G1 group) and abnormal (ABNL) coronary endothelial function group (G2 group), where different colors represent different groups, where the horizontal axis represents different ferroptosis molecules and the vertical axis represents the gene expression. The vertical coordinate represents *p < 0.05, **p < 0.01, ***p < 0.001, and the asterisk represents the degree of significance (*p). Two groups of samples were significant by the Wilcoxon test. B, C Expression distribution of TRFC and GPX4 genes in different groups. The horizontal axis represents different groups of samples, whereas the vertical axis represents the gene's expression distribution. The G1 group represents the control group and the G2 group represents the ABNL group
Overlapping genes and FRGs correlation
To further understand the relationship between overlapping genes and FRGs, two sets of gene correlation analyses were conducted and found that TFRC was positively correlated with CCNA2, PBK, PCNA, CDK1, RRM2, with CDK1 being the strongest correlation (P = 0.000537831). GPX4 was negatively correlated with CCNA2, PBK, PCNA, CDK1, RRM2, among which CCNA2 was the strongest correlation (P = 0.002340535) (Fig. 7).
The correlation between overlapping genes and FRGs. Multi-gene correlation: heat map of multiple genes and multiple genes (or one) correlation, both horizontal and vertical coordinates represent genes, where different colors depict correlation coefficients (red represents positive correlation and blue reflects negative correlation), darker colors represent stronger correlation between the two, *p < 0.05, **p < 0.01, and asterisks represent the degree of importance (*p)
Validation of gene expression
Expression of overlapping genes and FRGs (CCNA2, CDK1, GPX4, TFRC) were verified using ELISA in normal and CA patients. The Elisa results showed that the levels of CCNA2, CDK1, and TFRC in the CA group were significantly increased. GPX4 is lowly expressed in patients with CA and highly expressed in controls (Fig. 8).
Comparison of GPX4, TFRC, CDK1, and CCNA2 in aortic tissue of mice
As shown in Fig. 9, the protein expressions of GPX4, TFRC, CDK1, and CCNA2 in aortic tissue were statistically different in both control and CA model groups of mice (P < 0.01). Compared with the control group, GPX4 expression was lower in the model group (P = 0.0003). TFRC, CDK1, and CCNA2 protein expression were high and statistically different in the model group compared with the control group (P < 0.0001; P = 0.0009; P = 0.0003).
GPX4, TFRC, CCNA2, and CDK1 expression in aortic tissue of mice. GPX4, TFRC, CCNA2, and CDK1 in protein levels were detected by Western blots (A). The grayscale values of the bands were quantified by using Image J software (B). The data shown are the mean ± SD of at least three independent experiments. **p < 0.01 versus Control group
Discussion
Coronary artery disease, caused by CA, is the major cause of mortality in patients with CVD. It is a common chronic disease that, if left untreated, has a serious impact on a patient's life safety and quality of life. Therefore, the search for vulnerable disease markers to reveal the initial dysregulation and underlying mechanisms of CA is a problem that needs to be resolved in current research. In this investigation, 102 DEGs were extracted from the dataset GSE132651. 48 up-regulated genes and 54 down-regulated genes were identified in this study. Further GO enrichment analysis revealed that DEGs may influence the response to positive regulation of cell proliferation, signal transduction, extracellular space, endopeptidase inhibitor activity, endopeptidase regulator activity, and peptidase inhibitor activity. KEGG enrichment analysis of DEGs revealed that they were mainly enriched in MAPK signaling pathway, HLTV-1 infection, osteoclast differentiation, and Cytokine-cytokine receptor interaction signaling pathway. After constructing the PPI network, we selected twenty hub genes. Furthermore, five overlapping genes were observed between hub genes and CA genes, including CCNA2, RRM2, PBK, PCNA, CDK1.
CCNA2 is located on human chromosome 4, region q27, with a gene length of 7489 bp. This gene encodes a protein that belongs to a family of highly conserved cell cycle proteins and is expressed in almost all human tissues. It plays a major role in the G1/S and G2/M phases in cell cycles [16]. CCNA2 is over-expressed in a variety of human malignancies, suggesting its potential role in cancer transformation and progression. The study found that CCNA2 expression was significantly high in carcinoma tissues than in normal controls, and CCNA2 was considered as a potential immunotherapy marker in breast cancer [17]. Meanwhile, some studies have shown that CCNA2 can be used for the early detection and prognosis of colon cancer [18]. The protein CCNA2 is thought to be essential in epithelial-mesenchymal transition and metastasis [19, 20]. At present, the relevance of CCNA2 to cardiovascular diseases, especially coronary artery disease, has not yet been reported.
RRM2 is a ribonucleotide reductase subunit (RR), the only rate-limiting enzyme for intracellular DNA synthesis, and it is essential for nucleic acid metabolism. The reduction of nucleoside diphosphates to deoxyribonucleotide reductase is catalyzed by RR. RR catalyzes the rate-limiting step in the biosynthesis of deoxyribonucleoside triphosphate, the reduction of nucleoside diphosphate to deoxyribonucleoside diphosphate [21]. RRM2 is involved in the biosynthesis of deoxyribonucleoside triphosphate (dNTP) and is an indispensable enzyme in the process of DNA synthesis and repair. It is a key enzyme in DNA synthesis and repair. In recent years, RRM2 has gradually been discovered to be overexpressed in cancers, including breast cancer, non-small cell lung cancer, bladder cancer, and colorectal cancer [22,23,24,25,26,27,28].
PBK also known as lymphokine-activated killer T-cell-derived protein kinase (TOPK), has a significant impact on mitotic regulation. PBK is highly expressed in a variety of tumors, including lung, colorectal, ovarian, and prostate cancers, and patients with tumors accompanied by high PBK expression often have a worse prognosis [29, 30]. PBK serves an essential regulatory role in tumor cell proliferation, invasion, and metastasis. Knockdown of PBK leads to G2/M cell cycle arrest and apoptosis [31]. Yang et al. showed that PBK is extensively expressed in hepatocellular carcinoma cells and promotes hepatocellular carcinoma cell invasion and migration via the ETV4-uPAR signaling pathway [32, 33].
PCNA is a non-histone protein with a molecular weight of 36kD found in the nucleus and is an auxiliary protein of DNA polymerase [34]. It is rarely found in quiescent cells or is only synthesized and expressed in proliferating cells and can be used to evaluate the proliferative status and activity of cells [35, 36]. PCNA can be regarded as a marker of smooth muscle cell (SMC) proliferation, mainly assisting in the synthesis of DNA guide and follower strands during cell division. Enhanced expression of PNCA can be an important factor influencing carotid atherosclerotic plaque formation [37].
Cell cycle protein-dependent kinase 1 (CDK1) belongs to the serine/threonine kinase family of proteins and is essential for driving each phase of the cell cycle [38]. Several previous studies have demonstrated that CDK1 is crucial in both cardiac biological processes and the pathology of cardiovascular diseases. CDK1 regulates cell cycle processes, which facilitate the proliferation and migration of vascular smooth muscle cells [39]. However, the above-mentioned genes are less studied in cardiovascular diseases and still need further clinical validation.
Atherosclerosis is caused by the accumulation of lipid-rich macrophages in the subendothelial region of the vascular system. Macrophage accumulation in the subendothelial area of the arterial wall, and the lipid-rich cells promote an inflammatory response and leads to a variety of fatal pathological consequences [40,41,42]. Ferroptosis has been connected to a range of cardiovascular diseases, including heart failure, ischemia–reperfusion injury, and adriamycin cardiotoxicity [43,44,45,46]. The accumulation of iron ions in atherosclerotic plaques, along with the occurrence of intraplaque hemorrhage can promote the rapid development of atherosclerotic plaques and lead to serious cardiovascular events. According to a study conducted by Bai et al., the ferroptosis inhibitor ferrostatin-1 (Fer-1) could inhibit iron accumulation, lipid peroxidation, and lessen AS lesion in HFD-fed ApoE−/− mice [47]. Therefore, ferroptosis may be engaged in the pathological process of AS and may be a potential treatment for the disease. A comparison of ferroptosis differences gene was performed in CA and normal groups, and significant differences were found for GPX4 and TFR in this study.
Glutathione peroxidase 4 (GPX4) can be used as a reference marker for determining cellular iron death. GPX4 protein has the function of scavenging lipid peroxides, and inactivation of GPX4 leads to disruption of oxidative homeostasis, disruption of membrane structure by lipid peroxides, and initiation of iron death. GPX4 has been reported to be closely associated with tumors and tumor resistance [48]. The role of GPX4 in cardiovascular disease is also gaining attention. Our study results show that GPX4 was highly expressed in the normal group and low expressed in the abnormal (ABNL) coronary endothelial function group. In mouse aortic endothelial cells (MAECs), ox-LDL causes mitochondrial damage and decreases the expression of SLC7A11 and GPX4, whereas Fer-1 can suppress ox-LDL induced lipid peroxidation and endothelial dysfunction [47].
TFRC, as transferrin on the cell membrane, can bind extracellular iron ions intracellularly, therefore, the expression of TFRC can indirectly reflect the degree of intracellular iron loading [49, 50]. TFRC knockdown suppresses elastin-induced ferroptosis, according to previous research Gao et al. discovered that inhibiting TFRC prevents ferroptosis caused by amino acid/cysteine deprivation [51]. The study shows that Ferrostatin-1, an iron death inhibitor, significantly increased the gene and protein expression of iron death inhibitors GPX4 and SLC7A11 and significantly decreased the iron ion receptor TFRC in the cell injury model group after its intervention [52]. Studies have shown that YAP pathway activation in mesothelioma tissue enhances iron death sensitivity through ACSL4 and TFRC, and YAP, ACSL4, and TFRC are expected to be biological markers for clinical prediction of iron death sensitivity in mesothelioma [50]. In our study, we found that TFRC was low expressed in the normal group and highly expressed in the ABNL group.
To verify the expression of the four genes, we used ELISA experiments on coronary artery disease and normal subjects, respectively, and found that GPX4 was low expressed in the coronary artery disease group and highly expressed in the control group, while TFRC, CCNA2, and CDK1 were highly expressed in the coronary artery disease group and low expressed in the control group. The same trend was also obtained in animal experiments, with low expression in the GPX4 group and high expression in the control group in CA model mice, and the opposite trend for TFRC, CCNA2, and CDK1. This is consistent with our predicted results. Recent studies have shown that in the treatment of CA, a combination strategy of statins and non-statin drugs (ezetimibe or everolimus) has been shown to promote the regression of coronary atherosclerosis and improve the prognosis of patients at moderate to high cardiovascular risk [53,54,55]. Ferroptosis as a new model of cell death, drugs targeting ferroptosis are bound to provide a new therapeutic strategy for CA treatment. Studies have shown that GPX4 activators can reduce lipid peroxide production and inhibit inflammation, thus GPX4 activators are expected to be potential therapeutic agents for the treatment of CA [47].
Because most of the genes identified in this study have never been linked to CA, additional large-scale validation studies and molecular mechanisms are needed to investigate their role. Furthermore, this study used only one data set, and additional data sets will be necessary for future studies to verify these findings.
Conclusions
Ferroptosis-related genes GPX4 and TFRC were closely correlated with the identified overlapping genes CCNA2 and CDK1, which may serve as targeted therapies for the treatment of CA.
Abbreviations
- CA:
-
Coronary atherosclerosis
- FRGs:
-
Ferroptosis-related genes
- DEGs:
-
Differentially expressed genes
- GO:
-
Gene ontology
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- PPI:
-
Protein–Protein interaction
- CVD:
-
Cardiovascular disease
- BOEC:
-
Blood outgrowth endothelial cells
- ABNL:
-
Abnormal coronary endothelial function samples
- BP:
-
Biological process
- CC:
-
Cellular component
- MF:
-
Molecular function
References
Virani SS, Alonso A, Benjamin EJ, Bittencourt MS, Callaway CW, Carson AP, Chamberlain AM, Chang AR, Cheng S, Delling FN, et al. Heart disease and stroke statistics-2020 update: a report from the American heart association. Circulation. 2020;141(9):e139–596.
Hirschhorn T, Stockwell BR. The development of the concept of ferroptosis. Free Radic Biol Med. 2019;133:130–43.
Hu H, Chen Y, Jing L, Zhai C, Shen L. The link between ferroptosis and cardiovascular diseases: a novel target for treatment. Front Cardiovasc Med. 2021;8: 710963.
Sönmez Aydın F, Hukkamlı B, Budak H. Coaction of hepatic thioredoxin and glutathione systems in iron overload-induced oxidative stress. J Biochem Mol Toxicol. 2021;35(4): e22704.
Wang Y, Wei Z, Pan K, Li J, Chen Q. The function and mechanism of ferroptosis in cancer. Apoptosis. 2020;25(11–12):786–98.
Capelletti MM, Manceau H, Puy H, Peoc’h K. Ferroptosis in liver diseases: an overview. Int J Mol Sci. 2020;21(14):4908.
Ouyang S, You J, Zhi C, Li P, Lin X, Tan X, Ma W, Li L, Xie W. Ferroptosis: the potential value target in atherosclerosis. Cell Death Dis. 2021;12(8):782.
Li S, Zheng L, Zhang J, Liu X, Wu Z. Inhibition of ferroptosis by up-regulating Nrf2 delayed the progression of diabetic nephropathy. Free Radic Biol Med. 2021;162:435–49.
Hao S, Yu J, He W, Huang Q, Zhao Y, Liang B, Zhang S, Wen Z, Dong S, Rao J, et al. Cysteine dioxygenase 1 mediates erastin-induced ferroptosis in human gastric cancer cells. Neoplasia. 2017;19(12):1022–32.
Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013;41(Database issue):D991-995.
Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–7.
Liu Z, Zhao Q, Zuo ZX, Yuan SQ, Yu K, Zhang Q, Zhang X, Sheng H, Ju HQ, Cheng H, et al. Systematic analysis of the aberrances and functional implications of ferroptosis in cancer. iScience. 2020;23(7):101302.
Iglesia MD, Parker JS, Hoadley KA, Serody JS, Perou CM, Vincent BG. Genomic analysis of immune cell infiltrates across 11 tumor types. J Natl Cancer Inst. 2016;108(11):djw144.
Hoadley KA, Yau C, Wolf DM, Cherniack AD, Tamborero D, Ng S, Leiserson MDM, Niu B, McLellan MD, Uzunangelov V, et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014;158(4):929–44.
Zhou T, Cai Z, Ma N, Xie W, Gao C, Huang M, Bai Y, Ni Y, Tang Y. A novel ten-gene signature predicting prognosis in hepatocellular carcinoma. Front Cell Dev Biol. 2020;8:629.
Ko E, Kim Y, Cho EY, Han J, Shim YM, Park J, Kim DH. Synergistic effect of Bcl-2 and cyclin A2 on adverse recurrence-free survival in stage I non-small cell lung cancer. Ann Surg Oncol. 2013;20(3):1005–12.
Wang Y, Zhong Q, Li Z, Lin Z, Chen H, Wang P. Integrated profiling identifies CCNA2 as a potential biomarker of immunotherapy in breast cancer. Onco Targets Ther. 2021;14:2433–48.
Li J, Zhou L, Liu Y, Yang L, Jiang D, Li K, Xie S, Wang X, Wang S. Comprehensive analysis of cyclin family gene expression in colon cancer. Front Oncol. 2021;11: 674394.
Bendris N, Arsic N, Lemmers B, Blanchard JM. Cyclin A2 Rho GTPases and EMT. Small GTPases. 2012;3(4):225–8.
Li J, Ying Y, Xie H, Jin K, Yan H, Wang S, Xu M, Xu X, Wang X, Yang K, et al. Dual regulatory role of CCNA2 in modulating CDK6 and MET-mediated cell-cycle pathway and EMT progression is blocked by miR-381-3p in bladder cancer. Faseb j. 2019;33(1):1374–88.
Herrick J, Sclavi B. Ribonucleotide reductase and the regulation of DNA replication: an old story and an ancient heritage. Mol Microbiol. 2007;63(1):22–34.
Li C, Zheng J, Chen S, Huang B, Li G, Feng Z, Wang J, Xu S. RRM2 promotes the progression of human glioblastoma. J Cell Physiol. 2018;233(10):6759–67.
Wang S, Wang XL, Wu ZZ, Yang QC, Xiong HG, Xiao Y, Li H, Sun ZJ. Overexpression of RRM2 is related to poor prognosis in oral squamous cell carcinoma. Oral Dis. 2021;27(2):204–14.
Rahman MA, Amin AR, Wang D, Koenig L, Nannapaneni S, Chen Z, Wang Z, Sica G, Deng X, Chen ZG, et al. RRM2 regulates Bcl-2 in head and neck and lung cancers: a potential target for cancer therapy. Clin Cancer Res. 2013;19(13):3416–28.
Zhuang S, Li L, Zang Y, Li G, Wang F. RRM2 elicits the metastatic potential of breast cancer cells by regulating cell invasion, migration and VEGF expression via the PI3K/AKT signaling. Oncol Lett. 2020;19(4):3349–55.
Yang Y, Li S, Cao J, Li Y, Hu H, Wu Z. RRM2 regulated by LINC00667/miR-143-3p signal is responsible for non-small cell lung cancer cell progression. Onco Targets Ther. 2019;12:9927–39.
Morikawa T, Maeda D, Kume H, Homma Y, Fukayama M. Ribonucleotide reductase M2 subunit is a novel diagnostic marker and a potential therapeutic target in bladder cancer. Histopathology. 2010;57(6):885–92.
Chang CC, Lin CC, Wang CH, Huang CC, Ke TW, Wei PL, Yeh KT, Hsu KC, Hsu NY, Cheng YW. miR-211 regulates the expression of RRM2 in tumoral metastasis and recurrence in colorectal cancer patients with a k-ras gene mutation. Oncol Lett. 2018;15(5):8107–17.
Zhang Y, Yang X, Wang R, Zhang X. Prognostic value of PDZ-binding kinase/T-LAK cell-originated protein kinase (PBK/TOPK) in patients with cancer. J Cancer. 2019;10(1):131–7.
Xu M, Xu S. PBK/TOPK overexpression and survival in solid tumors: a PRISMA-compliant meta-analysis. Medicine (Baltimore). 2019;98(10): e14766.
Stauffer S, Zeng Y, Zhou J, Chen X, Chen Y, Dong J. CDK1-mediated mitotic phosphorylation of PBK is involved in cytokinesis and inhibits its oncogenic activity. Cell Signal. 2017;39:74–83.
Yang QX, Zhong S, He L, Jia XJ, Tang H, Cheng ST, Ren JH, Yu HB, Zhou L, Zhou HZ, et al. PBK overexpression promotes metastasis of hepatocellular carcinoma via activating ETV4-uPAR signaling pathway. Cancer Lett. 2019;452:90–102.
Zhou Z, Li Y, Hao H, Wang Y, Zhou Z, Wang Z, Chu X. Screening hub genes as prognostic biomarkers of hepatocellular carcinoma by bioinformatics Analysis. Cell Transplant. 2019;28(1_suppl):76s–86s.
Johnson A, Yao NY, Bowman GD, Kuriyan J, O’Donnell M. The replication factor C clamp loader requires arginine finger sensors to drive DNA binding and proliferating cell nuclear antigen loading. J Biol Chem. 2006;281(46):35531–43.
Cardano M, Tribioli C, Prosperi E. Targeting proliferating cell nuclear antigen (PCNA) as an effective strategy to inhibit tumor cell proliferation. Curr Cancer Drug Targets. 2020;20(4):240–52.
Wang SC. PCNA: a silent housekeeper or a potential therapeutic target? Trends Pharmacol Sci. 2014;35(4):178–86.
Lavezzi AM, Milei J, Grana DR, Flenda F, Basellini A, Matturri L. Expression of c-fos, p53 and PCNA in the unstable atherosclerotic carotid plaque. Int J Cardiol. 2003;92(1):59–63.
Haneke K, Schott J, Lindner D, Hollensen AK, Damgaard CK, Mongis C, Knop M, Palm W, Ruggieri A, Stoecklin G. CDK1 couples proliferation with protein synthesis. J Cell Biol. 2020;219(3):e201906147.
Liu S, Yang Y, Jiang S, Xu H, Tang N, Lobo A, Zhang R, Liu S, Yu T, Xin H. MiR-378a-5p regulates proliferation and migration in vascular smooth muscle cell by targeting CDK1. Front Genet. 2019;10:22.
Liu Y, Sun Y, Hu C, Liu J, Gao A, Han H, Chai M, Zhang J, Zhou Y, Zhao Y. Perivascular adipose tissue as an indication, contributor to, and therapeutic target for atherosclerosis. Front Physiol. 2020;11: 615503.
Zhu Y, Zhang Y, Huang X, Xie Y, Qu Y, Long H, Gu N, Jiang W. Z-Ligustilide protects vascular endothelial cells from oxidative stress and rescues high fat diet-induced atherosclerosis by activating multiple NRF2 downstream genes. Atherosclerosis. 2019;284:110–20.
Wolf D, Ley K. Immunity and inflammation in atherosclerosis. Circ Res. 2019;124(2):315–27.
Wu X, Li Y, Zhang S, Zhou X. Ferroptosis as a novel therapeutic target for cardiovascular disease. Theranostics. 2021;11(7):3052–9.
Fang X, Wang H, Han D, Xie E, Yang X, Wei J, Gu S, Gao F, Zhu N, Yin X, et al. Ferroptosis as a target for protection against cardiomyopathy. Proc Natl Acad Sci U S A. 2019;116(7):2672–80.
Yan HF, Tuo QZ, Yin QZ, Lei P. The pathological role of ferroptosis in ischemia/reperfusion-related injury. Zool Res. 2020;41(3):220–30.
Tadokoro T, Ikeda M, Ide T, Deguchi H, Ikeda S, Okabe K, Ishikita A, Matsushima S, Koumura T, Yamada KI, et al. Mitochondria-dependent ferroptosis plays a pivotal role in doxorubicin cardiotoxicity. JCI Insight. 2020;5(9):e132747.
Bai T, Li M, Liu Y, Qiao Z, Wang Z. Inhibition of ferroptosis alleviates atherosclerosis through attenuating lipid peroxidation and endothelial dysfunction in mouse aortic endothelial cell. Free Radic Biol Med. 2020;160:92–102.
Yang WS, SriRamaratnam R, Welsch ME, Shimada K, Skouta R, Viswanathan VS, Cheah JH, Clemons PA, Shamji AF, Clish CB, et al. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156(1–2):317–31.
Masaldan S, Clatworthy SAS, Gamell C, Meggyesy PM, Rigopoulos AT, Haupt S, Haupt Y, Denoyer D, Adlard PA, Bush AI, et al. Iron accumulation in senescent cells is coupled with impaired ferritinophagy and inhibition of ferroptosis. Redox Biol. 2018;14:100–15.
Wu J, Minikes AM, Gao M, Bian H, Li Y, Stockwell BR, Chen ZN, Jiang X. Intercellular interaction dictates cancer cell ferroptosis via NF2-YAP signalling. Nature. 2019;572(7769):402–6.
Gao M, Monian P, Quadri N, Ramasamy R, Jiang X. Glutaminolysis and transferrin regulate ferroptosis. Mol Cell. 2015;59(2):298–308.
Shi G, Chen W, Zhang Y, Dai X, Zhang X, Wu Z. An antifouling hydrogel containing silver nanoparticles for modulating the therapeutic immune response in chronic wound healing. Langmuir. 2019;35(5):1837–45.
Gragnano F, Calabrò P. Role of dual lipid-lowering therapy in coronary atherosclerosis regression: evidence from recent studies. Atherosclerosis. 2018;269:219–28.
Tsujita K, Sugiyama S, Sumida H, Shimomura H, Yamashita T, Yamanaga K, Komura N, Sakamoto K, Oka H, Nakao K, et al. Impact of dual lipid-lowering strategy with ezetimibe and atorvastatin on coronary plaque regression in patients with percutaneous coronary intervention: the multicenter randomized controlled PRECISE-IVUS trial. J Am Coll Cardiol. 2015;66(5):495–507.
Nicholls SJ, Puri R, Anderson T, Ballantyne CM, Cho L, Kastelein JJ, Koenig W, Somaratne R, Kassahun H, Yang J, et al. Effect of evolocumab on progression of coronary disease in statin-treated patients: the GLAGOV randomized clinical trial. JAMA. 2016;316(22):2373–84.
Acknowledgements
Many thanks to all colleagues who helped with this study.
Funding
This study was supported by the National Natural Science Foundation of China (No.81960663), Open Project Fund of Hainan Provincial Key Laboratory of Tropical Brain Research and Transformation (JCKF2021001), Youth Incubation Fund Project of Hainan Medical College (No. HYPY201912), Natural Science Foundation of Hainan Province Youth Project (No. 821QN401).
Author information
Authors and Affiliations
Contributions
Qingwen Meng, Yiqian Xu, Xuebin ling are co-first authors. MQW designed the study and had written the article. XYQ, WHL, FXY helped to perform the experiments. Among them, XYQ was responsible for completing the western blot experimental part in the article revision process. LXB is responsible for the collection of blood specimens. LHJ, DS, YDM were responsible for data collection and statistical analysis. LTF, LQB had revised the article. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
All animal experimental protocols mentioned in this study were carried out in compliance with the Guide for the Care and Use of Laboratory Animals (NIH publication, 8th Edition, 2011). The study on animals was in accord with the ARRIVE guidelines. The experiments that used animals were approved by the Institutional Animal Care and Use Committee of Hainan Medical University (Number: HYLL-2021–140). The ethics committee of the First Affiliated Hospital of Hainan Medical University gave its approval to the experiment of involved human blood specimens (Number: No. 1, 2022). All participants have signed written an informed consent.
Consent for publication
Not applicable.
Competing interests
The authors declare that there are no competing interests in the publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
Up-regulated and down-regulated DEGs obtained from GSE132651 dataset. Table S2. The correlation coefficients between overlapping genes and FRG.
Additional file 2: Table S3.
Raw data on the correlation of overlapping genes and FRG.
Additional file 3: Fig. S1.
CCNA2, GPX4 and CDK1 expression in aortic tissue of mice. Fig. S2. TFRC and β-actin expression in aortic tissue of mice. Fig. S3. Expression of CCNA2 in mouse aortic tissue at different exposure times. Fig. S4. Expression of GPX4 in mouse aortic tissue at different exposure times. Fig. S5. Expression of CDK1 in mouse aortic tissue at different exposure times. Fig. S6. Expression of TFRC in mouse aortic tissue at different exposure times. Fig. S7. Expression of β-actin in mouse aortic tissue at different exposure times.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Meng, Q., Xu, Y., ling, X. et al. Role of ferroptosis-related genes in coronary atherosclerosis and identification of key genes: integration of bioinformatics analysis and experimental validation. BMC Cardiovasc Disord 22, 339 (2022). https://doi.org/10.1186/s12872-022-02747-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12872-022-02747-x