Abstract
Background
Whether adiponectin (ADIPOQ) polymorphisms are associated with coronary artery disease (CAD) remain controversial. Therefore, we performed this meta-analysis to better explore potential roles of ADIPOQ polymorphisms in CAD.
Methods
PubMed, Web of Science, Embase and CNKI were searched for eligible studies. Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated.
Results
Totally 45 studies were included for pooled analyses. A significant association with the susceptibility to CAD was detected for rs2241766 (dominant model: p = 0.0009, OR = 0.82, 95%CI 0.73–0.92; recessive model: p = 0.04, OR = 1.29, 95%CI 1.02–1.64; allele model: p < 0.0001, OR = 0.80, 95%CI 0.73–0.88) polymorphism in overall population. Further subgroup analyses by ethnicity showed that rs1501299 polymorphism was significantly associated with the susceptibility to CAD in East Asians, whereas rs2241766 polymorphism was significantly associated with the susceptibility to CAD in Caucasians, East Asians and South Asians.
Conclusions
Our findings indicated that rs1501299 and rs2241766 polymorphisms both affect the susceptibility to CAD in certain populations.
Similar content being viewed by others
Background
Coronary artery disease (CAD) is the leading cause of death and disability worldwide [1, 2]. To date, the exact pathogenesis of CAD remains largely unknown. Nevertheless, plenty of evidences demonstrated that genetic factors are crucial for the development of CAD. First, family clustering of CAD was observed extensively, and past twin studies showed that the heredity grade of CAD was over 50 % [3, 4]. Second, numerous genetic variants were found to be associated with an increased susceptibility to CAD by previous genetic association studies, and screening of common causal variants was also proved to be an efficient way to predict the individual risk of developing CAD [5, 6]. Overall, these findings jointly indicated that genetic predisposition to CAD is important for its occurrence and development.
Adiponectin (ADIPOQ), an adipocytokine that regulates energy and material metabolism, is implicated in the development of multiple metabolic disorders including obesity and type II diabetes. And it was evident that these two common metabolic disorders were associated with an increased risk of CAD [7]. Furthermore, previous studies demonstrated that adipoenctin have both anti-atherogenic and anti-inflammatory property [8, 9]. Moreover, the expression level of adiponectin was also significantly decreased in CAD patients [10, 11]. Overall, these evidences jointly suggested that adipoenctin might exert favorable protection effects against CAD. Therefore, functional ADIPOQ genetic polymorphisms, which may alter the expression level of adiponectin, may also affect individual susceptibility to CAD. Recently, some pilot studies already investigated associations of two common functional ADIPOQ polymorphisms, rs1501299 and rs2241766, with the susceptibility to CAD. However, the results of these studies were not consistent, especially when they were conducted in different populations [12,13,14,15,16,17,18,19]. Previous studies failed to reach a consensus regarding associations between ADIPOQ polymorphisms and CAD partially because of their relatively small sample sizes. Thus, we performed the present meta-analysis to explore the relationship between ADIPOQ polymorphisms and CAD in a larger pooled sample size. Additionally, we also aimed to elucidate the potential effects of ethnic background on associations between ADIPOQ polymorphisms and CAD.
Methods
The current meta-analysis followed the Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) checklist [20,21,22].
Literature search and inclusion criteria
The combination of following terms: (adiponectin OR ADIPOQ) AND (polymorphism OR variant OR mutation OR genotype OR allele) AND (coronary heart disease OR coronary artery disease OR angina pectoris OR acute coronary syndrome OR myocardial infarction) was used to searched for potentially eligible articles that were published prior to December 1, 2018 in PubMed, Web of Science, Embase and China National Knowledge Infrastructure (CNKI). We also reviewed the reference lists of all retrieved articles for other potentially eligible studies.
To test the research hypothesis of this meta-analysis, included studies must meet all the following criteria: (1) case-control study on associations between ADIPOQ polymorphisms (rs1501299 and rs2241766) and CAD; (2) provide genotypic and/or allelic frequency of investigated polymorphisms; (3) full text in English or Chinese available. Studies were excluded if one of the following criteria was fulfilled: (1) not relevant to ADIPOQ polymorphisms and CAD; (2) case reports or case series; (3) abstracts, reviews, comments, letters and conference presentations. In the case of duplicate reports by the same authors, we only included the most recent study.
Data extraction and quality assessment
We extracted the following information from eligible studies: 1. name of the first author; 2. year of publication; 3. country and ethnicity of participants; 4. sample size; and 5. genotypic distributions of ADIPOQ polymorphisms in cases and controls. The probability value (p value) of Hardy-Weinberg equilibrium (HWE) was also calculated.
We used the Newcastle-Ottawa scale (NOS) to evaluate the quality of eligible studies [23]. The NOS has a score range of zero to nine, and studies with a score of more than seven were thought to be of high quality.
Two reviewers conducted data extraction and quality assessment independently (Xia Zhang and YanJun Cao). When necessary, we wrote to the corresponding authors for extra information. Any disagreement between two reviewers was solved by discussion until a consensus was reached.
Statistical analyses
In the current study, Review Manager Version 5.3.3 was used to perform statistical analyses. We calculated ORs and 95% CIs to estimate potential associations between ADIPOQ polymorphisms and CAD in all possible genetic models, and a p value of 0.05 or less was defined as statistically significant. Between-study heterogeneities were evaluated by I2 statistic. Random-effect models (REMs) would be used for analyses if I2 was greater than 50%. Otherwise, analyses would be performed with fixed-effect models (FEMs). Subgroup analyses by ethnicity and type of disease were subsequently carried out. Stabilities of synthetic results were tested in sensitivity analyses. Publication biases were assessed by funnel plots.
Results
Characteristics of included studies
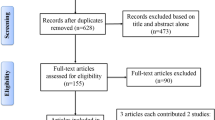
We found 442 potential relevant articles. Among these articles, totally 45 eligible studies were finally included for pooled analyses (see Fig. 1). Baseline characteristics of included studies were shown in Table 1.
Overall and subgroup analyses
Results of overall and subgroup analyses were summarized in Table 2. To be brief, a significant association with the susceptibility to CAD was detected for rs2241766 (dominant model: p = 0.0009, OR = 0.82, 95%CI 0.73–0.92; recessive model: p = 0.04, OR = 1.29, 95%CI 1.02–1.64; allele model: p < 0.0001, OR = 0.80, 95%CI 0.73–0.88) polymorphism in overall analyses. Further subgroup analyses by ethnicity revealed that rs1501299 polymorphism was significantly associated with the susceptibility to CAD in East Asians, whereas rs2241766 polymorphism was significantly associated with the susceptibility to CAD in Caucasians, East Asians and South Asians. No any other positive results were observed in overall and subgroup analyses (see Table 2 and Fig. 2).
Forest plots for overall analyses of investigated polymorphisms. a Forest plot of rs1501299 polymorphism and CAD under dominant comparison; b Forest plot of rs1501299 polymorphism and CAD under recessive comparison; c Forest plot of rs1501299 polymorphism and CAD under overdominant comparison; d Forest plot of rs1501299 polymorphism and CAD under allele comparison; e Forest plot of rs2241766 polymorphism and CAD under dominant comparison; f Forest plot of rs2241766 polymorphism and CAD under recessive comparison; g Forest plot of rs2241766 polymorphism and CAD under overdominant comparison. h Forest plot of rs2241766 polymorphism and CAD under allele comparison
Sensitivity analyses
We performed sensitivity analyses by excluding studies that deviated from HWE. No alterations of results were detected in sensitivity analyses, which suggested that our findings were statistically reliable.
Publication biases
Publication biases were evaluated with funnel plots. We did not find obvious asymmetry of funnel plots in any comparisons, which indicated that our findings were unlikely to be impacted by severe publication biases.
Discussion
Based on combined analyses of 45 eligible studies, our study showed that rs1501299 and rs2241766 polymorphisms were both significantly associated with the susceptibility to CAD in certain populations, which suggested that these two polymorphisms may be used to identify individuals with higher susceptibility to CAD. There are two possible explanations for our positive findings. First, genetic variations of the ADIPOQ gene may lead to alternations in gene expression or changes in ADIPOQ protein structure, which may subsequently affect biological functions of ADIPOQ and ultimately impact individual susceptibility to CAD. Second, it is also possible that ADIPOQ polymorphisms may be linked to each other or even linked to other unidentified genes, which could also impact individual susceptibility to CAD.
There are several points that should be noted about this meta-analysis. Firstly, previous experimental studies demonstrated that mutant alleles of investigated polymorphisms could lead to decreased adiponectin generation, which may partially explain our positive findings [12,13,14,15,16,17,18,19]. Secondly, it is also worth noting that for rs1501299 polymorphism, the trends of associations in different ethnicities were not always consistent, and this may be attributed to ethnic differences in genotypic distributions of investigated polymorphisms. However, it is also that these inconsistent findings may be resulted from a complex interaction of both genetic and environmental factors. Thirdly, it should be noted that significant between-study heterogeneities were observed in all genetics comparisons of overall analyses, which may partially attributed to ethnic and racial differences of eligible studies. To overcome between-study heterogeneities, REMs were used for pooled analyses, and in further subgroup analyses, we noticed that between-study heterogeneities among studies that were conducted in Caucasians were relatively small, which also supported that ethnic background could impact individual susceptibility to CAD. Fourthly, a recent meta-analyses conducted by Hou et al. [24] also tried to explore potential associations between ADIPOQ polymorphisms and CAD. However, our findings should be considered as more conclusive compared to that of previous meta-analysis since many related studies were published in the last three years, which warranted an update meta-analysis. Totally 10 more eligible studies were enrolled in our pooled analyses, and the sample sizes of our analyses were also significantly larger than that of previous meta-analyses, which could significantly reduce the risk of obtaining false positive or false negative results. Compared with the previous meta-analysis, similar positive results were detected for rs2241766 polymorphism in overall and subgroup analyses. However, positive results in Caucasians for rs1501299 polymorphism were no longer observed in our meta-analysis. Instead, we found that rs1501299 polymorphism could impact individual susceptibility to CAD in East Asians under recessive genetic model. Therefore, future studies with larger sample sizes are still needed to test the potential associations between ADIPOQ polymorphisms and CAD, especially for rs1501299 polymorphism. Fifthly, our study only focused on two mostly investigated ADIPOQ polymorphisms, and future meta-analyses should try to investigate the associations between CAD and other common ADIPOQ polymorphisms such as rs266729, rs822395 and rs17300539. These polymorphisms were not analyzed by us because we failed to find any additional eligible studies compared to the previous meta-analysis conducted by Hou et al. [24].
Some limitations of this meta-analysis should also be acknowledged when interpreting our findings. First, our pooled analyses were based on unadjusted estimations due to lack of raw data, and failure to perform further adjusted analyses may impact the reliability of our findings [25, 26]. Second, since our pooled analyses were based on retrospective case-control studies, despite our positive findings, future perspective studies are still needed to examine whether there is direct causal relationship between ADIPOQ polymorphisms and CAD [27, 28]. Third, associations between ADIPOQ polymorphisms and CAD may also be modified by gene-gene and gene-environmental interactions. However, due to lack of raw data, we could not conduct relevant analyses [29, 30]. Fourth, our analyses were based on retrospective case-control studies. Thus, despite the relatively high NOS score, it was still possible that our findings might be impacted by potential selection, measurement and confounding biases. Taking the above mentioned limitations into consideration, our findings should be interpreted with caution.
Conclusions
In conclusion, our meta-analysis suggested that rs1501299 and rs2241766 polymorphisms were both significantly associated with the susceptibility to CAD in certain populations. However, further well-designed studies are still warranted to confirm our findings.
Abbreviations
- ADIPOQ :
-
Adiponectin
- CAD:
-
Coronary artery disease
- HWE:
-
Hardy-Weinberg equilibrium
- NOS:
-
Newcastle-Ottawa scale
References
Moran AE, Forouzanfar MH, Roth G, Mensah GA, Ezzati M, Flaxman A, et al. The global burden of ischemic heart disease in 1990 and 2010: the global burden of disease 2010 study. Circulation. 2014;129:1493–501.
Global Burden of Disease Study 2013 Collaborators. Global, regional, and national incidence, prevalence, and years lived with disability for 301 acute and chronic diseases and injuries in 188 countries, 1990-2013: a systematic analysis for the global burden of disease study 2013. Lancet. 2015;386:743–800.
Mayer B, Erdmann J, Schunkert H. Genetics and heritability of coronary artery disease and myocardial infarction. Clin Res Cardiol. 2007;96:1–7.
Evans A, Van Baal GC, McCarron P, DeLange M, Soerensen TI, De Geus EJ, et al. The genetics of coronary heart disease: the contribution of twin studies. Twin Res. 2003;6:432–41.
Sayols-Baixeras S, Lluís-Ganella C, Lucas G, Elosua R. Pathogenesis of coronary artery disease: focus on genetic risk factors and identification of genetic variants. Appl Clin Genet. 2014;7:15–32.
Dai X, Wiernek S, Evans JP, Runge MS. Genetics of coronary artery disease and myocardial infarction. World J Cardiol. 2016;8:1–23.
Wang ZV, Scherer PE. Adiponectin, the past two decades. J Mol Cell Biol. 2016;8:93–100.
Katsiki N, Mantzoros C, Mikhailidis DP. Adiponectin, lipids and atherosclerosis. Curr Opin Lipidol. 2017;28:347–54.
Ohashi K, Ouchi N, Matsuzawa Y. Anti-inflammatory and anti-atherogenic properties of adiponectin. Biochimie. 2012;94:2137–42.
Persson J, Lindberg K, Gustafsson TP, Eriksson P, Paulsson-Berne G, Lundman P. Low plasma adiponectin concentration is associated with myocardial infarction in young individuals. J Intern Med. 2010;268:194–205.
Piestrzeniewicz K, Luczak K, Komorowski J, Maciejewski M, Piechowiak M, Jankiewicz-Wika J, Goch JH. Obesity and adiponectin in acute myocardial infarction. Cardiol J. 2007;14:29–36.
Zhang Z, Li Y, Yang X, Wang L, Xu L, Zhang Q. Susceptibility of multiple polymorphisms in ADIPOQ, ADIPOR1 and ADIPOR2 genes to myocardial infarction in Han Chinese. Gene. 2018;658:10–7.
Zhong C, Zhen D, Qi Q, Genshan M. A lack of association between adiponectin polymorphisms and coronary artery disease in a Chinese population. Genet Mol Biol. 2010;33:428–33.
Liang C, Yawei X, Qinwan W, Jingying Z, Aihong M, Yanqing C. Association of AdipoQ single-nucleotide polymorphisms and smoking interaction with the risk of coronary heart disease in Chinese Han population. Clin Exp Hypertens. 2017;39:748–53.
Mohammadzadeh G, Ghaffari MA, Heibar H, Bazyar M. Association of two common single nucleotide polymorphisms (+45T/G and +276G/T) of ADIPOQ gene with coronary artery disease in type 2 diabetic patients. Iran Biomed J. 2016;20:152–60.
Rizk NM, El-Menyar A, Marei I, Sameer M, Musad T, Younis D, et al. Association of adiponectin gene polymorphism (+T45G) with acute coronary syndrome and circulating adiponectin levels. Angiology. 2013;64:257–65.
Ambroziak M, Kolanowska M, Bartoszewicz Z, Budaj A. Adiponectin gene variants and decreased adiponectin plasma levels are associated with the risk of myocardial infarction in young age. Gene. 2018;642:498–504.
Chiodini BD, Specchia C, Gori F, Barlera S, D'Orazio A, Pietri S, et al. Adiponectin gene polymorphisms and their effect on the risk of myocardial infarction and type 2 diabetes: an association study in an Italian population. Ther Adv Cardiovasc Dis. 2010;4:223–30.
Hegener HH, Lee IM, Cook NR, Ridker PM, Zee RY. Association of adiponectin gene variations with risk of incident myocardial infarction and ischemic stroke: a nested case-control study. Clin Chem. 2006;52:2021–17.
Moher D, Liberati A, Tetzlaff J, Altman DG. PRISMA group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med. 2009;151:264–9.
Li YY, Wang H, Qian J, Kim HJ, Wu JJ, Wang LS, Zhou CW, Yang ZJ, Lu XZ. PRISMA-combined myeloperoxidase -463G/a gene polymorphism and coronary artery disease: a meta-analysis of 4744 subjects. Medicine (Baltimore). 2017;96:e6461.
Ma WQ, Han XQ, Wang X, Wang Y, Zhu Y, Liu NF. Associations between XRCC1 gene polymorphisms and coronary artery disease: a meta-analysis. PLoS One. 2016;11:e0166961.
Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25:603–5.
Hou H, Ge S, Zhao L, Wang C, Wang W, Zhao X, et al. An updated systematic review and meta-analysis of association between adiponectin gene polymorphisms and coronary artery disease. OMICS. 2017;21:340–51.
Xie X, Shi X, Liu M. The roles of TLR gene polymorphisms in atherosclerosis: a systematic review and meta-analysis of 35,317 subjects. Scand J Immunol. 2017;86:50–8.
Sun H, Li Q, Jin Y, Qiao H. Associations of tumor necrosis factor-α polymorphisms with the risk of asthma: a meta-analysis. Exp Mol Pathol. 2018;105:411–6.
Dong J, Ping Y, Wang Y, Zhang Y. The roles of endothelial nitric oxide synthase gene polymorphisms in diabetes mellitus and its associated vascular complications: a systematic review and meta-analysis. Endocrine. 2018;62:412–22.
Shi X, Xie X, Jia Y, Li S. Associations of insulin receptor and insulin receptor substrates genetic polymorphisms with polycystic ovary syndrome: a systematic review and meta-analysis. J Obstet Gynaecol Res. 2016;42:844–54.
Zhu Y, Zheng G, Hu Z. Association between SERT insertion/deletion polymorphism and the risk of irritable bowel syndrome: a meta-analysis based on 7039 subjects. Gene. 2018;679:133–7.
Xie X, Shi X, Xun X, Rao L. Endothelial nitric oxide synthase gene single nucleotide polymorphisms and the risk of hypertension: a meta-analysis involving 63,258 subjects. Clin Exp Hypertens. 2017;39:175–82.
Acknowledgments
None.
Funding
None.
Availability of data and materials
The current study was based on results of relevant published studies.
Author information
Authors and Affiliations
Contributions
XZ, HC and JZ conceived of the study, participated in its design. XZ and YC conducted the systematic literature review. HZ performed data analyses. XZ, HC and JZ drafted the manuscript. All gave final approval and agree to be accountable for all aspects of work ensuring integrity and accuracy.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Zhang, X., Cao, Y.J., Zhang, H.Y. et al. Associations between ADIPOQ polymorphisms and coronary artery disease: a meta-analysis. BMC Cardiovasc Disord 19, 63 (2019). https://doi.org/10.1186/s12872-019-1041-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12872-019-1041-3