Abstract
Background
The aim of our study was the identification of genetic variants associated with postoperative complications after cardiac surgery.
Methods
We conducted a prospective, double-blind, multicenter, randomized trial (RIPHeart). We performed a genome-wide association study (GWAS) in 1170 patients of both genders (871 males, 299 females) from the RIPHeart-Study cohort. Patients undergoing non-emergent cardiac surgery were included. Primary endpoint comprises a binary composite complication rate covering atrial fibrillation, delirium, non-fatal myocardial infarction, acute renal failure and/or any new stroke until hospital discharge with a maximum of fourteen days after surgery.
Results
A total of 547,644 genotyped markers were available for analysis. Following quality control and adjustment for clinical covariate, one SNP reached genome-wide significance (PHLPP2, rs78064607, p = 3.77 × 10− 8) and 139 (adjusted for all other outcomes) SNPs showed promising association with p < 1 × 10− 5 from the GWAS.
Conclusions
We identified several potential loci, in particular PHLPP2, BBS9, RyR2, DUSP4 and HSPA8, associated with new-onset of atrial fibrillation, delirium, myocardial infarction, acute kidney injury and stroke after cardiac surgery.
Trial registration
The study was registered with ClinicalTrials.gov NCT01067703, prospectively registered on 11 Feb 2010.
Similar content being viewed by others
Background
Coronary heart disease is the leading cause of death and disability worldwide and is responsible for about 7.2 million deaths every year. Cardiac surgery is one of the most common cardiac procedures, performed annually in about 1.5 million patients worldwide. In spite of advances in surgical techniques, the incidence of complications after cardiac surgery using cardiopulmonary bypass is still high. The most common complications following cardiac surgery are atrial fibrillation (AF), delirium, myocardial infarction (MI), acute renal failure and stroke, all of which increase mortality and lead to a prolonged stay on intensive care unit (ICU) of patients. The restoration of regional blood flow after a period of ischemia during cardiac surgery frequently causes further cellular organ injury and thereby potentially limiting the recovery of function. Reperfusion is often associated with microvascular dysfunction. Activated endothelial cells produce excessive reactive oxygen species (ROS), but less nitric oxide, leading to release of inflammatory mediators, mitochondrial dysfunction, oxidative stress and finally cell death [1].
Yet, the inflammatory mediators released as a consequence of reperfusion also appear to activate endothelial cells in remote organs not initially exposed to the ischemic stress, i.e. the kidney and the central nervous system. This distant response to ischemia/ reperfusion (I/R) can result in leukocyte-dependent microvascular injury that is characteristic of the “systemic inflammatory response syndrome”, potentially leading to delirium, stroke and acute kidney dysfunction [2].
Hypercholesterolemia, diabetes and hypertension, the occurrence and extent of complications after cardiac surgery could have a genetic basis. A genetic bias is strongly suggested by observations that the wide variability concerning incidence and severity of complications after cardiac surgery could not be explained by clinical or interventional risks. As postoperative organ dysfunction is common after cardiac surgery, several previous studies have performed preoperative genomic characterization of patients to identify genotypes, which render the patients vulnerable for the development of a specific organ dysfunction. Kertai et al. identified genetic variants in patients exhibiting AF and MI after cardiac surgery [3, 4]. Another recent study reported genetic variants concerning acute kidney injury after cardiac surgery [5]. All of these studies investigated one or two complications after cardiac surgery but none examined the complications in all, nor investigated a composite of all main complications. Therefore, we conducted a comprehensive genome-wide association study (GWAS) to identify common genetic variants associated with the main complications after cardiac surgery as new-onset postoperative AF, MI, delirium, stroke and acute renal failure.
Methods
We performed a GWAS using in total 1170 DNA samples from the RIPHeart (Remote Ischemic Preconditioning for Heart Surgery) study cohort [6, 7] (871 males, 299 females) in the dataset testing 547,644 variants to identify candidate genes that predetermine main complications after cardiac surgery. Various samples were excluded for the different subanalyses. See below (statistics) for more information.
The initial objective of our prospective, double-blind, multicentre, randomized controlled RIPHeart study was to investigate whether upper limb remote ischemic preconditioning compared to sham intervention reduced the incidence of the primary endpoint including death, MI, stroke, and acute renal failure until hospital discharge in adults scheduled for elective cardiac surgery requiring cardiopulmonary bypass (for further information please read the english synopsis and study protocol of the RIPHeart study in Additional file 1: Figure S3 and Additional file 2: Figure S4 in the supplementary). As the initial intervention study did not show any group differences, this predefined secondary analysis of genome-wide association now includes all patients irrespective of the initial group assignment.
Patient populations
The cohort comprised 1204 patients who underwent an elective cardiac surgery requiring cardiopulmonary bypass (e.g. coronary artery bypass graft, valve surgery, ascending aorta replacement) between January 2011 and May 2014 and were analysed. 1170 (871 males, 299 females) patients met eligibility criteria after applying quality control and excluding patients with missing genotypes or phenotypic information.
Outcome measures
In this GWAS-study, the primary endpoint comprises a composite complication rate covering AF, delirium, MI, acute renal failure, and/or any new stroke. Non-fatal myocardial infarction was defined by
-
biomarker values more than five times the 99th percentile of the normal reference range combined with new pathological Q-waves
-
or new left bundle branch block (LBBB) within the first 72 h
-
standard clinical criteria for myocardial infarction from 72 h on
-
new ischemic finding by echocardiography/angiography
-
or myocardial infarction diagnosed at autopsy.
Some patients present with ST elevation or new LBBB, and suffer sudden cardiac death before cardiac biomarkers become abnormal or pathological signs of myocardial necrosis become evident at autopsy. These patients should be classified as having had a fatal myocardial infarction [8].
A blinded clinical endpoint committee assessed all available electrocardiograms for reference reading. Stroke was defined by any new, temporary or permanent, focal or global neurological deficit, or evidence of stroke on autopsy, and was evaluated according to the National Institutes of Health Stroke Scale (≥ 4 points) [9]. Acute renal failure was defined by any serum creatinine greater than or equal to two-fold increase from baseline, urine output ≤0.5 mL/kg/h for 12 h [10], use of renal replacement therapy, or evidence of renal failure on autopsy. New onset of AF was recorded by electrocardiograms. Delirium was assessed with the CAM-ICU score [11].
While delirium and AF were recorded within 4 days after surgery, MI, stroke and acute renal failure were analyzed until hospital discharge with a maximum of 14 days after surgery. Additionally several other clinical variables were recorded. For details see Table 1.
Genotyping
Genomic DNA was isolated from whole blood of patients using standard procedures. Genotyping was performed for 1224 samples (500 cases and 724 controls) on the Illumina Human CoreExome-24 BeadChip (Illumina, Inc., San Diego, CA, USA) comprising 547,644 markers.
Quality control of genotyped data
Markers were called using zCall [12], an algorithm specifically designed for calling low frequency variants. Samples with the following criteria were excluded: Call rate < 98%, heterozygosity > mean + 3x standard deviation (SD), duplicate samples and related samples (IBD > 0.185). Equally, markers with the following criteria were excluded: Call rate < 95%, failing Hardy-Weinberg-equilibrium tests in controls (p < 0.00001), markers mapping to chromosome 0, markers with differential missingness between batches (p < 10− 5) duplicate and triallelic markers as well as INDELs. For the principal components analysis (HAPMAP PCA), data were merged to the HAPMAP Phase III CEU, CHB and YRI populations. Variants with a minor allele frequency (MAF)-threshold smaller than 0.05 and with a p-value of the statistical test for Hardy Weinberg Equilibrium (HWE) in control samples p < 10− 5 were excluded. A batch QC was performed on the remaining samples. Data were again analyzed using PCA and flashpca [13]. Samples outside the rectangle [median + − 3* standard deviation] were excluded from the analysis. This left 1170 samples and 522,502 variants for analysis.
Phasing and imputation
Data were then phased using SHAPEIT v2 [14], excluding all variants not matching to the 1000 Genomes Phase I variants according to their allelic information and using only chromosomes 1 to 22. Data were phased using 100 states, 7 burn, 8 prune and 20 main iterations as well as an effective cohort size of 11,418. Data were then imputed using IMPUTE2 v3.0 [15] in fragments of 5 Mb. Fragments comprising less than 200 variants were merged before imputation. Imputation was performed using the 1000 Genomes Phase I cohort as reference with 20,000 Ne, 250 buffer and 10 burnin and 30 main iterations as well as 80 k and 500 k_hap, 3 outdp. Genotyped data were kept and not overwritten by IMPUTE2. Following imputation markers with an imputation INFO-Score < 0.8 were excluded, leaving 9,007,469 variants on chromosomes 1 to 22 for analysis.
Statistical analysis
Analysis was performed on 1163 samples. 7 of the above samples were excluded with missing phenotype data. Association tests were performed using PLINK v1.9 [16]. Statistical analysis of phenotypic data was performed using R (v3.0.2). Fisher’s exact test was used for the analysis of categorical and binary data, Student’s t-tests were used for the analysis of continuous data. Also, to take into account possible combined effects of the investigated clinical outcomes, we performed a variable selection algorithm using a logistic regression and a stepwise procedure (forwards and backwards) based on the Akaike Information Criterion (AIC) in R as well as a random forest approach using r2VIM [17] with 500 trees, mtry = 1/3 node size proportion of 10% and 5 runs. Random forest, when using regression mode for a binary outcome, will return probabilities of class membership. The overlap of these three methods was calculated. Variables that occurred more than 2 times were used.
r2vim is a feature selection method that uses relative importance measures to estimate the predictive power of, in this case, covariates in respect to the outcome (here disease) using a random forest approach and is described further in [17]. Using these relative importance measures, features that are correlated with the outcome are selected. r2vim is a package that is available in R. In a stepwise regression approach the stepAIC function of R can be used to build statistical models (starting from a null model that contains no variables in forward selection and from the full model containing all covariates in backwards selection) that contain any number of covariates. The final model then contains all variables that are most predictive for the outcome. In each step a covariate is added or deleted and the improvement of the model is tested against a previous model using the AIC. The AIC is a statistical measure of the goodness of fit of a model to a particular dataset (in this case disease outcome and clinical phenotypes).
Logistic regression analysis was performed on AF, delirium, MI, renal failure, stroke and the composite. The composite is a “compound” phenotype stating whether or not a sample had a phenotype or not. Genotypes were presented as dosage data between 0 and 2 and the p-value of association as well as the Odds ratios and their respective 95% confidence intervals were recorded. All analyses were adjusted for EuroSCORE (European System for Cardiac Operative Risk Evaluation) [18], age, diabetes mellitus, type of surgery, and baseline creatinine. Analysis of MI was adjusted for use of any cardiac assist device; stroke for severe sepsis/ septic shock; AF for cholesterol lowering drugs and smoking; delirium for any delirium medications, re-thoracotomy, cholesterol lowering drugs, center, NYHA (New York Heart Association) class, severe sepsis/ septic shock; renal failure was not adjusted for any additional covariate. Since one individual might have multiple of the outcomes, two types of analyses were performed: adjust each phenotype for each of the other outcomes to identify variants unique for each phenotype or without adjusting for other outcomes. Here, variants that might overlap between the outcomes are analyzed. A complete list of all adjustments is provided in the supplementary (Additional file 3: Table S1).
In this analysis we considered the widely excepted p-value threshold of 5 × 10− 8 as genome-wide significant and 1 × 10− 5 as nominally significant.
In total, 882 samples were analyzed for AF, 967 for delir, 1026 for MI, 1072 for renal failure and 1106 for stroke.
Results
Baseline characteristics
A total of 1403 patients underwent randomization in the RIPHeart study, 1170 of these patients were included in the GWA analysis (Fig. 1). Baseline demographic and clinical characteristics are shown in Table 1. A wide range of cardiac surgical procedures were included. More than 27% of the original patients had congestive heart failure with NYHA class III or higher and 31% of patients had an EuroSCORE ≥6 or higher before surgery representing a high proportion of high-risk patients [6].
Postoperative AF was seen in 27.4% (242/882) of the patients. Delirium occurred in 15.3% (148/967), MI in 8.5% (87/1026), acute renal failure in 4.9% (52/1072) and stroke in 1.5% (17/1106).
Explorative genotype analysis
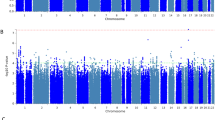
A total 9,007,469 imputed markers with an imputation info score higher than 0.8 were available for analysis. We performed analyses with and without an adjustment for all other outcomes and a composite. The GWAS results are depicted using Manhattan plots (Additional file 4: Figure S1) and quantile-quantile (Q-Q) plots (Additional file 5: Figure S2) for each analysis and each complication. In this analysis we considered the widely excepted p-value threshold of 5 × 10− 8 as genome-wide significant and 1 × 10− 5 as nominally significant. Results presented were performed with adjustment for all other outcomes and the composite.
Only one SNP reached genome-wide significance of p < 5 × 10− 8(rs78064607, located in an intron of PH domain and leucine rich repeat protein phosphatase 2 gene (PHLLP2), p = 3.77 × 10− 8, (OR_L95 = 0.01, OR_U95 = 0.09, OR with 95% CI 0.02) and was associated with renal failure.
The complete list of SNPs showing associations with genes and exhibit the pre-defined p-value of p < 1 × 10− 5 from the GWAS with regard to AF, MI, delirium, and renal failure, and stroke is shown in the Additional file 6: Table S2. SNPs with lowest p-values for each complication are shown in Table 2.
SNPs with lowest p-values located in regions associated with genes in patients with complications after cardiac surgery, adjusted for each outcome are: STON1 (stonin1, rs7557600, p = 7.25 × 10− 7), ZBTB20 (Zinc finger and BTB domain containing 20, rs115155878, p = 8.45 × 10− 7), LINC00371 (Long intergenic non-protein coding RNA 371, rs9563027, p = 9.82 × 10− 7) and GPR98 (G protein-coupled receptor 98, rs4574581, p = 2.43 × 10− 6) for AF, AC074391.1 (rs13008718, p = 9.99 × 10− 7), LINC00871 (Long intergenic non-protein coding RNA 871, rs1886223516, p = 4.02 × 10− 6) for delirium, FHIT (Fragile histidine triad, rs727476, p = 1.70 × 10− 6) and SUGCT (Succinyl-CoA-glutarate-CoA-transferase, rs9690969, p = 1.91 × 10− 6) for MI, SNORA40 (Small nucleolar RNA, H/ACA Box 40, rs189437718, p = 3.60 × 10− 7), EIF4G3 (Eukaryotic Translation Initiation Factor 4 Gamma 3, rs72654815, p = 6.79 × 10− 7) for renal failure and BBS9 (Bardet-Biedl Syndrome 9, rs79995619, p = 3.35 × 10− 7), TP63 (Tumor protein p63, rs181832941, p = 3.65 × 10− 7), RNU6-443P (RNA, U6 small nuclear 443, rs140914711, p = 4.07 × 10− 7) and (RyR2 (Ryanodine receptor 2, rs192540202, p = 6.33 × 10− 7) for stroke.
Besides adjusted analysis, the composite, meaning a “compound” phenotype stating whether or not a sample had a phenotype or not, comprises five SNPs, three with association with genes: RP5-968 J1.1 (rs200890 p = 1.19 × 10− 6), DUSP4 (Dual Specificity Phosphatase 4, rs4732926, p = 5.53 × 10− 6, OR_L95 = 0.47, OR_U95 = 0.74) and WLS (Wntless Wnt Ligand Secretion Mediator) and GNG12-AS1 (GNG Antisense RNA 1), (rs74081211, p = 6.25 × 10− 6).
Loci of SNPs with lowest p-values in genes are shown in Fig. 2.
Discussion
As postoperative organ dysfunction is common after cardiac surgery, previous studies have performed preoperative genomic characterization of patients to identify genotypes, which render the patients vulnerable for the development of a specific organ dysfunction. Kertai et al. identified genetic variants in patients exhibiting AF and MI after cardiac surgery [3, 4]. Another recent study reported genetic variants concerning acute kidney injury after cardiac surgery [5]. To identify common genetic variants associated with the main complications AF, delirium, MI, acute renal failure and stroke, we performed a GWAS study with DNA samples of 1170 patients after cardiac surgery. We identified one SNP reaching genome-wide significance (p < 5 × 10− 8) and nearly 150 SNPs which reached the a priori defined discovery threshold of p < 1 × 10− 5. Since one individual might have multiple of the outcomes, we performed two types of analyses. Either we adjusted each phenotype of the other outcomes to identify variants unique for each phenotype or performed analyses without adjusting for each other. Here, variants might overlap between the outcomes and influenced genes could have effects to several organs. Besides, we identified a “compound” phenotype stating whether or not a sample has a phenotype or not.
DUSP4 (rs4732926, Dual Specificity Phosphatase 4) was identified as a compound phenotype. DUSP4 is an inducible nuclear phosphatase that is involved in regulating cardiovascular function under oxidative stress [19]. DUSP4 −/− mice showed an increase of I/R-induced infarct caused by an over activation of p38, a stress-activated and pro-inflammatory kinase. In accordance to this, overexpression of DUSP4 in endothelial cells prevents hypoxia/ reoxygenation-induced apoptosis via the upregulation of eNOS [20]. Therefore, it would be worthwhile to have a closer look inside the exact role of DUSP4 during cardiac surgery.
rs78064607, the only SNP with genome-wide significance in our study is located in PHLPP2 (PH Domain and Leucine Rich Repeat Protein Phosphatase 2) and was associated with increased risk of acute kidney injury. PHLPP2 is a phosphatase, important for the regulation of Akt kinases and PKC isoforms [21, 22]. Akt belongs to the so called pro-survival kinases, involved in the protective pathway during myocardial ischemia/ reperfusion [23]. Akt controls the balance between cell survival and apoptosis, as well as proliferation and cellular quiescence. Activation of PI3K/Akt seems to be protective against I/R injury. PHLPP2 dephosphorylates Akt (precisely Akt1 and Akt3) and therefore inactivates Akt. PHLPP2 inhibition leads to neuronal protection after cerebral ischemia/reperfusion injury in rats [24, 25]. The imbalance of cell pro-death and pro-survival signaling pathways determines the neuronal fate during ischemia/reperfusion injury. In a rat model of I/R injury it was shown that inhibition of PHLPP2 attenuates cell death in I/R injury. Very recently, it was demonstrated that PHLPP2 plays a pivotal role in acetaminophen induced oxidative renal toxicity by influencing Nrf2 stability via Akt1/Gsk3b/Fyn kinase axis [26]. Down regulation of PHLPP2 by morin, a bioflavonoid, significantly prevented the toxicity induced renal damages. In this respect, down regulation of PHLPP2 may provide positive effects in the kidney and in the brain, two vital organs affected by cardiac surgery. In line with our GWAS of complications after cardiac surgery we found the SNP rs78064607 located in the PHLPP2 gene with a genome-wide significance. Because the SNP is located in the intron region or intergenic region, respectively, of the gene, the exact effect on the PHLPP2 gene cannot be evaluated. Probably an enhancer/ silencer region is affected, leading to up or down regulation of the PHLPP2 gene. Association of possible changes in the expression of PHLPP2 with increased occurrence of complications, especially RENFAIL after cardiac surgery could be a hind of an increased expression of PHLPP2. But this assumption is highly speculative and has to be confirmed by further gene expression analyses.
ZBTB20 (Zinc finger and BTB domain containing 20, rs115155878 is related to this gene) is widely expressed in human hematopoietic cells, including DCs, monocytes, B cells and T cells. ZBTB20 deficiency in mice attenuated TLR-triggered production of pro inflammatory cytokines in macrophages [27]. This could have an influence of mechanisms within the scope of I/R.
For stroke, rs4098926, the SNP with the lowest p-value for this complication is located in the BBS9 gene. BBS9 is also known as parathyroid hormone-responsive B1 gene (PTHB1). Mutations of BBS9 are related with the Bardet-Biedl syndrome, a rare genetic disorder with highly variable symptoms. The underlying cause is malfunction of primary cilia, a key component of cellular communication function as signal transduction antennae. Kidney disease is a key feature and major cause of early mortality of patients with Bardet-Biedl syndrome. Intact cilia are critical under kidney injury conditions caused by ischemia/ reperfusion, because cilia are sensors of damages and activate cell proliferation probably to promote renal recovery [28, 29]. Changes in BBS9 could contribute to higher complication rate concerning the kidney in patients undergoing cardiac surgery and further investigations of involvement of BBS9 in postoperative renal injuries are worthwhile. Additionally, the exact role of BBS9 in the pathogenesis of stroke after cardiac surgery has to be evaluated.
rs192540202 is located in an intron region of RyR2 (ryanodine receptor 2). RyR2 is primarily found in cardiac muscle and forms a Ca2+ release channel on the membrane of the sarcoplasmic reticulum. Abnormal RyR2 function is recognized as an important part of the pathophysiology of heart failure, especially contractile dysfunction, arrhythmia and sudden death [30, 31]. Numerous studies revealed that abnormal Ca2+ homeostasis may play an important role in the electric and contractile remodeling accompanying sustained atrial fibrillation [32, 33]. Very recently, Xie et al. identified a link between oxidative stress and RyR2 [34]. Mice with mutations in the RyR2 receptor exhibited mitochondrial dysfunction, increased reactive oxygen species production and increased AF susceptibility. Because oxidative stress and mitochondria dysfunction plays a pivotal role in the pathogenesis of I/R injury of organs, changes in RyR2 receptor associated with disturbed Ca2+ homeostasis could contribute to higher risk of complication after cardiac surgery.
RyR2 also has an impact on Ca2+ homeostasis in the brain during cerebral ischemia [35]. In a rat model of brain ischemia, Bull et al. could demonstrate that amplification of Ca2+ by RyR2 entry signals may contribute to cortical neuronal death.
Very interestingly, knockdown of RyR2 in a spinal cord injury model in rats inhibited the increase of pro-inflammatory cytokines, improved mitochondrial dysfunction and reduced oxidative stress [36]. Because release of pro-inflammatory cytokines, massive ROS production and mitochondrial dysfunction are the main causes of I/R injury of organs, examination of the exact role of RyR2 could be very interesting.
rs77876049 is located in the HSPA8 gene. Although this SNP has a higher p-value, it is reasonable to have a closer look at this protein because of its interesting involvement in the mechanisms of I/R injury. HSPA8 (Heat Shock Protein 8, also known as Hsc70 or Hsp73) is a member of the heat shock protein 70 family and facilitates the correct folding of newly translated or misfolded proteins. HSPA8 plays an important role in signal transduction, apoptosis, protein homeostasis, cell growth and differentiation. Zou et al. could demonstrate that HSPA8, constitutively expressed in the myocardium, is released during ischemia/ reperfusion and induces the myocardial inflammatory response and modulates cardiac function [37]. Acute myocardial ischemia can lead to a cascade of cellular and ischemic tissue, causing irreversible damage. In myocardial ischemia and reperfusion, the myocardial cells release HSPA8 and reduce myocardial cell injury [38]. Thus, HSPA8 plays a critical role in regulating the myocardial innate immune system and cardiac function after ischemia/ reperfusion. Probably, HSPA8 specifically has a protective effect in patients undergoing open heart surgery [39]. It has also been shown that Chaperone-mediated autophagy (CMA), under involvement of HSPA8, of damaged or leaky RyR2 receptors after I/R may play a protective role after I/R injury and could contribute to myocardial remodeling [40]. A combined functional impairment of HSPA8 and RyR2 in patients undergoing cardiac surgery could contribute to increased myocardial complications because of lack of functioned RyR2 receptors and the inability to remove damaged RyR2 receptors by CMA. HSPA8 also plays a protective role in the process of ischemic stroke by protection of nerve cells and inhibition of neuronal apoptosis [41,42,43]. HSPA8 seems to play an important role in the regulation of cellular processes after I/R injury both in the heart and in the brain. Therefore, patients with variants in this gene might have an increased complication rate after cardiac surgery. So, HSPA8 might be a prognostic factor, but validation of these findings will require additional studies with independent subject panels.
None of the further identified genetic variants associated with atrial fibrillation [44, 45], myocardial infarction [46, 47], stroke [48] or renal dysfunction [49] in non-surgical patients was found in our analyses. In contrast, some of the described genetic variants associated with complications after cardiac surgery, namely BBS9 in renal dysfunction [5], were found in our study. This indicates a unique pathogenesis in the subset of ischemia/ reperfusion after cardiac surgery that differs from pathogenesis in non-surgical patients. Surprisingly, neither Kertai et al. [3] nor our study could replicate previously reported associations between common genetic variants at the 9p21 locus and risk for myocardial infarction after cardiac surgery [50, 51]. Probably, variations in study design or differences in data analysis could be reasons for these variations and further studies are needed to explain these discrepancies.
Conclusions
Here we report the first GWAS in a cohort of patients at risk of AF, delirium, MI, acute renal failure and stroke after cardiac surgery. We identified several polymorphisms associated with these complications. In most cases, loci are noncoding, and many loci are far from discovered genes in non-coding regions, the effects of SNPs on genes are completely unknown or the functions of the influenced genes are unknown. Furthermore, GWAS almost exclusively detects the effects of common SNPs, any rare variants will not be detected. Nevertheless, we identified some very interesting potential correlations between genetic polymorphisms and the occurrence of complications. In particular, the described concurrence of HSPA8 and RyR2 for atrial fibrillation and myocardial infarction, the involvement of DUSP4 in I/R injury, the role of PHLPP2 in developing complications after cardiac surgery and the involvement of BBS9 in renal dysfunction could be interesting for further future examinations. Follow-up studies are needed to transfer these findings into biological insights that could result in predictive and therapeutic advances in the perioperative care of cardiac surgery patients.
Abbreviations
- AF:
-
Atrial Fibrillation
- GWAS:
-
Genome Wide Association Analysis
- ICU:
-
Intensive Care Unit
- MI:
-
Myocardial Infarction
- SNP:
-
Single Nucleotide Polymorphism
References
Carden DL, Granger DN. Pathophysiology of ischaemia-reperfusion injury. J Pathol. 2000;190(3):255–66.
Kraft F, Schmidt C, van Aken H, Zarbock A. Inflammatory response and extracorporeal circulation. Best Pract Res Clin Anaesthesiol. 2015;29(2):113–23.
Kertai MD, Li Y-J, Li Y-W, Ji Y, Alexander J, Newman MF, et al. Genome-wide association study of perioperative myocardial infarction after coronary artery bypass surgery. BMJ Open. 2015;5(5):e006920.
Kertai MD, Li Y-J, Ji Y, Qi W, Lombard FW, Shah SH, et al. Genome-wide association study of new-onset atrial fibrillation after coronary artery bypass grafting surgery. Am Heart J. 2015;170(3):580–90.e28.
Stafford-Smith M, Li Y-J, Mathew JP, Li Y-W, Ji Y, Phillips-Bute BG, et al. Genome-wide association study of acute kidney injury after coronary bypass graft surgery identifies susceptibility loci. Kidney Int. 2015;88(4):823–32.
Meybohm P, Bein B, Brosteanu O, Cremer J, Gruenewald M, Stoppe C, et al. A multicenter trial of remote ischemic preconditioning for heart surgery. N Engl J Med. 2015;373(15):1397–407.
Meybohm P, Zacharowski K, Cremer J, Roesner J, Kletzin F, Schaelte G, et al. Remote ischaemic preconditioning for heart surgery. The study design for a multi-center randomized double-blinded controlled clinical trial--the RIPHeart-study. Eur Heart J. 2012;33(12):1423–6.
Thygesen K, Alpert JS, White HD. Universal definition of myocardial infarction. J Am Coll Cardiol. 2007;50(22):2173–95.
Wityk RJ, Pessin MS, Kaplan RF, Caplan LR. Serial assessment of acute stroke using the NIH stroke scale. Stroke. 1994;25(2):362–5.
Bellomo R, Ronco C, Kellum JA, Mehta RL, Palevsky P. Acute renal failure - definition, outcome measures, animal models, fluid therapy and information technology needs: the second international consensus conference of the acute dialysis quality initiative (ADQI) group. Crit Care. 2004;8(4):R204–12.
Ely EW, Inouye SK, Bernard GR, Gordon S, Francis J, May L, et al. Delirium in mechanically ventilated patients: validity and reliability of the confusion assessment method for the intensive care unit (CAM-ICU). JAMA. 2001;286(21):2703–10.
Goldstein JI, Crenshaw A, Carey J, Grant GB, Maguire J, Fromer M, et al. zCall: a rare variant caller for array-based genotyping: genetics and population analysis. Bioinformatics. 2012;28(19):2543–5.
Abraham G, Inouye M. Fast principal component analysis of large-scale genome-wide data. PLoS One. 2014;9(4):e93766.
Delaneau O, Marchini J, Zagury J-F. A linear complexity phasing method for thousands of genomes. Nat Methods. 2011;9(2):179–81.
Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39(7):906–13.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75.
Szymczak S, Holzinger E, Dasgupta A, Malley JD, Molloy AM, Mills JL, et al. r2VIM: a new variable selection method for random forests in genome-wide association studies. BioData Min. 2016;9:7.
Nashef SA, Roques F, Michel P, Gauducheau E, Lemeshow S, Salamon R. European system for cardiac operative risk evaluation (EuroSCORE). Eur J Cardiothorac Surg. 1999;16(1):9–13.
Barajas-Espinosa A, Basye A, Angelos MG, Chen C-A. Modulation of p38 kinase by DUSP4 is important in regulating cardiovascular function under oxidative stress. Free Radic Biol Med. 2015;89:170–81.
Dougherty JA, Kilbane Myers J, Khan M, Angelos MG, Chen C-A. Dual-specificity phosphatase 4 overexpression in cells prevents hypoxia/Reoxygenation-induced apoptosis via the upregulation of eNOS. Front Cardiovasc Med. 2017;4:22.
Cai J, Fang L, Huang Y, Li R, Yuan J, Yang Y, et al. miR-205 targets PTEN and PHLPP2 to augment AKT signaling and drive malignant phenotypes in non-small cell lung cancer. Cancer Res. 2013;73(17):5402–15.
Xu J, Wang Y, Hua X, Xu J, Tian Z, Jin H, et al. Inhibition of PHLPP2/cyclin D1 protein translation contributes to the tumor suppressive effect of NFkappaB2 (p100). Oncotarget. 2016;7(23):34112–30.
Kunuthur SP, Mocanu MM, Hemmings BA, Hausenloy DJ, Yellon DM. The Akt1 isoform is an essential mediator of ischaemic preconditioning. J Cell Mol Med. 2012;16(8):1739–49.
Jiao S, Zhu H, He P, Teng J. Betulinic acid protects against cerebral ischemia/reperfusion injury by activating the PI3K/Akt signaling pathway. Biomed Pharmacother. 2016;84:1533–7.
Wei X-E, Zhang F-Y, Wang K, Zhang Q-X, Rong L-Q. Assembly of the FKBP51-PHLPP2-AKT signaling complex in cerebral ischemia/reperfusion injury in rats. Brain Res. 2014;1566:60–8.
Mathur A, Rizvi F, Kakkar P. PHLPP2 down regulation influences nuclear Nrf2 stability via Akt-1/Gsk3beta/Fyn kinase axis in acetaminophen induced oxidative renal toxicity: protection accorded by morin. Food Chem Toxicol. 2016;89:19–31.
Liu X, Zhang P, Bao Y, Han Y, Wang Y, Zhang Q, et al. Zinc finger protein ZBTB20 promotes toll-like receptor-triggered innate immune responses by repressing IkappaBalpha gene transcription. Proc Natl Acad Sci U S A. 2013;110(27):11097–102.
Weimbs T. Polycystic kidney disease and renal injury repair: common pathways, fluid flow, and the function of polycystin-1. Am J Physiol Renal Physiol. 2007;293(5):F1423–32.
Wang S, Dong Z. Primary cilia and kidney injury: current research status and future perspectives. Am J Physiol Renal Physiol. 2013;305(8):F1085–98.
Jiang D, Xiao B, Yang D, Wang R, Choi P, Zhang L, et al. RyR2 mutations linked to ventricular tachycardia and sudden death reduce the threshold for store-overload-induced Ca2+ release (SOICR). Proc Natl Acad Sci U S A. 2004;101(35):13062–7.
Belevych AE, Radwanski PB, Carnes CA, Gyorke S. ‘Ryanopathy’: causes and manifestations of RyR2 dysfunction in heart failure. Cardiovasc Res. 2013;98(2):240–7.
Sun H, Gaspo R, Leblanc N, Nattel S. Cellular mechanisms of atrial contractile dysfunction caused by sustained atrial tachycardia. Circulation. 1998;98(7):719–27.
Goette A, Honeycutt C, Langberg JJ. Electrical remodeling in atrial fibrillation. Time course and mechanisms. Circulation. 1996;94(11):2968–74.
Xie W, Santulli G, Reiken SR, Yuan Q, Osborne BW, Chen BX, et al. Mitochondrial oxidative stress promotes atrial fibrillation. Sci Rep. 2015;5:11427.
Bull R, Finkelstein JP, Gálvez J, Sánchez G, Donoso P, Behrens MI et al.. Ischemia enhances activation by Ca2+ and redox modification of ryanodine receptor channels from rat brain cortex. J Neurosci. 2008; 28(38):9463–9472. Available from: URL: http://www.jneurosci.org/content/jneuro/28/38/9463.full.pdf.
Liao B, Zhang Y, Sun H, Ma B, Qian J. Ryanodine receptor 2 plays a critical role in spinal cord injury via induction of oxidative stress. Cell Physiol Biochem. 2016;38(3):1129–37.
Zou N, Ao L, Cleveland JC Jr, Yang X, Su X, Cai G-Y, et al. Critical role of extracellular heat shock cognate protein 70 in the myocardial inflammatory response and cardiac dysfunction after global ischemia-reperfusion. Am J Physiol Heart Circ Physiol. 2008;294(6):H2805–13.
Ao L, Zou N, Cleveland JC Jr, Fullerton DA, Meng X. Myocardial TLR4 is a determinant of neutrophil infiltration after global myocardial ischemia: mediating KC and MCP-1 expression induced by extracellular HSC70. Am J Physiol Heart Circ Physiol. 2009;297(1):H21–8.
Demidov ON, Tyrenko VV, Svistov AS, Komarova YY, Karpishenko AI, Margulis BA, et al. Heat shock proteins in cardiosurgery patients. Eur J Cardiothorac Surg. 1999;16(4):444–9.
Pedrozo Z, Torrealba N, Fernandez C, Gatica D, Toro B, Quiroga C, et al. Cardiomyocyte ryanodine receptor degradation by chaperone-mediated autophagy. Cardiovasc Res. 2013;98(2):277–85.
Liu Y, Jiang S, Yang PY, Zhang YF, Li TJ, Rui YC. EF1A1/HSC70 cooperatively suppress brain endothelial cell apoptosis via regulating JNK activity. CNS Neurosci Ther. 2016;22(10):836–44.
Chen S, Brown IR. Translocation of constitutively expressed heat shock protein Hsc70 to synapse-enriched areas of the cerebral cortex after hyperthermic stress. J Neurosci Res. 2007;85(2):402–9.
McLaughlin B, Hartnett KA, Erhardt JA, Legos JJ, White RF, Barone FC, et al. Caspase 3 activation is essential for neuroprotection in preconditioning. Proc Natl Acad Sci U S A. 2003;100(2):715–20.
Darbar D, Roden DM. Genetic mechanisms of atrial fibrillation: impact on response to treatment. Nat Rev Cardiol. 2013;10(6):317–29.
Ellinor PT, Lunetta KL, Albert CM, Glazer NL, Ritchie MD, Smith AV, et al. Meta-analysis identifies six new susceptibility loci for atrial fibrillation. Nat Genet. 2012;44(6):670–5.
Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, et al. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357(5):443–53.
McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316(5830):1488–91.
Ikram MA, Seshadri S, Bis JC, Fornage M, DeStefano AL, Aulchenko YS, et al. Genomewide association studies of stroke. N Engl J Med. 2009;360(17):1718–28.
Zhao B, Lu Q, Cheng Y, Belcher JM, Siew ED, Leaf DE, et al. A Genome-Wide Association Study to Identify Single Nucleotide Polymorphisms for Acute Kidney Injury. Am J Respir Crit Care Med. 2017;195(4):482–90.
Liu KY, Muehlschlegel JD, Perry TE, Fox AA, Collard CD, Body SC, et al. Common genetic variants on chromosome 9p21 predict perioperative myocardial injury after coronary artery bypass graft surgery. J Thorac Cardiovasc Surg. 2010;139(2):483.
Muehlschlegel JD, Liu K-Y, Perry TE, Fox AA, Collard CD, Shernan SK, et al. Chromosome 9p21 variant predicts mortality after coronary artery bypass graft surgery. Circulation. 2010;122(11 Suppl):S60–5 References.
Acknowledgements
We thank all the patients who participated in the study and the clinical and research staff at all trial sites, without whose assistance the study would never have been completed.
RIPHeart-Study Collaborators.
Aachen (Department of Anaesthesiology, Medical Faculty RWTH Aachen, University Aachen, Germany): Ana Stevanovic, Rolf Rossaint, Marc Felzen, (Department of Thoracic and Cardiovascular Surgery): Andreas Goetzenich; 195 patients;
Berlin (Department of Anaesthesiology and Intensive Care Medicine, Charité-Universitätsmedizin Berlin, Campus Charité Mitte, Berlin, Germany): Tobias Moormann, Katharina Chalk; 37 patients;
Bonn (Department of Anaesthesiology and Intensive Care Medicine, University Hospital Bonn, Bonn, Germany): Pascal Knuefermann, Olaf Boehm, Andreas Hoeft; 73 patients;
Duesseldorf (Department of Anaesthesiology and Intensive Care Medicine, University Hospital Duesseldorf, Germany): Michael Winterhalter; 65 patients;
Frankfurt am Main (Department of Anaesthesiology, Intensive Care Medicine and Pain Therapy, University Hospital Frankfurt, Frankfurt am Main, Germany): Sonja Iken, Christian Weber, Carolin Wiedenbeck, Gerhard Schwarzmann, Karin Pense, (Department of Thoracic and Cardiovascular Surgery): Andreas Zierer, (Internal Medicine III: Cardiology, Angiology, Nephrology): Stephan Fichtlscherer; 117 patients;
Giessen (Department of Cardiovascular Surgery, University of Giessen, Germany): Gerold Goerlach, Matthias Wollbrueck, Ursula Boening; (Department of Anesthesiology): Markus Weigand; 148 patients;
Goettingen (Department of Anaesthesiology and Intensive Care Medicine, University Hospital Goettingen, Germany): Julia Strauchmann, Konrad August; 91 patients;
Jena (Department of Anesthesiology and Intensive Care Medicine, Jena University Hospital, Jena, Germany): Kai U. Morsbach, Markus Paxian, Konrad Reinhard; 76 patients;
Kiel (Department of Anaesthesiology and Intensive Care Medicine, University Hospital Schleswig-Holstein, Campus Kiel, Germany): Matthias Gruenewald, Jens Scholz, Ole Broch, Helga Francksen, Martin Albrecht, Bernd Kuhr; 237 patients;
Luebeck (Department of Anaesthesiology, University Hospital Luebeck, Luebeck, Germany): Hermann Heinze, Hauke Paarmann; (University Heart Center Luebeck, Medical Clinic II (Cardiology/Angiology/Intensive Care Medicine), University Hospital Schleswig-Holstein, Luebeck): Hans-Hinrich Sievers, Stefan Klotz; 56 patients;
Magdeburg (Department of Anaesthesiology, University Hospital Magdeburg, Germany); Thomas Hachenberg; 14 patients;
Mainz (Department of Anesthesiology, Medical Center of Johannes Gutenberg-University, Mainz, Germany): Christian Werner, Susanne Mauff; 116 patients;
Rostock (Clinic of Anaesthesiology and Intensive Care Medicine, University Hospital Rostock, Rostock, Germany): Angela Alms, Stefan Bergt; 146 patients;
Wuerzburg (Department of Anaesthesiology, University Hospital Wuerzburg, Wuerzburg, Germany): Norbert Roewer; 32 patients
Funding
This work was supported by a grant (ME 3559/1–1) from the German Research Foundation with funds for staff and required gene chips for analysis. The funding had no impact on the design of the study and the collection and the scientific analysis and interpretation of data as well as in writing the manuscript.
Availability of data and materials
The present manuscript was not published anywhere. This manuscript is a sub analysis of the RIPHeart study “A multicentre trial of remote ischemic preconditioning for heart surgery” that has been published in NEJM (2015 Oct 8; 373 [15]).
Supplementary material will be provided with the manuscript or are already published in NEJM. No further additional unpublished data are available.
Author information
Authors and Affiliations
Consortia
Contributions
PM, BB, JC, DH, OB and KZ designed the study. CS, BB, MG, JRe, JC, MC, GS, AB, BN, FK, JRo, US, CR, RLF, MF, IFB, MB, AK, SNS, MW, GB, TMT, PK, MH, JS, MS, ST, TS, EW, TS, GF, PM, KZ and the RIPHeart collaborators made acquisition of data. SW, FD, AF, SM and LT made analysis and interpretation of data. SW, RS, MK and PM were involved in drafting the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The ethics committees of the Medical Faculty of Christian-Albrechts-University of Kiel, Germany, and of all participating centers approved the study protocol, patient information and informed consent. Each patient gave written informed consent for DNA analysis. The study was registered with ClinicalTrials.gov NCT01067703, prospectively registered on 11 Feb 2010.
Consent for publication
Not applicable.
Competing interests
No competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Figure S3. English synopsis of study protocol. (PDF 105 kb)
Additional file 2:
Figure S4. Study protocol of the RIPHeart study. (PDF 4311 kb)
Additional file 3:
Table S1. List of adjustment variables. (PDF 65 kb)
Additional file 4:
Figure S1. Manhattan Plot of genome-wide association with AF, delirium, MI, renal failure and stroke. The x-axis represents the chromosomes in physical order, the y axis showing –log10(p) for all single nucleotide polymorphisms (SNPs). a) Adjustment for all other outcomes: one SNP reached genome-wide significance (p < 5 × 10− 8, red line) and 139 SNPs reached the predefined threshold of p < 1 × 10− 5 (blue line). b) No adjustment for all other outcomes and composite: 132 SNPs reached the predefined threshold of p < 1 × 10− 5. (PDF 498 kb)
Additional file 5:
Figure S2. Quantile-quantile plot showing the expected distribution of association test statistics across the SNPs compared to the observed values for AF, delirium, MI, renal failure and stroke. a) Adjustment for all other outcomes. b) No adjustment for all other outcomes and composite. (PDF 305 kb)
Additional file 6:
Table S2. Complete table of SNPs reaching the predefined threshold of p < 1 × 10− 5 with and without adjustment for all other complications. (PDF 1177 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Westphal, S., Stoppe, C., Gruenewald, M. et al. Genome-wide association study of myocardial infarction, atrial fibrillation, acute stroke, acute kidney injury and delirium after cardiac surgery – a sub-analysis of the RIPHeart-Study. BMC Cardiovasc Disord 19, 26 (2019). https://doi.org/10.1186/s12872-019-1002-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12872-019-1002-x