Abstract
Background
Floating bamboo (Hygroryza aristata) is an endangered species with a narrow native distribution and is renowned for its unique aesthetic qualities, which holds significant ecological and ornamental value. However, the lack of genetic information research, with only one complete plastome available, significantly hampers conservation efforts and further research for this species.
Results
In this research, we sequenced and assembled the organelle genomes of floating bamboo, including the mitogenome (587,847 bp) and plastome (135,675 bp). The mitogenome can recombine into various configurations, which are mediated by 25 repeat pairs (13 SRs, 6 MRs, 1 LR, and 5 CRs). LR1 and SR5 are particularly notable as they have the ability to combine with other contigs, forming complex repeat units that facilitate further homologous recombination. The rate of homologous recombination varies significantly among species, yet there is still a pronounced positive correlation observed between the length of these repeat pairs and the rate of recombination they mediate. The mitogenome integrates seven intact protein-coding genes from the chloroplast. The codon usage patterns in both organelles are similar, with a noticeable bias towards C and T on the third codon. The gene map of Poales shows the entire loss of rpl6, succinate dehydrogenase subunits (sdh3 and sdh4). Additionally, the BOP clade retained more variable genes compared to the PACMAD clade.
Conclusions
We provided a high-quality and well-annotated mitogenome for floating bamboo and demonstrated the presence of diverse configurations. Our study has revealed the correlation between repeat length and their corresponding recombination rate despite variations among species. Although the mitogenome can potentially exist in the form of a unicircular in vivo, this occurrence is rare and may not be stable.
Similar content being viewed by others
Backgroud
Hygroryza aristata, also known as floating bamboo, is an aquatic, perennial grass renowned for its ability to float on the surface of slowly flowing freshwater bodies due to its spongy, inflated leaf sheaths [1, 2]. H. aristata is native only to the tropical regions of Southeast Asia [1, 2]. However, due to the limited native distribution range, habitat destruction and fragmentation, as well as extensive use as feed for fish and livestock [3, 4], the rarity of this species is increasing rapidly [5]. In 2021, it was classified as a second-level nationally protected wild plant in China. Meanwhile, owing to its unique aesthetic qualities, this species is widely favored by aquarium enthusiasts worldwide, posing some invasive risks outside of its native range. Increasing concerns about water pollution in recent years have brought unprecedented attention to H. aristata as a natural material for water purification [6, 7]. Additionally, it serves as a source of chemical compounds, such as lignan and indole alkaloids, which are utilized in traditional medicine for their anti-inflammatory and antioxidant properties [8]. However, research on its genetic information is severely lacking. Currently, there is only one complete chloroplast record of H. aristata available [9], severely hampering conservation and research efforts for this species. Poaceae is the fifth-largest family in angiosperm (~ 12,000 species in 12 subfamilies) and holds the most significant economic importance for humans [10]. It includes essential food sources such as rice, maize, wheat, as well as numerous forage and biofuel species. Members of the Oryzoideae subfamily (such as Oryza and Hygroryza) are more adapted to aquatic habitats, unlike most other grasses that thrive in dry ecosystems [11].
Organelles believed to have originated from ancient endosymbiotic events are present in most eukaryotes [12, 13]. These organelles retain their genomes which originated independently from nuclear genomes. However, only a limited set of genes remains in these genomes to maintain primary biogenesis during long-term symbiosis [12]. Nonetheless, the biogenesis of these organelles also relies on the coordinated expression of genes in the nucleus, rendering them semiautonomous. Plants possess two organelles: the chloroplast (cp) and the mitochondrion (mt). Chloroplast can be considered the cornerstone of ecosystems as an essential organelle for carbon fixation via photosynthesis. Studies have confirmed that plant mitochondria are crucial for respiration, metabolism, programmed cell death (similar to animal mitochondria) and play a significant role in cytoplasmic male sterility breeding [14,15,16], making them an indispensable component in breeding research.
As of January 2024, NCBI has published complete plastomes of 6,116 land plants, while the number of complete mitogenomes is only 272 (this count is slightly lower as it includes only mitogenomes with a single chromosomal structure; Table 1). This stark contrast is attributed to the complexity of the mitogenome in embryophyta. In comparison to the relatively compact and conserved genomes of plant chloroplasts and animal mitochondria, plant mitogenomes exhibit unique features, including extensive variation in size (Table 1), frequent genome rearrangements, the incorporation of DNA from various sources through intracellular and horizontal transfer, and extensive RNA editing sites [17,18,19]. The variability of plant mitochondrial genomes can even fluctuate significantly within a single species [20, 21]. Among the major branches of eukaryotes, embryophyta mitochondria have the largest genome size and the highest GC content (~ 44.16%; Table 1). These distinctive features among plant mitogenomes may elucidate why so few have been appropriately assembled and characterized. However, despite the structural variability of plant mitochondria, the coding sequences within their genomes are highly conserved in terms of both number and genetics [12, 22].
Although most mitogenomes can be mapped as a unicircular structure on physical maps, referred to as the “master circle” model, more experimental evidence has confirmed that this is not the dominant structure of mitochondria in vivo [22, 23]. Instead, they are constantly involved in dynamic homologous recombination (HR) mediated by repeat pairs, resulting in a dynamic mixture of small-sized forms, including branched linear, circular, linear, degraded, comet, and branched circular structures, among others [23, 24]. HR is an important and evolutionarily conserved homology-dependent DNA repair process used to eliminate potentially harmful lesions, particularly double-stranded breaks (DSBs) [18]. HR also exists in the plastome, albeit at a lower frequency. Fluorescence in situ hybridization (FISH) shows that reorganized cpDNA represent between 0.8% and 2% of all [25]. Most published plant mitogenomes contain at least one pair of repeats that can serve as sites for inter- or intramolecular recombination, resulting in multiple alternative arrangements (isoforms). For now, the only exception is Brassica hirta; restriction mapping studies have revealed that, unlike other cogeneric species, the mitochondrion of B. hirta exists in vivo as a slightly smaller single circular chromosome [26]. It has been proposed that the large repeats can frequently recombine intra- or intermolecularly, while the small repeats may lead to subtle rearrangements [18]. However, quantitative studies based on long sequencing reads show significant variations among species [27,28,29,30,31,32,33,34,35,36].
In this research, we successfully: (1) utilized tissue culture techniques for the conservation of floating bamboo germplasm resources; (2) sequenced and assembled the mitogenome of floating bamboo; (3) verified the positive correlation between the length of repeat sequences and the mitogenome recombination rate; (4) further investigated sequence transfer between mitochondrion and chloroplast, revealing similarities in codon usage bias between these two organelles; (5) explored gene loss pattern in the mitogenome of Poales.
Methods
Plant material and genome sequencing
The sample of Hygroryza aristata was collected from Wuhan, China and preserved at the Institute of Hydrobiology, Chinese Academy of Sciences through tissue culture (Fig. S1). Genomic DNA was extracted from fresh leaves using a modified CTAB method [37]. The purified DNA was used to prepare libraries for both Illumina (VAHTS Universal Plus DNA Library Prep Kit, Vazyme Biotech, Nanjing, China) and Nanopore (SQK-LSK110 Ligation Sequencing Kit, Oxford Nanopore Technologies, Oxford, UK) following the manufacturers’ protocols. The Illumina library was sequenced on the Illumina Hiseq4000 platform (Illumina, San Diego, USA). Raw reads were filtered by Fastp v0.23.2 [38] with the parameters “qualified_quality_phred 20; --unqualified_percent_limit 50; --length_required 35; --detect adapter_for_pe”. The Nanopore library was sequenced on the PromethION P48 sequencer (Oxford Nanopore Technologies, Oxford, UK), base calling and prefiltering were performed by Guppy v6.3.8 (Oxford Nanopore Technologies, Oxford, UK). To ensure that our data were sufficient for the assembly of organelles, we utilized Jellyfish v2.2.10 [39] and GenomeScope v2.0 [40] to assess the whole genome size based on k-mer.
Assembly and annotation of organelle genomes
The chloroplast genome was assembled using GetOrangelle v1.7.7.0 [41] with Illumina whole-genome sequencing (WGS) data as input.
To detect repeat sequences in the mitogenome of H. aristata that could potentially facilitate recombination and provide insights into its variable conformations within the organism, we employed a sophisticated assembly strategy. 1). We utilized both Flye v2.9.3 [42] and Unicycler v0.5.0 [43] with default parameters to initially assemble the Nanopore long reads. 2). All contigs were searched against the complete set of mitochondrial protein-coding genes encoded by angiosperm using BLASTn v2.13.0+ [44] with an e-value cutoff of 1e-5. 3). Then, we used 44 candidate contigs as references to identify potential mitochondrial reads from the Illumina WGS data using BWA-MEM2 v2.2.1 [45]. 4). Following this, a de novo assembly was performed on the extracted Illumina reads using SPAdes v3.15.5 (k-mer = 27, 53, 71, 87, 99, 111, 119, 127) [46]. 5). The assembled graph was then simplified by removing edges with less than 10× coverage depth. Additionally, chloroplast- and nuclear-derived nodes were manually removed using the interactive visualization interface of Bandage v0.8.1 [47]. During this process, we detected several repeats that potentially mediated HR, as they exhibited multiple connections and doubled coverage depth. 6). Finally, with the assistance of Nanopore long reads, we successfully obtained the complete mitogenome of H. aristata as a single circular molecule (i.e., the “master circle” model).
The annotation for the cp and mt genomes was conducted using GeSeq [48], with reference to the previously released cp genome of H. aristata (NCBI accession number NC_058302.1) and the available mitogenomes of Poales (Table S1), respectively. Additionally, tRNAs were validated by tRNAscan-SE with default settings [49]. The PCGs underwent manual verification and editing using Geneious Prime R9.0.2 [50]. Finally, both circular organelles were visualized using OGDRAW [51].
Detection of genome recombination
During the process of assembling the mitogenome, we detected 25 repeat sequences that have the potential to facilitate HR. Although a “master circle” mitogenome was obtained with the assistance of Nanopore long reads, it is essential to acknowledge that this assembly merely represents one possible configuration. To confirm the occurrence of HR and to evaluate the correlation between the length of repeat sequences and their associated recombination frequency, Nanopore long reads were mapped to these presumed conformations. Each repeat had two paths indicating the primary conformation (m1 and m2) and two paths representing the secondary conformation (s1 and s2). Utilizing minimap2 v2.26 [52], Nanopore long reads were individually aligned to these four hypothetical conformations. Each hypothetical path included the repeat sequence and its neighboring contigs. If the adjacent contig was shorter than 1 kb, an additional contig was included to ensure alignment accuracy. Only reads that aligned with the entire repeat sequence and also included regions extending at least 100 bp on both flanks of the repeat sequence were deemed supportive of that specific configuration. If two paths supported the same conformation (m1 and m2, s1 and s2), only the path with the highest count was acknowledged. Recombination frequency of m1/m2 (Fm) is calculated as max(m1|m2). Similarly, the recombination frequency of s1/s2 (Fs) is calculated as max(s1|s2). The recombination rate is determined by min(Fm|Fs)/(Fm+Fs). Therefore, according to this calculation method, the recombination rate should range from 0 to 50%. Meanwhile, we have included ten other angiosperm species that have been subject to qualitative assessment of recombination frequency in prior studies [27,28,29,30,31,32,33,34,35,36] while investigating the inter-species variations in repeat sequences.
Detection of intracellular gene transfer
Currently, only the two organelle genomes are available for identifying intracellular sequence migration in H. aristata due to the lack of a published nuclear genome. To identify homologous sequences that could potentially be transferred among the organelles, we compared the cpDNA and mtDNA using BLASTn with the following parameters: “-evalue 1e-5; -word_size 7; -max_hsps 10”. The identified transferred DNA fragments were extracted and annotated using GeSeq. Subsequently, these results were visualized using Tbtools v2.065 [53].
Analysis of repeat elements
We analyzed three different types of repeat sequences in both organelles. (1) Simple sequence repeats (SSRs), also known as microsatellites, were identified using MISA [54], with parameter settings of 10, 5, 4, 3, 3, and 3 for mono-, di-, tri-, tetra-, penta-, and hexanucleotide repeats, respectively. (2) An online tool REPuter [55] was utilized to identify four types of dispersed repeats, namely forward (F), palindromic (P), reverse (R), and complement (C) repeats (Hamming Distance = 3; Minimal Repeat Size = 30 bp). (3) Tandem repeat elements were identified using TRF v4.09 [56] (Match = 2; Mismatch = 7; Delta = 7; PM = 80; PI = 10; Minscore = 50; MaxPeriod = 500).
Codon usage bias
To investigate the bias pattern of codon usage in these two organelles, we extracted a total of 51 cp CDS and 34 mt CDS, following the criteria outlined by Zhang et al. [57]. The basic compositional properties of these genes, including overall GC content, GC1/2/3 (the GC content at the first, second, and third base position of the codons), GC12 (the mean of GC1 and GC2), as well as GC3s (the GC content at the third base position of synonymous codons, excluding Met, Trp, and the three stop codons), were calculated using EMBOSS v6.6.0 [58]. Subsequently, CodonW v1.4.2 [59] was employed to conduct the codon usage analysis, which included calculating the effective number of codons (ENC) and determining the relative synonymous codon usage (RSCU). The ENC value ranges from 20 (extreme bias) to 61 (no bias), indicating the degree of codon usage bias. Typically, a threshold of 35 is used to evaluate the strength of codon preference [60]. Neutrality plot analysis, ENC-plot analysis, and Parity rule 2 (PR) plot analysis were performed following the methods described by Liu et al. (2020) [61].
Identification of RNA editing sites
To identify RNA editing sites occurring in PCGs, we downloaded a transcriptomic data from NCBI (SRR16192102). We then mapped the RNA-sequencing data to the PCGs extracted from the organelle genomes using BWA-MEM2. Subsequently, we utilized REDItools v2.0 [62] to analyze the base composition and coverage of each site, with the following parameters: --min-read-length 50; --min-read-quality 35. For high-copy chloroplast PCGs, a minimum coverage of 20× and at least 10% read support were required to consider as an RNA editing site. However, for mitochondrial PCGs with low copy number and low expression, the coverage threshold was relaxed to 10×.
Phylogenetic and synteny analyses
Phylogenetic analyses were conducted using all available mitogenomes of Poales (Table S1). Due to incomplete annotation information in these genomes and the absence of many core genes, we conducted a reannotation of them. Subsequently, we obtained a dataset consisting of 32 genes, including 24 core genes and 8 variable genes (rps1, rps2, rps3, rps4, rps7, rps12, rps13, and rpl16). Each gene was aligned using MAFFT v7.508 [63] and manually trimmed before being concatenated. We utilized IQ-TREE v2.2.0.3 [64] to conduct phylogenetic analysis, with the TVM + F + I + I + R2 model selected as the best-fit nucleotide substitution model.
Based on the phylogenetic results, we selected two species from Oryza (O. rufipogon and O. sativa) that are most closely related to H. aristata to study the structural changes of their mitogenomes. First, the genomes were pairwise compared using the BLASTn program. Then, only homologous regions with a length exceeding 1,000 bp were retained. Finally, the visualization was conducted using RIdeogram [65].
Results
Mitochondrial genome assembly and annotation
In total, we generated 23.60 Gb Illumina reads and 22.54 Gb Nanopore reads through whole-genome sequencing (Table S2). The whole genome size of floating bamboo was estimated to be around 319.08 Mb (Fig. S2). A subset of 6.43 Gb Illumina short reads was identified as potentially derived from the mitochondrial genome and was subsequently used for de novo assembly. Due to the presence of numerous repeat sequences, the initial assembly graph exhibited a complex conformation (Fig. S4). However, leveraging ONT long reads enabled us to represent the mitogenome as a single circular molecule (587,847 bp; Fig. 1), which is significantly larger than the average length of mitogenomes in land plants (404.02 kb; Table 1). The coverage depth of the chloroplast and mitochondrial genomes is 822.3× and 159.5×, respectively (only Illumina reads were considered). The GC content of the mitogenome was 44.63% (Fig. 1), noticeably higher than the GC content of the chloroplast genome of the same species (39.03%; Fig. S3).
A representative mitochondrial genome map of Hygroryza aristata. The outer circle represents gene locations, while the inner circle displays GC content. Genes belonging to different functional groups are color-coded for easy identification. The red arrow indicates the starting point of the mitogenome. Genes transcribed clockwise and counter-clockwise are depicted on the inside and outside of the circles, respectively. Complete gene transfers from chloroplast DNA are highlighted in red
In the mitogenome of H. aristata, we annotated a complete set of 24 core genes (Fig. 1), including five ATP synthase genes (atp1, atp4, atp6, atp8, and atp9), nine NADH dehydrogenase genes (nad1, nad2, nad3, nad4, nad4L, nad5, nad6, nad7, and nad9; in addition to a chloroplast-derived ndhB), four cytochrome C biogenesis genes (ccmB, ccmC, ccmFc, and ccmFn), three cytochrome C oxidase genes (cox1, cox2, and cox3), ubiquinol cytochrome c reductase (cob), a transport membrane protein (mttB), and a maturase (matR). The variable genes in the mitogenome include three large subunits of ribosome proteins (rpl2, rpl5, and rpl16; in addition to chloroplast-derived rpl2 and rpl23), ten small subunits of ribosome proteins (rps1, rps2, rps3, rps4, rps7, rps11, rps12, rps13, rps14, and rps19; in addition to a chloroplast-derived rps7).
Repeats mediate the homologous recombination
During the assembly of Illumina short reads, we detected 20 repeat sequences in the initial graph (SR1-SR13, MR1-MR6, LR1) with multiple connections and doubled coverage depth, resulting in a complex mitochondrial genome structure (Fig. S4). Among them, except for LR1 and MR3, the rest of the repeat sequences have low recombination frequencies below 15% (Table 2). Furthermore, since there was no supporting evidence from ONT reads confirming the secondary configuration for SR8 and SR10, both of them were considered false positives or inactive in mediating recombination.
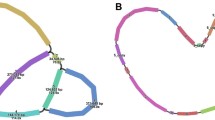
The two configurations resulting from LR1 (major: contig7-LR1-contig8/contig31-LR1-contig32; secondary: contig7-LR1-contig32/contig31-LR1-contig8) occurred with almost equal frequency (385/363) (Fig. 2; Fig. S5; Table 2). In either configuration, LR1 can combine with other contigs to form component repeats (Fig. 2; Fig. S5). For example, in our master circle model, contig7-LR1-contig8 (CR1) and contig31-LR1-contig32 (CR2) (Fig. 2A), or contig8-LR1-contig31 (CR3) and contig7-LR1-contig32 (CR4) (Fig. 2B). The sequencing depth of the four aforementioned contigs is approximately twice the average depth (contig7: 278.3×; contig8: 252.0×; contig31: 248.3×; contig32: 226.9×). However, the depth of LR1 is four times the average (474.8×). Meanwhile, SR5-contig29 also forms a component repeat, namely CR5 (Fig. 2).
There is a clear positive correlation between the length of repeat sequences and their corresponding genome recombination rate (r = 0.58, p < 0.01; Fig. 3). Within floating bamboo, this correlation is even more pronounced (r = 0.74, p < 0.01). The six repeats exceeding 1000 bp (LR1, CR1-CR5) all exhibit high recombination rates (> 0.3; Table 2). Theoretically, these six repeats can lead to 64 different high-frequency configurations.
Sequence exchange between genomes: MTCPs
During the process of evolution, mitochondrial genomes tend to incorporate segments from other intracellular genomes and integrate them into their genomes. Specific contigs of the mitochondrial genome display higher-than-average depth (contig25: 1723.0×; contig28: 1776.6×; contig29: 1492.7×; contig48: 1679.5×; Fig. S4), which are derived from the chloroplast genome. This suggests significant sequence transfer between these two organelles. By comparing the two organelles of H. aristata, we identified six mitochondrial chloroplast DNA sequences (MTCPs) (Fig. 4; Table S3), four of which originated from the inverted repeat (IR) regions of the chloroplast and corresponded to the high-depth contigs mentioned above. This explains why the depth of these contigs is greater than the sum of the average depths of the mt and cp genomes. The total length of these six MTCPs is 20,684 bp, accounting for 3.52% and 15.25% of the total length of the mt and cp genomes, respectively. Many intact genes inside these six long chloroplast fragments are transferred to the mitogenome. These transfers include nine protein-coding genes (rpl2, rpl23, rps7, ndhI, ndhB, rbcL, ycf2, atpE, and atpB), among which atpE and atpB have become pseudo-genes due to the introduction of stop codons through mutations. However, despite the presence of seven other complete cp PCGs in mtDNA, we have found no evidence of their expression in transcriptomic data.
Demonstration of the homologous sequences between the two organelle genomes of H. aristata. The outer circle in blue and pink represents the mitogenome (mtDNA) and the plastome (cpDNA), respectively. Repeat sequences within the mitogenome and mitochondrial chloroplast DNAs (MTCPs) are indicated by gray and yellow arcs, respectively
Repeat elements analysis
We identified 23 simple sequence repeats (SSRs) in the plastome and 145 in the mitogenome (Table S4). Dispersed repeats are more frequent than SSRs in both organelles, with 67 in the plastome and 681 in the mitogenome. Among them, there are 89 (13.07%) dispersed repeats larger than 1 kb in the mitogenome. Additionally, no reverse (R) or complement (C) repeats were detected. Relative to the high-frequency occurrence in the nuclear genome, tandem repeat elements have been found only 35 times in the mitochondrial genome of H. aristata, accounting for 0.51% of the total length of the mtDNA.
Codon usage bias and RNA editing
The GC content of the PCGs in the mitogenome is as follows: GC1 (48.21%) > GC2 (42.84%) > GC3 (38.07%) (Table S5). Moreover, GC1 is notably higher than the average GC content of the entire genome (44.63%; Fig. 1). Although the two organelles share the same most frequent codon for each amino acid (Table S6), the effective number of codons (ENC) in the mitogenome remains significantly higher than that in the plastome (55.45 > 50.61; Table S5), indicating that the mitogenome exhibits a weaker codon preference. All PCGs are located near the expected curve in the GC3S-ENC plot, without significant outliers that deviate excessively from the expected values (Fig. S6B). In addition, there is a significant bias towards T/C bases at the third position of codons in the PCGs of both organelles (Fig. S6C).
When the RNA-sequencing data was mapped to the PCGs, it was observed that the average coverage of mt PCGs was only 3.72, while the corresponding value of cp PCGs was 59.90. This indicates a significantly lower expression level of mt PCGs compared to cp PCGs. Due to the low expression level of mt PCGs, only 2 mt genes with 3 editing sites were identified, which is much fewer than in chloroplast genes (14 genes with 61 editing sites; Table S7). Furthermore, the editing efficiency of these three editing sites in mt is all 1, suggesting the possibility of gene mutations between individuals cannot be ruled out.
Phylogenetic and synteny analyses
The topology of the BOP clade, to which H. aristata belongs, was recovered as [Oryzoideae, (Bambusoideae, Pooideae)] with strong support (BS = 97) (Fig. 5). The majority of analyzed species contain a full set of 24 core unique mitochondrial genes. Furthermore, all species have retained the following genes: 3 rRNA genes, rps3, rps4, rps7, rps12, rps13, and rpl16. However, there is no presence of sdh3 and sdh4. Cynodon dactylon × C. transvaalensis have lost the most genes, including five core genes (atp1, atp4, matR, nad4L, and nad9), and 10 variable genes, also exhibiting the fastest substitution rate in Poaceae. Regarding tRNA, only eight tRNA genes (trnC-GCA, trnD-GUC, trnF-GAA, trnK-UUU, trnM-CAU, trnP-UGG, and trnS-GCU) have not been lost in any of the species.
The phylogenetic relationships and gene map of Poales. (A) Maximum likelihood phylogeny of Poales inferred from IQ-TREE analysis of the concatenated 32 mitochondrial gene supermatrix. Branch lengths are in number of substitutions per site. All nodes have maximal support (IQ-TREE UFBoot = 100) unless noted with red-colored numbers. (B) Mitochondrial gene map of Poales where color-filled squares represent the presence of at least one complete copy. Species belonging to different subfamilies are color-coded for easy identification. Genes without absence in all species are highlighted in red. All tRNAs have scores exceeding 30.0 in tRNAscan-SE
Despite significant structural variations and rearrangements, there are abundant homologous sequences between the mitogenomes of Oryza sativa and O. rufipogon, with essentially no unique segments found in the latter (Fig. 6). Although Hygroryza and Oryza belong to the same subfamily, only 27.10% of sequences in the mitogenome of H. aristata show homology with Oryza, with the majority being in protein-coding regions.
Discussion
In this study, the final assembly of the mitogenome was based solely on Illumina short reads, and Nanopore long reads were used to quantify the homologous recombination rate of each repeat pair (Table 2). As pointed out by Yang et al. (2022) [66], this approach can help avoid incorrect bases, especially in protein-coding regions, that may arise when constructing the scaffold using long reads [67]. While Unicycle [43] implements a similar strategy to the one used in this study for small genomes, it may overlook some repeat sequences with low recombination rates during the assembly process. Floating bamboo, as an endangered species, holds significant ecological and economic value. However, the lack of genetic information about this species hinders its conservation and research. Our presentation of its first complete mitochondrial genome enhances our understanding of this species and provides a critical case study for mitochondrial genomes in aquatic higher plants.
We detected 25 repeat pairs (13 SRs, 6 MRs, 1 LRs, and 5 CRs; Table 2) in the mitogenome of floating bamboo. Among these, except for SR8 and SR10, their activity in mediating homologous recombination has been proven. In addition to serving as sites for inter- and intra-molecular recombination, repeats also significantly inflate the estimated size of mtDNA. Except for CR3 and CR4, which do not appear in the master circle conformation, the length of the other repeat pairs reaches 81,633 bp, accounting for 13.88% of its total length. Many studies have confirmed that repeat pairs do not need to be identical to mediate HR [28, 34, 35]. However, we have not encountered this situation in floating bamboo. We have not found any precedent in former studies for the complex repeats we identified (CR1-CR5), undoubtedly adding to the complexity of plant mitogenomes. As one of the most important features in the mitochondria of land plants, homologous recombination mediated by repeats has been found in nearly all studied species. However, by losing two repeats, the mtDNA in Brassica hirta presents as a single circular chromosome with a smaller size compared to other Brassica species [26]. At the individual level, it seems preferable to keep the genome absolutely intact. However, a degree of instability is essential for species evolution, as genomic reshaping leads to genetic diversity. Disrupting the mechanisms would ultimately compromise the resilience and adaptability of species to changing environmental conditions [18].
Based on a review of previous studies, Marechal and Brisson (2010) [18] concluded that fairly large repeated sequences (> 1 kb) could frequently undergo intra- or intermolecular recombination, while homologous recombination also occurs sporadically between shorter repeats (> 100 bp, < 1 kb). As seen in Fig. 3, quantitative studies based on long sequencing reads generally support this conclusion. Most repeats with a recombination rate greater than 0.3 are longer than 1 kb. Except for Aeginetia indica, all seven repeats in this species range from 100 to 250 bp in length, yet their corresponding recombination ratios are greater than 0.4. In contrast, Nymphaea colorata exhibits a different pattern, as all ten repeats (4 SRs, 4 MRs, and 2 LRs) exhibit a low recombination rate (< 0.1). The differences between species are substantial, Salvia miltiorrhiza, compared to its congener S. officinalis, possesses more and longer repeat sequences and undergoes more frequent HR. Despite that, overall, the positive correlation between the length of repeat sequences and their corresponding genome recombination rate remains pronounced (r = 0.58, p < 0.01).
While horizontal gene transfer (HGT) between plant mitochondrial genomes and other intra- or extracellular genomes is frequently reported and contributes to the large size of plant mitogenomes [27, 68], there is currently no evidence confirming that these transferred exogenous genes can be expressed and functional within mitochondria. Fields et al. (2022) [69] reported a complete sequence insertion of 641 kb in the mtDNA of Arabidopsis thaliana from its nuclear genome. Li et al. (2022) [27] identified 28 MTCPs in okra, including only three complete chloroplast genes (rps7, psaA, and psbJ). Although we only identified six MTCP fragments in floating bamboo (Fig. 4), due to their long length, these fragments contain nine complete chloroplast protein-coding genes, with atpE and atpB undergoing pseudogenization (Table S3).
Our analysis confirmed that both organelle genomes of H. aristata exhibit the same pattern of codon usage bias. However, the PCGs in mtDNA show a more balanced usage of synonymous codons (Table S5 and S6). Furthermore, PCGs are distributed around the expected curve in the GC3S-ENC plot (Fig. S6B), consistent with the pattern summarized by Wang et al. (2011) [70]. They reported that during the evolution of the Streptophyta lineage, from Charophyta algae to the earliest land plant group (bryophytes) and finally to the highly derived and successful angiosperms (monocots and eudicots), the codon usages in their mitochondrial genomes have become less biased. They also suggested that the GC-accumulating strategy in the mitogenome of land plants may provide an advantage for DNA by reducing damage from strong ultraviolet radiation, as plants have gradually adapted to dry habitats over the course of their evolution.
It is a consensus that PCGs in mitogenomeare highly conserved in both quantity and genetics, contrasting with the complex and variable structure of its genome (Fig. 6). They are considered the most conservative among the three sets of genetic materials in angiosperms. The phylogenetic position of H. aristata based on them remains clear (Fig. 5) and is supported by previous studies based on nuclear and chloroplast datasets [9, 71].
Among a complete set of 41 PCGs in the mitogenome, 12 of them had been lost at least once in the 35 studied species (excluding Cynodon dactylon × C. transvaalensis). Gene loss is even more common in the chloroplasts of Poales. Wu et al. (2024) [72] observed that 30 out of 80 cp PCGs have been lost at least once in 93 Poales species, they speculate that the loss of the photosynthetic ndh gene may be an adaptation to the aquatic environment. Additionally, we observed that the aquatic lineage within Oryzoideae (Oryza and H. aristata) retains more genes related to ribosomal synthesis (rpl and rps). The reduction of cytoplasmic genomes is one of the prominent themes in eukaryotic genome evolution. However, it is rare for mitogenomes and plastomes to be lost entirely despite their ancient origins approximately 1–2 billion years ago. The parasitic plant Viscum scurruloideum possesses the smallest known mitochondrial genome among land plants, measuring only 66 kb [73]. With the reduction in size, there is a significant decrease in gene content. It has lost all nine mitochondrial nad genes, which are responsible for encoding the respiratory complex I (NADH dehydrogenase), an unprecedented occurrence in a multicellular organism. By comparing mitogenome content across the extant diversity of eukaryotes, researchers have inferred the evolutionary timing of mitochondrial gene loss events in different lineages [74,75,76,77]. In the most common ancestor of eukaryotes, the number of mitochondrial genes has reduced from over 1,000 genes estimated to have been present in the bacterial progenitor of mitochondria to 69 [12]. Subsequently, heterogeneous reduction occurred with variation both through time and across phylogenetic lineages, resulting in extant eukaryotes differing greatly in organelle gene content [74, 76]. In the sampled species we studied, Cynodon dactylon × C. transvaalensis has lost the highest number of genes, including atp1, atp4, matR, nad4L, nad9, rps1, trnN-GUU, trnQ-UUG, and trnY-GUA, which are unique to the Poales gene loss profile (Fig. 5). The mitogenome of the somatic hybrid Solanum commersonii × S. tuberosum also contains fewer genes than both parental species. Cho et al. (2022) [78] speculated that it may have originated from homologous recombination between the two parents.
Conclusions
We have successfully cultured Hygroryza aristata using tissue culture, providing a new approach to conserving the germplasm resources of this endangered species. With the assistance of Nanopore long reads, we quantified the homologous recombination rate mediated by 25 repeat pairs (13 SRs, 6 MRs, 1 LR, and 5 CRs) and represented the mtDNA of floating bamboo as a master circle model. By incorporating former quantified studies from 10 other angiosperms, despite the variation among species, we confirmed a pronounced positive correlation between the length of repeats and their corresponding recombination rate. Among these species, floating bamboo exhibits the most complex conformations, with six repeat pairs exceeding 1000 bp in length showing an HR rate exceeding 0.3. Meanwhile, the complex repeat pairs discovered in this study are unprecedented and shed light on the evolution of mitochondrial genomes in aquatic higher plants, allowing deeper insights into the repeat-mediated recombination pattern in plant mitogenomes. The chloroplast insertions in H. aristata largely originate from the inverted repeat region and collectively contain nine complete protein-coding genes, among which two have undergone pseudogenization, while the remaining seven show no sign of expression in the mitogenome. Both organelle genomes exhibit similar codon usage bias, with the PCGs in mtDNA showing a more balanced usage of synonymous codons. The gene map of the Poales mitogenome indicates the complete loss of rpl6, sdh3, and sdh4 genes, with Oryzoideae retaining a greater number of variable genes compared to Pooideae.
Data availability
The assembled organelle genome sequences have been deposited in NCBI with accession number: PP534167 (mtDNA); PP534166 (cpDNA). The raw sequence data have been deposited in the Short Read Achieve (SRA) database of NCBI (PRJNA1090843).
Abbreviations
- HR:
-
Homologous recombination
- PCG:
-
Protein-coding gene
- cpDNA:
-
Chloroplast DNA
- mtDNA:
-
Mitochondrial DNA
- MTCP:
-
Mitochondrial chloroplast DNA sequence
- ENC:
-
Effective number of codons
- SR:
-
Small repeat (< 500 bp)
- MR:
-
Middle repeat (> 500 bp, < 1 kb)
- LR:
-
Large repeat (> 1 kb)
- CR:
-
Complex repeat
References
Sun XZ, Simpson DA. Flora of China. Volume 9. Beijing: Science Press and Missouri Botanical Garden; 1992.
World Grass Species. descriptions, identification, and information retrieval. http://www.kew.org/data/grasses-db.html.
Hossain MM, Shahjahan M, Khan S, Juraimi AS, Uddin MK, Hasan M. Potentialities of the Asian watergrass (Hygroryza aristata) as feed in aquaculture. Sustainability. 2021;13(12):14.
Hossain MM, Rahman MH, Ali ML, Khan S, Haque MM, Shahjahan M. Development of a low-cost polyculture system utilizing Hygroryza aristata floating grass in the coastal wetlands of Bangladesh. Aquaculture 2020, 527.
Malik V, Mohammad I, Pranita. Ecology and conservation of rare Hygroryza aristata (Retz.) Nees Ex Wight & Arn. Biol Forum 2014, 6(1).
Xie DZ. Physiological responses to cadmium and high-temperature stress of Hygroryza aristata. Zhejiang: Zhejiang A&F University; 2019.
Cai JG. Ecological habits and landscape design of wetland plants in Hangzhou City. Beijing Forestry University; 2006.
Chung Y-M, Lan Y-H, Hwang T-L, Leu Y-L. Anti-inflammatory and antioxidant components from Hygroryza aristata. Molecules. 2011;16(3):1917–27.
Wang H, Wu Z, Fan G, Zheng G, Gu C, Tembrock LR, Yang Z, Yang J, Wang J. Characterization of the complete chloroplast genome of Hygroryza aristata (Retz.) Nees Ex Wight & Arn. (Zizaniinae, Poaceae). Mitochondrial DNA Part B-Resources. 2021;6(7):1949–50.
Soreng RJ, Peterson PM, Zuloaga FO, Romaschenko K, Clark LG, Teisher JK, Gillespie LJ, Barbera P, Welker CAD, Kellogg EA, et al. A worldwide phylogenetic classification of the Poaceae (Gramineae) III: an update. J Syst Evol. 2022;60(3):476–521.
Zhang T, Huang W, Zhang L, Li D-Z, Qi J, Ma H. Phylogenomic profiles of whole-genome duplications in Poaceae and landscape of differential duplicate retention and losses among major Poaceae lineages. Nat Commun. 2024;15(1):3305.
Sloan DB, Warren JM, Williams AM, Wu ZQ, Abdel-Ghany SE, Chicco AJ, Havird JC. Cytonuclear integration and co-evolution. Nat Rev Genet. 2018;19(10):635–48.
Gray MW. Mitochondrial evolution. Cold Spring Harb Perspect Biol 2012, 4(9).
Kim Y-J, Zhang D. Molecular control of male fertility for crop hybrid breeding. Trends Plant Sci. 2018;23(1):53–65.
Siqueira JA, Hardoim P, Ferreira PCG, Nunes-Nesi A, Hemerly AS. Unraveling interfaces between energy metabolism and cell cycle in plants. Trends Plant Sci. 2018;23(8):731–47.
Van Akent O, Van Breusegem F. Licensed to kill: mitochondria, chloroplasts, and cell death. Trends Plant Sci. 2015;20(11):754–66.
Wu ZQ, Liao XZ, Zhang XN, Tembrock LR, Broz A. Genomic architectural variation of plant mitochondria—A review of multichromosomal structuring. J Syst Evol. 2020;60(1):160–8.
Marechal A, Brisson N. Recombination and the maintenance of plant organelle genome stability. New Phytol. 2010;186(2):299–317.
Rice DW, Alverson AJ, Richardson AO, Young GJ, Virginia Sanchez-Puerta M, Munzinger J, Barry K, Boore JL, Zhang Y, dePamphilis CW, et al. Horizontal transfer of entire genomes via mitochondrial fusion in the angiosperm Amborella. Science. 2013;342(6165):1468–73.
Wu Z, Sloan DB. Recombination and intraspecific polymorphism for the presence and absence of entire chromosomes in mitochondrial genomes. Heredity. 2019;122(5):647–59.
Wu Z, Cuthbert JM, Taylor DR, Sloan DB. The massive mitochondrial genome of the angiosperm Silene noctiflora is evolving by gain or loss of entire chromosomes. Proc Natl Acad Sci U S A. 2015;112(33):10185–91.
Kozik A, Rowan BA, Lavelle D, Berke L, Schranz ME, Michelmore RW, Christensen AC. The alternative reality of plant mitochondrial DNA: one ring does not rule them all. PLoS Genet 2019, 15(8).
Manchekar M, Scissum-Gunn K, Song D, Khazi F, McLean SL, Nielsen BL. DNA recombination activity in soybean mitochondria. J Mol Biol. 2006;356(2):288–99.
Bendich AJ. Structural analysis of mitochondrial DNA molecules from fungi and plants using moving pictures and pulsed-field gel electrophoresis. J Mol Biol. 1996;255(4):564–88.
Lilly JW, Havey MJ, Jackson SA, Jiang J. Cytogenomic analyses reveal the structural plasticity of the chloroplast genome in higher plants. Plant Cell. 2001;13(2):245–54.
Palmer JD, Herbon LA. Unicircular structure of the Brassica hirta mitochondrial genome. Curr Genet. 1987;11(6–7):565–70.
Li J, Li J, Ma Y, Kou L, Wei J, Wang W. The complete mitochondrial genome of okra (Abelmoschus esculentus): using nanopore long reads to investigate gene transfer from chloroplast genomes and rearrangements of mitochondrial DNA molecules. BMC Genomics 2022, 23(1).
Dong S, Zhao C, Chen F, Liu Y, Zhang S, Wu H, Zhang L, Liu Y. The complete mitochondrial genome of the early flowering plant Nymphaea colorata is highly repetitive with low recombination. BMC Genomics 2018, 19(1).
Zhong Y, Yu R, Chen J, Liu Y, Zhou R. Highly active repeat-mediated recombination in the mitogenome of the holoparasitic plant Aeginetia indica. Front Plant Sci 2022, 13.
Liu J, Ni Y, Liu C. Polymeric structure of the Cannabis sativa L. mitochondrial genome identified with an assembly graph model. Gene 2023, 853.
Yang H, Chen H, Ni Y, Li J, Cai Y, Wang J, Liu C. Mitochondrial genome sequence of Salvia officinalis (Lamiales: Lamiaceae) suggests diverse genome structures in cogeneric species and finds the stop gain of genes through RNA editing events. Int J Mol Sci 2023, 24(6).
Fang B, Li J, Zhao Q, Liang Y, Yu J. Assembly of the complete mitochondrial genome of Chinese Plum (Prunus salicina): characterization of genome recombination and RNA editing sites. Genes 2021, 12(12).
Cao P, Huang Y, Zong M, Xu Z. De Novo assembly and comparative analysis of the complete mitochondrial genome of Chaenomeles speciosa (Sweet) Nakai revealed the existence of two structural isomers. Genes 2023, 14(2).
Dong S, Chen L, Liu Y, Wang Y, Zhang S, Yang L, Lang X, Zhang S. The draft mitochondrial genome of Magnolia biondii and mitochondrial phylogenomics of angiosperms. PLoS ONE 2020, 15(4).
Yang H, Chen H, Ni Y, Li J, Cai Y, Ma B, Yu J, Wang J, Liu C. De Novo hybrid assembly of the Salvia miltiorrhiza mitochondrial genome provides the first evidence of the multi-chromosomal mitochondrial DNA structure of Salvia species. Int J Mol Sci 2022, 23(22).
Liu D, Guo H, Zhu J, Qu K, Chen Y, Guo Y, Ding P, Yang H, Xu T, Jing Q et al. Complex physical structure of complete mitochondrial genome of Quercus acutissima (Fagaceae): a significant energy plant. Genes (Basel) 2022, 13(8).
Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987;19(1):11–5.
Chen S, Zhou Y, Chen Y, Gu J. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):884–90.
Marcais G, Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27(6):764–70.
Ranallo-Benavidez TR, Jaron KS, Schatz MC. GenomeScope 2.0 and Smudgeplot for reference-free profiling of polyploid genomes. Nat Commun 2020, 11(1).
Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol 2020, 21(1).
Kolmogorov M, Yuan J, Lin Y, Pevzner PA. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol. 2019;37(5):540–.
Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 2017, 13(6).
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinformatics 2009, 10.
Vasimuddin M, Misra S, Li H, Aluru S. Efficient architecture-aware acceleration of BWA-MEM for multicore systems. In: 33rd IEEE international parallel and distributed processing symposium (IPDPS). Rio de Janeiro, Brazil; 2019. pp. 314–324.
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Son P, Prjibelski AD, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–77.
Wick RR, Schultz MB, Zobel J, Holt KE. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 2015;31(20):3350–2.
Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017;45(W1):W6–11.
Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25(5):955–64.
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647–9.
Greiner S, Lehwark P, Bock R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019;47(W1):W59–64.
Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34(18):3094–100.
Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020;13(8):1194–202.
Beier S, Thiel T, Munch T, Scholz U, Mascher M. MISA-web: a web server for microsatellite prediction. Bioinformatics. 2017;33(16):2583–5.
Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29(22):4633–42.
Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27(2):573–80.
Zhang P, Xu W, Lu X, Wang L. Analysis of codon usage bias of chloroplast genomes in Gynostemma species. Physiol Mol Biology Plants. 2021;27(12):2727–37.
Rice P, Longden I, Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000;16(6):276–7.
Peden JF. Analysis of codon usage. Nottingham, UK: University of Nottingham; 2000.
Wright F. The effective number of codons used in a gene. Gene. 1990;87(1):23–9.
Liu H, Lu Y, Lan B, Xu J. Codon usage by chloroplast gene is bias in Hemiptelea Davidii. J Genet 2020, 99(1).
Picardi E, Pesole G. REDItools: high-throughput RNA editing detection made easy. Bioinformatics. 2013;29(14):1813–4.
Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–80.
Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020;37(5):1530–4.
Hao Z, Lv D, Ge Y, Shi J, Weijers D, Yu G, Chen J. RIdeogram: drawing SVG graphics to visualize and map genome-wide data on the idiograms. PeerJ Comput Sci 2020, 6.
Yang Z, Ni Y, Lin Z, Yang L, Chen G, Nijiati N, Hu Y, Chen X. De novo assembly of the complete mitochondrial genome of sweet potato (Ipomoea batatas [L.] Lam) revealed the existence of homologous conformations generated by the repeat-mediated recombination. BMC Plant Biol 2022, 22(1).
Delahaye C, Nicolas J. Sequencing DNA with nanopores: troubles and biases. PLoS ONE 2021, 16(10).
Schneider AC, Chun H, Stefanovic S, Baldwin BG. Punctuated plastome reduction and host-parasite horizontal gene transfer in the holoparasitic plant genus Aphyllon. Proc Royal Soc B-Biol Sci. 2018;285(1887).
Fields PD, Waneka G, Naish M, Schatz MC, Henderson IR, Sloan DB. Complete sequence of a 641-kb insertion of mitochondrial DNA in the Arabidopsis thaliana nuclear genome. Genome Biol Evol 2022, 14(5).
Wang B, Yuan J, Liu J, Jin LA, Chen JQ. Codon usage bias and determining forces in green plant mitochondrial genomes. J Integr Plant Biol. 2011;53(4):324–34.
Huang W, Zhang L, Columbus JT, Hu Y, Zhao Y, Tang L, Guo Z, Chen W, McKain M, Bartlett M, et al. A well-supported nuclear phylogeny of Poaceae and implications for the evolution of C4 photosynthesis. Mol Plant. 2022;15(4):755–77.
Wu H, Li D-Z, Ma P-F. Unprecedented variation pattern of plastid genomes and the potential role in adaptive evolution in Poales. BMC Biol. 2024;22(1):97.
Skippington E, Barkman TJ, Rice DW, Palmer JD. Miniaturized mitogenome of the parasitic plant Viscum Scurruloideum is extremely divergent and dynamic and has lost all nad genes. Proc Natl Acad Sci U S A. 2015;112(27):E3515–24.
Janouskovec J, Tikhonenkov DV, Burki F, Howe AT, Rohwer FL, Mylnikov AP, Keeling PJ. A new lineage of eukaryotes illuminates early mitochondrial genome reduction. Curr Biol. 2017;27(23):3717–.
Johnston IG, Williams BP. Evolutionary inference across eukaryotes identifies specific pressures favoring mitochondrial gene retention. Cell Syst. 2016;2(2):101–11.
Kannan S, Rogozin IB, Koonin EV. MitoCOGs: clusters of orthologous genes from mitochondria and implications for the evolution of eukaryotes. BMC Evol Biol 2014, 14.
Huynen MA, Duarte I, Szklarczyk R. Loss, replacement and gain of proteins at the origin of the mitochondria. Biochim Et Biophys Acta-Bioenergetics. 2013;1827(2):224–31.
Cho K-S, Lee H-O, Lee S-C, Park H-J, Seo J-H, Cho J-H, Park Y-E, Choi J-G, Yang T-J. Mitochondrial genome recombination in somatic hybrids of Solanum commersonii and S. Tuberosum. Sci Rep 2022, 12(1).
Acknowledgements
We would like to express our gratitude to Wuhan Benagen Tech Solutions Company Limited (www.benagen.com) for providing DNA-seq service and to the MitoRun research group for their technical support in this project.
Funding
This work was supported by the Strategic Priority Research Program of Chinese Academy of Sciences [grant numbers XDB31000000] and the Key Research and Development Project of Hubei Province [grant numbers 2023BCB107].
Author information
Authors and Affiliations
Contributions
HJ.W., study conception and design, data analysis, data visualization, manuscript preparation, manuscript revision. ZG.W., sample collection, data visualization, manuscript revision. JD.Z., study conception and design, manuscript revision. T.L., study conception and design, supervision. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This study’s material collections and experimental research were obtained in accordance with relevant institutional, national and international guidelines and legislation.
Competing interests
The authors declare no competing interests.
Consent for publication
Not applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Wang, H., Wu, Z., Li, T. et al. Highly active repeat-mediated recombination in the mitogenome of the aquatic grass Hygroryza aristata. BMC Plant Biol 24, 644 (2024). https://doi.org/10.1186/s12870-024-05331-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-024-05331-x