Abstract
Subsistence farmers and global food security depend on sufficient food production, which aligns with the UN's “Zero Hunger,” “Climate Action,” and “Responsible Consumption and Production” sustainable development goals. In addition to already available methods for early disease detection and classification facing overfitting and fine feature extraction complexities during the training process, how early signs of green attacks can be identified or classified remains uncertain. Most pests and disease symptoms are seen in plant leaves and fruits, yet their diagnosis by experts in the laboratory is expensive, tedious, labor-intensive, and time-consuming. Notably, how plant pests and diseases can be appropriately detected and timely prevented is a hotspot paradigm in smart, sustainable agriculture remains unknown. In recent years, deep transfer learning has demonstrated tremendous advances in the recognition accuracy of object detection and image classification systems since these frameworks utilize previously acquired knowledge to solve similar problems more effectively and quickly. Therefore, in this research, we introduce two plant disease detection (PDDNet) models of early fusion (AE) and the lead voting ensemble (LVE) integrated with nine pre-trained convolutional neural networks (CNNs) and fine-tuned by deep feature extraction for efficient plant disease identification and classification. The experiments were carried out on 15 classes of the popular PlantVillage dataset, which has 54,305 image samples of different plant disease species in 38 categories. Hyperparameter fine-tuning was done with popular pre-trained models, including DenseNet201, ResNet101, ResNet50, GoogleNet, AlexNet, ResNet18, EfficientNetB7, NASNetMobile, and ConvNeXtSmall. We test these CNNs on the stated plant disease detection and classification problem, both independently and as part of an ensemble. In the final phase, a logistic regression (LR) classifier is utilized to determine the performance of various CNN model combinations. A comparative analysis was also performed on classifiers, deep learning, the proposed model, and similar state-of-the-art studies. The experiments demonstrated that PDDNet-AE and PDDNet-LVE achieved 96.74% and 97.79%, respectively, compared to current CNNs when tested on several plant diseases, depicting its exceptional robustness and generalization capabilities and mitigating current concerns in plant disease detection and classification.
Similar content being viewed by others
Introduction
Agriculture, as a significant driver of the global economy, serves as the primary provider of food, income, revenue, and employment opportunities. Different human societies have been capable of producing food to adequately cater to the current and growing population using advanced technology in the agricultural sector [1]. However, depending on the season or environmental factors, plant pests and diseases are caused by nematodes, fungi, viruses, protozoa, and bacteria [2, 3]. These severely influence plant health, structure quality, production, quantity, and the economy. One of the highly complex tasks regarding plant protection is the timely identification of plant symptoms, pests, and diseases [4]. Traditional approaches used in underdeveloped or developing nations are through human eye inspection, which is inaccurate, tedious, and time-consuming. Furthermore, smart agricultural gadgets are costly, and understanding these obtained classifications and detection on large farms needs agronomists and specialists is expensive [5].
Employing intelligent technologies capable of automatically detecting plant pests and diseases presents a promising approach to reducing total expenses in agriculture [6]. Therefore, academia and industry have used transfer learning (TL) and CNNs, particularly in the agricultural sector, for instance, in plant leaves, fruit, and disease classification, among other applications [7]. However, deep learning (DL) demands an increased number of parameters, thus increasing the training time and resulting in implementing small devices becoming complex and impractical [8]. Furthermore, properly extracting relevant characteristics from any given dataset is vital to the CNN-based model performance; for example, the studies utilized the widely used PlantVillage dataset, with various species of plant diseases across distinct categories [9, 10].
There has been a growing emphasis on rapid plant disease identification and classification using TL architectures. The complexity and required parameters of the TL model are determined by the level of model sophistication and the number of filters utilized [11]. Although TL methods often require advanced image processing techniques, they have simplified the procedure, making it more efficient in terms of time, especially when the model has no starting weights [12]. In addition, TL models require minimal computational resources compared to traditional learning approaches. However, implementing these models on small devices with limited resources can be challenging and limitation of the traditional learning approach [13].
Several studies demonstrate that some current models are developed using the idea of TL to attain better results compared to other well-developed approaches using DL architectures through potent computing equipment, such as graphics processing units (GPUs) and servers [14]. Because of the high cost, it is not practical to use advanced equipment that includes GPUs in the agriculture field that traditional farmers cannot afford. Therefore, there is a need for applications with a reduced number of parameters and reduced levels of computation and power consumption [15].
A survey on adopting computer vision and soft computing methods for disease identification and classification from plant leaves was conducted. It demonstrated that Computer vision techniques enhance plant growth, increasing productivity, quality, and economic value [16]. They are critical in medical, defense, agriculture, remote sensing, and business analysis applications. Digital image processing methods simulate human visual capabilities, providing automatic monitoring, disease management, and water management [17].
Another proposed system used a neural network to segment mango leaves for disease. It involved real-time images, preprocessing, feature extraction, training, and extraction of diseased regions. The system achieved high-level accuracy for anthracnose disease segmentation, with an average Specificity of 0.9115 and Sensitivity of 0.9086. The system demonstrated an intuitive and user-friendly interface and is being developed for precision agriculture [18]. Similarly, a hybrid Fuzzy Competitive Learning-based Counter Propagation Network (FCPN) was proposed for image segmentation of natural scene images. Fuzzy Competitive Learning (FCL) was used to train the instar layer of FCPN, whereas Grossberg learning was used to train the Outstar layer. The region-developing method was utilized for seed point selection, clustering, and estimating the number of crop seeds. The FCPN method produced a lower convergence ratio and greater precision than alternative methods [19].
Pattern recognition and machine vision are indispensable for the resolution of complex problems. Combining conventional and optimization methods, like Nature-Inspired Algorithms (NIA) or Bio-inspired methods, can enhance precision and decrease computational time. One such application is image segmentation, for which the Bacterial Foraging Optimization Algorithm (BFOA) is a promising method [20]. The efficiency of the BFO-ANN method was demonstrated through comparison with other approaches. IPM was developed using an automated Radial Basis Function Neural Network (RBFNN) system to detect plant diseases. The system uses leaf images from the IPM agriculture database repository. The RBFNN achieves higher segmentation accuracy than other methods, making it a promising solution for detecting diseases in plants with biotic elements [21].
While a considerable number of studies availed some plant disease classification and detection models, there are notable deficiencies in these studies [4, 15, 17, 20], including training on limited dataset size leading to model overfitting and generalization complexity to diverse environments. Training models under controlled backgrounds and environmental conditions, in contrast to the natural setting that makes these models impractical in the natural environment, the accuracy and robustness of models. Computation-related issues, for example, overfitting and difficulties in accurately extracting fine features during training, have impacted the efficiency and usefulness of DL models in the identification and classification of plant diseases. The conventional laboratory diagnosis of plant diseases is expensive, laborious, and time-intensive, which restricts its feasibility for prompt prevention in agriculture. Several current models, like [22, 23], encounter challenges in terms of resilience and ability to apply them to diverse plant diseases since most are only trained on a single crop. Moreover, early classification and detection models proposed were done with restricted image constraints, like images containing colors. However, in the controlled environment, the background and the foreground are put in binary format [24, 25]. However, in this scenario, the approaches employed in many earlier experiments are unsuitable for real-world smart-based agricultural system deployment that employ images that vary with natural-world backdrops.
Such shortcomings leave a gap in the availability of a robust and generalized model trained on the big dataset to detect and classify plant diseases trained on images without restriction on the background to increase the growth truth, thus being practical and implementable on small devices. As illustrated in this study, it employs transfer learning with pre-trained CNNs to improve performance and solve the issue of data scarcity; fine-tuning and deep feature extraction techniques on current cutting-edge CNNs are used to cater to background complexities. Moreover, it tackles computational issues by introducing two models, namely early fusion and lead voting ensemble, that incorporate several pre-trained CNNs; these models assist in overfitting reduction and improving feature extraction.
This study proposes two plant disease detection (PDD) and classification for CNN architecture with considerably reduced parameters. The TL uses nine comparative models: EfficienceNetB7, NASNetMobile, ConvNeXtSmall, DenseNet201, DenseNet101, ResNet50, GoogleNet, ResNet18, and AlexNet. These architectures employ numerous convolutions with changing filter sizes, resulting in superior feature extraction. We have turned to residual connections to address early disease detection problems. We opted for depth-wise separable convolution over conventional convolution because it reduces computational complexity, size, and parameter set without compromising the performance. This study uses a real-time PlantVillage image containing natural image traits. Therefore, this research contributes the following:
-
Propose two detection models (PDDNet) CNN architectures integrating the top six common CNNs that extract significant features and perform better. These models are demonstrated concisely. Arithmetic average ensemble (PDDNet-AAE) integrated fine-tuned network outputs. For the use of ensemble feature attraction (PDDNet-AE), the early average fusion method is used. In this instance, we combined deep traits collected from multiple DNNs and then trained with the LR classifier on these combined features. Lead voting ensemble class labels (PDDNet-LVE).
-
The study uses a logistic regression classifier to assess the proposed model performance compared to its counterparts (nine pretrained CNNs) that were used to extract deep features. Ultimately, all class labels that were the highest (lead) were voted for, and the system's decision was the most predicted class label.
-
The suggested architecture needs minimal parameters and is faster than traditional ML models tested on DenseNet201, DenseNet101, ResNet50, GoogleNet, ResNet18, AlexNet, EfficientNet, NASNet, and ConvNet.
-
Since the PlantVillage dataset is the most significant plant disease dataset currently publicly available, it was used to assess the proposed approaches. PDDNet-LVE outperformed the other current network models.
-
The proposed models achieved 96.74% and 97.79% accuracy on the early fusion and lead (majority) voting methods for this plant disease detection and classification, respectively. The CNN-LR combination of PDDNet-AE and PDDNet-LVE outperformed the simple averages of CNNs and has demonstrated improved results.
The remainder of this research is arranged into four sections following the introduction. Section "Related Literature" sheds light on the related literature on plant pest and disease classification and detection utilizing TL models, demonstrating the classification techniques used, the type of crop studied, and the reported accuracy. Section "Material and Methods" illustrates the study materials and methodology, including the PlantVillage description and plant diseases within the dataset. Section "Obtained Results and Discussion" demonstrates the results, and related discussions are presented, illustrating performance evaluations on classifiers, sampled deep learning, PDDNet-AE, PDDNet-EA, and PDDNet-LVE, and a comparison based on state-of-art models. Finally, Section "Conclusion" discusses the research conclusion and future research directions.
Related literature
The section discusses some DL methods for plant pests and disease detection and classification. Traditional ML approaches are based on creating features and segmentation, and DL techniques are based on learning from data in its raw form.
Using pre-trained CNNs like GoogleNet and AlexNet could classify twenty-six pests and diseases within fourteen plant species [7]; 99.34% was obtained through GoogleNet. AlexNet, GoogleNet, VGG, feat, and AlexNetOWTBn could recognize 58 leaf diseases [9]. A nine-layered deep convolutional network was used for plant disease detection, and 96.46% achieved accuracy [26]. Similarly, AlexNet's fully connected layer with GoogLeNet's inception layer to classify four diseases of apple leaves, and the average accuracy score reported 97.62% [27]. For the model optimization, InceptionV3, VGG19, VGG16, and ResNet detected tomato leaf disease and obtained 93.70% field accuracy and 99.60% laboratory accuracy [28]. VGG16 identified eggplant diseases using the super vector machine classifier for red, green, and blue; YCbCr and HSV were tested for robustness with 99.4% RGB [29]. Entirely improved pretrained DL plant disease with 99.75% model classification accuracy.
The authors demonstrated that using the SVM classifier on rice leaf disease classification could categorize eleven deep CNN model features and obtain an average of 98.38% using ResNet50 depth SVM [1]. Authors in [30] identified ten diseases of four plant species using six pretrained TL architectures, VGG16 corrected 90% of test datasets, and the authors found three cassava plant diseases and two pest damages using InceptionV3 transfer learning noticed six cassava illnesses using mobile devices [31]. In [32], the study determined that a 50-layer residual neural network can detect three wheat diseases using the ReLU activation and batch normalization following convolution and pooling. Using the German real-time field images, they reported a 96% accuracy. SqueezeNet 227.6MB and SequenzeNet 2.9MB obtained four tea leaf diseases after being tested Cifar ten fast CNN model depthwise separable convolution [33].
A well-trained VGG model identified and classified rice and agricultural diseases [8]. Two inception layers replaced VGGNet's fully connected layers: corn with 80.38% and rice with 92%, respectively. Singh and Misra [34] detailed how the soft computing methods and segmentation of images aid in plant, pest, and disease identification and classification in mostly grown plants like Malus domestica (apple), Zea mays, and genus Vitis diseases using pre-trained CNNs like VGG16 model, some other metaheuristic-inspired algorithms like genetic algorithm.
Gray level co-occurrence matrix (GLCM) with a moveable client-to-server structure for leaf disease detection and their classifications through Gabor wavelet transform (GWT) was used. In the mobile disease diagnosis system, feature vectors represent disease regions that can indicate many resolutions and directions. The mobile client preprocesses leaf photos, segments, and the affected leaf sections and sends them to the Pathology Server, lowering transmission costs. The Server extracts GWT–GLCM features and classifies K-Nearest Neighbors. Short message service displayed results with 93% accuracy under ideal conditions [35]. Table 1 summarizes the conventional methods, datasets, and the reported performance accuracy corresponding to those methods. In most cases, to summarize this, these studies are presented in three primary stages:
-
Plant pests and disease image segmentation is based on applying techniques like mathematical morphology, edge detection, color transformation, and pattern classification.
-
Detection of plant pests and diseases using traditional ML techniques.
-
Representative feature extraction from the segmented images that were obtained utilizing approaches that were based on color, texture, and shape.
The presented models specified in Table 1 are classification algorithms that utilized minimal datasets to differentiate between a limited number of species. Some studies used datasets from apple, Solanum lycopersicum, R. groenlandicum, and maize plants, and most of the reported accuracy ranged from 84% to 97%. Several plant disease detection studies have employed DL as demonstrated. These systems, datasets, and outcomes are demonstrated in Table 2. Most of these experiments included deep network fine-tuning and pretrained CNN feature extraction. To illustrate this, Sabrol and Satish based their study on the tomato disease classification; they used TL to extract features from the images, for example, shape, texture, color, and features with constrained image appearance, and reported a 94% accuracy [40]. The algorithms described in the literature utilize varied datasets and categorize two to four plant species; hence, they cannot be compared directly.
Material and methods
This section entails the background of the deep learning techniques, the PlantVillage dataset, and the proposed methodology.
Deep learning techniques
Deep learning has been applied extensively in several arenas; its approach to plant disease detection and classification has been extensively used through pretrained deep networks [73,74,75,76]. Within this study, we use nine edge-cutting pretrained networks for deep feature extraction for our classification model to have a starting training weight. Table 3 demonstrates the nine pretrained deep CNNs (namely, EfficientNetB7, NASNet, ConvNet, DenseNet201, DenseNet101, ResNet50, GoogleNet, ResNet18, and AlexNet), showing their distinct characteristics on size, accuracy, parameters, depth, and GPU requirements.
PlantVillage dataset
There is a considerable number of plant pests and disease datasets publicly available, including strawberry [79], rice [80], NLB dataset for maize plant, Turkey-PlantDataset [81], apple, AES-CD9214, PlantVillage, among others. According to the available datasets, we consider the PlantVillage dataset since it has several plant species and over thirty categories with almost all plant characteristics from different datasets. The rest of the datasets checked were found to focus on a single crop that narrows the classification base, and the number of plant leaf images was limitedly low compared to the PlantVillage dataset. Using the pretrained CNNs on a big dataset like PlantVillage assumes proper deep feature extraction. DenseNet models are comparable to ResNet models, except that each layer receives information from all preceding layers. Each Densenet layer feeds forward as early as demonstrated [82]. This study employs DL with six models to extract features to categorize plant diseases. CNNs that have been previously trained and proficient at extracting features and training deep networks. This approach is exceptional since it is more precise in using the LR classifier as a substitute for the output layers of these CNNs.
The PlantVillage dataset was developed to provide effective methods for identifying 38 distinct plant disease classes. It comprises 61,486 plant images in three versions: color, gray-scaled, and segmented. However, we consider 15 categories containing 54,303 PlantVillage images for this experiment. The study considered the PlantVillage dataset with 15 categories since it is more evenly distributed across the different classes than 38 categories. Uneven data distribution can lead to class imbalance issues, where some classes have significantly fewer samples than others. This significantly impacts ML models' performance when accurately predicting the underrepresented classes. Notably, the source of this dataset (https://plantvillage.psu.edu) no longer exists. However, our open-source platforms, including Kaggle and GitHub, have datasets available as linked.
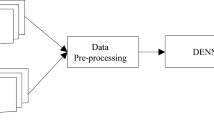
Deep features were extracted using nine different pretrained CNNs to make the dataset more diverse and show a wide range of details. During this process, numerous modifications were made employing three channels as well. These enhancements included gamma correction, principal component analysis, noise injection, scaling, image flipping, rotation, and color augmentation. In addition, scaling, rotation, and image flipping (RGB) were used. Figure 1 presents image samples from the PlantVillage plant disease species.
Plantvillage selected leaf image samples from the considered plant dataset in this study. (Legend: D1) Pepper bell bacterial spot, D2) potato early blight, D3) potato late blight, D4) Tomato bacterial spot, D5) Tomato early blight, D6) Tomato Lead mold, D7) Tomato Septoria leaf spot, D8) Spider mites Two-spotted spider mite, D9) Tomato target spot, D10) Tomato Yellow Leaf Curl Virus, D11) Tomato mosaic virus, D12) Apple Scrab, D13) Grape black rot, D14) Orange Huanglongbing (Citrus_greening), and D15) Squash powdery mildew
Methodology
To tackle the challenge of plant disease identification and classification, we consider feature extraction and fine-tuning approaches among the existing TL approaches, including the intermediate layers, fine-tuning, and feature extraction. The selected pre-trained CNNs are used as a feature extractor. The output of the last convolutional layer is used as a feature vector for the new task. Then, the CNNs are fine-tuned on the new dataset. The weights of the lower layers are frozen, and only the weights of the upper layers are updated. TL can save resources, as the model does not need to be trained from scratch.
Therefore, we consider TL for the nine most recent pretrained deep networks: DenseNet201, DenseNet101, ResNet50, ResNet18, GoogleNet, AlexNet, EfficientNetB7, NASNetMobile, and ConvNeXtSmall for feature extraction to aid in the classification problem process. Then, the LR classifier will evaluate the performance at an individual model level, utilizing the weights obtained from these networks. A comparison is then made based on the arithmetic average (AAE), initial (early), amalgamation or fusion (EA), and lead voting ensemble (LVE), commonly referred to as majority voting. Finally, we use the LR classifier to replace a superficial network block for fusion in the PDDNet technique coupled with the final layers of deep neural network in the PDDNet-LVE method.
The image input size is often different depending on the selected pretrained deep network architecture, as the second last column of Table 3 illustrates. For example, AlexNet and DesNet201 require different data inputs of 227 x 227 x 3 and 224 x 224 x 3, among others, at the input layer. Furthermore, due to the diverse CNNs selected for these experiments, the initial convolutional layer and the subsequent convolutional layers use different kernels; for instance, DenseNet201 with all convolutional layers use 3x3 kernels; ResNet101 utilized the initial convolutional layer uses a 7x7 kernel and the all-subsequent convolutional layers use 3x3 kernels.
ResNet50 at the initial convolutional layer uses a 7x7 kernel, and all subsequent convolutional layers use 3x3 kernels. GoogleNet uses 7x7 kernels at the initial convolutional layer uses, and most of the subsequent convolutional layers use 1x1 kernels, while a few layers use 3x3 kernels. AlexNet considers that the initial convolutional layer uses an 11x11 kernel, and the subsequent convolutional layers use 3x3 kernels. Lastly, ResNet18 at the initial convolutional layer uses a 7x7 kernel, and all subsequent convolutional layers use 3x3 kernels, as used during the experiment.
The proposed model approaches were executed using the MATLAB 2022b DL toolboxFootnote 1. The PlantVillage dataset was divided into training, validation, and testing. Adaptive Moment Estimation (Adam) is applied as the optimizer since it employs stochastic optimization, like ML and TL. The recursive nature of the method enables the efficient solving of noisy linear systems and the estimation of extreme values of functions that are only accessible over noisy annotations. Incorporating square propagation in stochastic gradient descent, adaptive gradient, and root mean, Adam combines the benefits of stochastic gradient descent with momentum and root mean square propagation. In addition, the batch sizes varied depending on a step size of 10 within the range of 10 to 100, and it was saturated at 10 epochs. The selected networks were configured with a 1 gradient threshold, and the learning rate ranged between 0.1 to 0.001.
PDDNet‑AAE
In this method, we experimented based on an arithmetic ensemble average that included late fusion. Initially, TL was applied to architectures, including DenseNet201, DenseNet101, ResNet50, ResNet18, GoogleNet, AlexNet, EfficientNetB7, NASNetMobile, and ConvNeXtSmall. In this instance, the focal contribution of this study is to substitute the last three layers of these CNNs, that is to say, a fully connected (designed to learn features from the images), a softmax (sometimes called a normalized exponential function that presents covert real numbers to probability function to approximate outcomes), and a classification (follows the softmax layer, it detects, classify mutually exclusive classes (categories) via the cross entropy function) layers with new layer definition. After fine-tuning procedure, the effectiveness of every transfer learning pretrained model was analyzed employing the data prepared for testing. Finally, the results of the PDDNet-AAE ensemble were agreed upon with the rest of the finely adjusted networks.
PDDN‑EA
For the early fusion, this model is trained with the LR classifier with features produced from numerous deep networks with fully connected layers and then concatenates these features using the methodology presented (Section "PDDNet‑AAE"). Figure 2 demonstrates an overview of the method's flow diagram.
Considering the demonstrated flow within Fig. 4, the classifier trains the deep features aggregated after being assembled from numerous pretrained networks. Additionally, we employed various combinations of six defined networks to ascertain the class label with the PDDNet model that we suggested. It is significant to mention that these pretrained networks were utilized in this ensemble.
PDDNet‑LVE
We started by extracting deep features from the layers of these fully connected architectures. Then, the final three layers were changed to the LR classifier of previously trained deep network architectures. The deep features accumulated from every architecture were utilized during classifier training. Finally, the approach of lead voting by a majority (LVE) was employed for all existing labels within the PlantVillage dataset. Only the class label considered to have the highest level of accuracy served as the final selection for the method (LVE), as depicted in Fig. 3.
Obtained results and discussion
This section mainly demonstrates the obtained results and the corresponding discussions of proposed models of an integrated ensemble LR model classifier that uses deep features and averages of the CNN models. The proposed models are based on deep feature extraction, and then we tested three model approaches, namely AAE, EA, and LVE, employing pretrained networks.
We used the PlantVillage dataset to test the suggested approach described in subsection "Methodology". This dataset includes color, gray-scaled, and segmented image categories, encompassing healthy and unhealthy plant left species collected and utilized in their natural ecological setting. Table 4 provides the dataset class literature according to disease names. Table 5 depicts the plant type and image sampling quantities used in this research's training and testing phases; the computer and simulation parameters are presented in Table 6.
We discuss the results and performance assessments in detail in the following subsections. The experiments were used using Matlab2022b simulatorFootnote 2 and NVIDIAFootnote 3 with GeForce RTX 2070 and DirectX runtime version 12.0.
Performance evaluation of classifiers
There are five standard classifiers, for instance, K-Nearest Neighbor (KNN), Support Vector Machine (SVM), Random Forest (RF), LR, and Naive Bayes (NB), that are often used in deep learning methodologies to evaluate these pretrained CNNs networks. Table 7 illustrates the testing accuracy for every class using different classifiers. Moreover, the model performances are further assessed in terms of F1 scores, accuracy, recall, and precision using False Positives (FP), False Negatives (FN), True Negatives (TN), and True Positives (TP).
TP represents the number of instances correctly predicted as positive by the model. In other words, it corresponds to the case where the model predicts the positive class correctly. TN represents the number of instances correctly predicted as negative by the proposed model. It corresponds to the case where the model predicted the negative class correctly. FP epitomizes the number of instances incorrectly predicted as positive by the model. It corresponds to the case where the model predicted the positive class when the actual class was negative. Finally, FN denotes the number of instances incorrectly predicted as negative by the model. It corresponds to the case where the model predicted the negative class when the actual class was positive.
Accuracy
The term "accuracy" is the proportion of correct predictions completed compared to the total number of data points collected (T). In scientific literature, it is referred to as recognition, correctness, or success rate and expressed as Eq. 1.
Precision
The proportion of actual positive samples found to the total samples anticipated to be positive calculated as presented in Eq. 2.
Sensitivity and recall
The term "sensitivity" or "recall" refers to the proportion of correctly anticipated positives to the total number of actual positive results (Eq. 3).
F1- scores
The F1-score refers to the harmonic mean of precision and sensitivity (recall), expressed in Eq. 4.
Based on the testing accuracies presented in Table 6, on average, LR obtained 93.88 %, NB with 70.98%, KNN with 84.93 %, SVM with 91.55%, and RF with an accuracy performance of 78.43%, thus making us select LR to be used during the experiments leading to the conclusion that increasing the data size improves exceptionally the performance accuracies. Table 7 presents the precision and recall values and F1 scores. Finally, Table 8 illustrates the accuracy scores obtained with different batch sizes in the LR classifier.
Performance evaluation on deep learning
We performed fine-tuning for previously trained CNN models using the DL methodology to evaluate these DL networks. The process of fine-tuning was accomplished by transferring new layers to our plant disease detection and classification problem to replace the deep CNN's last three layers, as described earlier. We examined the accuracy of fine-tuning to observe the effect of TL on the overall performance of the counterparts. After using the hyperparameter fine turning, we considered the minimum batch capacity to be sixteen, the max epochs were put to 10, 0.0001 on the weight decay adjustments, and the learning rate primarily ranged from 0.001 to 0.01. Similarly, for the learning optimization approach, a Mini Batch Stochastic Gradient Descent (MB-SGD) was applied for the deep neural networks to optimize their performance. As a result, 5000 iterations were fully completed for the training procedure, and the obtained accuracies are presented in Table 9. The bold figures within all tables denote the best-performing model.
According to Table 9, the DenseNet201 achieved the highest accuracy among pretrained models based on transfer learning, achieving 93.48%, while the AlexNet achieved the lowest performance with 86.93%. Both results can be compared to those attained using transfer learning on the DenseNet201 architecture. It is further observed that an increment in the complexity improves the accuracies. According to these reported results, the last layer of these models is replaced with the LR classifier. Consequently, the LR was fed with deep features extracted from pretrained CNN networks, presented in Table 10.
The LR classifier parameters used were quadratic kernel functions, cubic and tenfold cross-validation approach, and the "one versus all" strategy, which was proven to be the most effective evaluator. According to Table 11, the DenseNet201 model demonstrated an accuracy of 94.86% when detecting plant diseases. Depending on the results, this was the maximum level of accuracy that could be attained after several fine turns. More interestingly, the presented findings in Table 9 are improved to those in Table 10, demonstrating that utilizing the LR as the last layer is advantageous. As a result, we use LR with the other pretrained models with deep features for the remaining part of the experiments.
Performance evaluation on PDDNet‑ AAE model
To evaluate this proposed model, a combination of the above-mentioned pretrained CNNs is used by calculating the average scores from these networks for each class as early as demonstrated in [64]. The accuracy score was calculated using the score-based fusion technique of the deep CNNs with the finest performance, as Table 11 demonstrates. Based on the class distribution, the weighted average accuracy was 93.7%.
Performance evaluation on the PDDNet‑EA model
The early fusion that was hypothesized, the CNN-LR model, was initially developed based on an early fusion combining the information gathered from the deep CNNs (as Fig. 2 demonstrated). Through several combinations of the six selected CNNs, we achieved the outcomes provided in Table 12 in the subsequent columns, determined by the average accuracy and the standard deviation of those scores. For example, based on Table 13, the PDD-AAE model's maximum accuracy score was 96.79% using DenseNet201, ResNet101, AlexNet, ResNet50, and GoogleNet networks. Because of this, utilizing a pretrained version of ResNet18 in the presence of ResNet50 and ResNet101 is not productive, as most networks provide the most significant results without being used.
Performance evaluation on the PDDNet‑LVE model
The results were produced with the PDDNet-LVE model, based on the lead (majority) votes obtained from detecting the class labels acquired from the LR classifier with deep features presented in Fig. 3 and the last column of Table 12. Moreover, the maximum accuracy score possible with the PDD-LVE model was attained when a mixture of AlexNet, DenseNet201, ResNet50, ResNet101, and GoogleNet was used. This resulted in accuracy scores of 96.94% and 97.79% for the EA and LVE models, respectively. These findings are consistent with those seen in Table 13, which shows that the best outcomes were achieved with all CNNs.
Comparison with edge-cutting models
As demonstrated earlier, CNNs have widely been used in object class label classification, object recognition patterns, and objection detection most recently. Since the most pretrained deep networks were considered, DenseNet201, DenseNet101, ResNet50, GoogleNet, ResNet18, AlexNet, EfficientNetB7, NASNetMobile, and ConvNet have been compared based on the documented accuracy with the most recent published results about plant disease classification [83]. Table 13 demonstrates the accuracy of the models used during the experiment, and Table 14 shows the recently proposed model using some or all used pretrained models during the study.
The study considers tomato class with 16,703 plant images obtained from the PlantVillage dataset entailing 1,591 healthy leaves, 373 Mosaic Virus, 3,209 Yellow Leaf Curl Virus, 1,404 Target Spot, 1,676 Spider Mites Two Spotted Spider Mite, 1,771 Septoria leaf spot, 952 Leaf Mold, 1,909 Late Blight, 1,000 Late Blight, 2,127 Bacterial Spots images as presented within the dataset. After 10 epochs, the classification results are demonstrated in Fig. 4, utilizing some of the considered pre-trained models, namely ResNet101 and DesnseNet201. Figure 5 presents a confusion matrix after replacing the first and second modified layers (i.e., a fully connected and a softmax layer) of EfficientNet and ConvNet. Figure 6 presents a confusion matrix of the proposed two models. Note: 1 through 10 on the horizontal axis depict the ten tomato leaf image categories.
Conclusion
In this research, early fusion and lead voting ensembles were introduced, combined with nine pretrained CNNs, and fine-tuned for deep feature extraction. Using TL and 15 classes of PlantVillage Dataset, the models outperformed CNNs in plant and disease detection with 96.74% and 97.79% accuracy. These models are robust and generalizable, providing practical solutions to improve plant disease detection and classification accuracy and effectiveness, improving agricultural practices and sustainable food production as the population grows. The research's findings emphasize the significance of advanced technology in mitigating concerns associated with plant disease classification and detection.
In future research, focus on resolving issues related to real-time data collecting and creating a multi-object deep learning model capable of identifying plant illnesses based on a cluster of leaves rather than just a single leaf amidst comparative statistical analysis. Moreover, we are striving to implement a mobile application or web-enabled service utilizing the trained model derived from this research to support a wider plant disease research community to benefit the agricultural sector. Also, to move toward a more lightweight disease classification, model quantization, and object localization networks are critical to better spot the species leaves in a complex background using the trending vision transformers.
Availability of data and materials
The datasets generated during and analyzed during the current study are available at: https://www.kaggle.com/datasets/abdallahalidev/plantvillage-dataset.
References
Baheti H, Thakare A, Bhople Y, Darekar S, and Dodmani O. Tomato plant leaf disease detection using inception v3. Intell Syst Appl. 2023;959:49–60. Springer, Singapore.
Shin J, Mahmud M, Rehman TU, Ravichandran P, Heung B, et al. Trends and prospect of machine vision technology for stresses and diseases detection in precision agriculture. AgriEngineering. 2023;5(1):20–39.
Shamsul Kamar NA, Abd Rahim SK, Ambrose AA, Awing NH, Samdin Z, Hassan A, Saleh MN, Terhem R. Pest and disease incidence of coniferous species in Taman Saujana Hijau, Putrajaya urban park Malaysia. J Forest Res. 2023;34(6):2065–77.
Poyatos J, Molina D, Martinez AD, Del Ser J, Herrera F. EvoPruneDeepTL: An evolutionary pruning model for transfer learning based deep neural networks. Neural Networks. 2023;158:59–82.
Rangarajan Aravind K and Raja P. Automated disease classification in (selected) agricultural crops using transfer learning. Automatika: ˇcasopis za automatiku, mjerenje, elektroniku, raˇcunarstvo i komunikacije. 2020; 61(2): 260–272.
Praveen P, Nischitha M, Supriya C, Yogitha M, and Suryanandh A. To detect plant disease identification on leaf using machine learning algorithms. Intell Syst Design. 2023;494:239–49. Springer, Singapore.
Nigam S, Jain R, Marwaha S, Arora A, Haque MA, et al. Deep transfer learning model for disease identification in wheat crop. Ecol Inform. 2023;75:102068.
Li E, Wang L, Xie Q, Gao R, Su Z, Li Y. A novel deep learning method for maize disease identification based on small sample-size and complex background datasets. Ecol Inform. 2023;75:102011.
Dubey N, Bhagat E, Rana S, and Pathak K. A novel approach to detect plant disease using DenseNet-121 neural network. Smart Trends Comput Commun 2023;396:63–74. Springer, Singapore.
Eunice JJA, Popescu DE, Chowdary MK, Hemanth J. Deep learning-based leaf disease detection in crops using images for agricultural applications. Agronomy. 2022;12(10):2395.
Picon A, Alvarez-Gila A, Seitz M, Ortiz-Barredo A, Echazarra J, Johannes A. Deep convolutional neural networks for mobile capture device-based crop disease classification in the wild. Comput Electron Agric. 2019;161:280–90.
Geetharamani G, Pandian A. Identification of plant leaf diseases using a nine-layer deep convolutional neural network. Comp Electric Eng. 2019;76:323–38.
Shafik W, Tufail AA, Namoun A, De Silva, Apong RA. A Systematic Literature Review on Plant Disease Detection: Techniques, Dataset Availability, Challenges, Future Trends, and Motivations. IEEE Access. 2023; 11: 59174 – 59203.
Mokarram M, Pourghasemi HR, Pham TM. Identification of suitable location to cultivate grape based on disease infestation using multi-criteria decision-making (MCDM) and remote sensing. Ecol Informatics. 2023;76:102142.
Tao M, Ma X, Huang X, Liu C, Deng R, Liang K, Qi L. Smartphone-based detection of leaf color levels in rice plants. Comput Electron Agric. 2020;173:105431.
Chouhan SS, Singh UP, Jain S. Applications of computer vision in plant pathology: a survey. Arch Comput Methods Eng. 2020;27:611–132.
Thimmegowda TG, Jayaramaiah C. Cluster-based segmentation for tobacco plant detection and classification. Bull Electric Eng Inform. 2023;12(1):75–852023.
Chouhan SS, Singh UP, Jain S. Web facilitated anthracnose disease segmentation from the leaf of mango tree using radial basis function (RBF) neural network. Wireless Pers Commun. 2020;113:1279–96.
Chouhan SS, Kaul A, Singh UP. Image segmentation using fuzzy competitive learning based counter propagation network. Multimed Tools Appl. 2019;78:35263–87.
Chouhan SS, Kaul A and Sinzlr UP. Plants leaf segmentation using bacterial foraging optimization algorithm. In 2019 International Conference on Communication and Electronics Systems (ICCES). 2019: 1500-1505. IEEE.
Chouhan SS, Kaul A, and Singh UP. Radial Basis Function Neural Network for the Segmentation of Plant Leaf Disease. In 2019 4th International Conference on Information Systems and Computer Networks (ISCON). 2019: 713-716. IEEE.
Ferentinos KP. Deep learning models for plant disease detection and diagnosis. Comput Electron Agric. 2018;145:311–8.
Ramcharan A, Baranowski K, McCloskey P, Ahmed B, Legg J, et al. Deep learning for image-based cassava disease detection. Front Plant Sci. 2017;8:1852.
Lu Y, Yi S, Zeng N, Liu Y, Zhang Y. Identification of rice diseases using deep convolutional neural networks. Neurocomputing. 2017;267:378–84.
Iniyan S, Varma VA, Naidu CT. Crop yield prediction using machine learning techniques. Adva Eng Softw. 2023;175:103326.
Liu B, Zhang Y, He D, Li Y. Identification of apple leaf diseases based on deep convolutional neural networks. Symmetry. 2018;10(1):11.
Ahmad I, Hamid M, Yousaf S, Shah ST, and Ahmad MO. Optimizing pretrained convolutional neural networks for tomato leaf disease detection. Complexity. 2020;2020:1–6.
Rangarajan AK, Purushothaman R. Disease classification in eggplant using pre-trained vgg16 and msvm. Sci Rep. 2020;10(1):1–11.
Sethy PK, Barpanda NK, Rath AK, Behera SK. Deep feature based rice leaf disease identification using support vector machine. Comput Electron Agric. 2020;175:105527.
Gladston A and Sucithra B. Hand-crafted feature extraction and deep learning models for leaf image recognition. Handbook of Research on Computer Vision and Image Processing in the Deep Learning Era. 2023: 161-178. IGI Global.
Durmus H, Günes E. O, and Kırcı M.. Disease detection on the leaves of the tomato plants by using deep learning. 6th International Conference on Agro-Geoinformatics, Fairfax VA, USA. 2017: 1–5, IEEE.
Li Y, Nie J, Chao X. Do we really need deep cnn for plant diseases identification? Comput Electron Agric. 2020;178:105803.
Zeng W, Li M. Crop leaf disease recognition based on self-attention convolutional neural network. Comput Electron Agric. 2020;172:105341.
Singh V, Misra AK. Detection of plant leaf diseases using image segmentation and soft computing techniques. Inform Process Agric. 2017;4(1):41–9.
Prasad S, Peddoju SK and Ghosh D. Multi-resolution mobile vision system for plant leaf disease diagnosis. SIViP. 2016; 10(2) 379–388.
Hossain MS, Mou RM, Hasan MM, Chakraborty S, and Razzak MA. Recognition and detection of tea leaf's diseases using support vector machine. Proceedings of 14th International Colloquium on Signal Processing and Its Applications (CSPA), Penang, Malaysia. 2018: 150–154.
Kaur S, Pandey S, Goel S. Semi-automatic leaf disease detection and classification system for soybean culture. IET Image Process. 2018;12(6):1038–48.
Waghmare H, Kokare R and Dandawate Y. Detection and classification of diseases of grape plant using opposite colour local binary pattern feature and machine learning for automated decision support system. Proceedings of 3rd International Conference on Signal Processing and Integrated Networks (SPIN), Noida, India, 2016: 513–518,.
Athanikar G, Badar P. Potato leaf diseases detection and classification system. Int J Comput Sci Mobile Comput. 2016;5(2):76–88.
Sabrol H and Satish K. Tomato plant disease classification in digital images using classification tree. Proceedings of 2016 International Conference on Communication and Signal Processing (ICCSP), Melmaruvathur, India. 2016: 1242–1246.
Padol PB, and Yadav AA. SVM classifier based grape leaf disease detection. Proceedings of 2016 Conference on Advances in Signal Processing (CASP), Pune, India. 2016: 175–179.
Dubey SR, Jalal AS. Apple disease classification using color, texture and shape features from images. Signal Image Video Process. 2016;10(5):819–26.
Ramakrishnan M. Groundnut leaf disease detection and classification by using back probagation algorithm. IEEE International Conference on Communications and Signal Processing (ICCSP), Chennai, India. 2015: 964–968.
Yun S, Xianfeng W, Shanwen Z, Chuanlei Z. PNN based crop disease recognition with leaf image features and meteorological data. Int J Agric Biol Eng. 2015;8(4):60–8.
Y. Dandawate, and R. Kokare. An automated approach for classification of plant diseases towards development of futuristic Decision Support System in Indian perspective. Proceedings of IEEE international conference on advances in computing, communications and informatics (ICACCI), Bangalore, India. 2015: 794–799.
Pujari JD, Yakkundimath R, Byadgi AS. Classification of fungal disease symptoms affected on cereals using color texture features. Int J Signal Process Image Process Pattern Recogn. 2013;6(6):321–30.
Kurniawati NN, Abdullah SNHS, Abdullah S and Abdullah S. Investigation on image processing techniques for diagnosing paddy diseases. Proceedings of 2009 International Conference of Soft Computing and Pattern Recognition. 2009: 272–277.
Özgüven MM, Adem K. Automatic detection and classification of leaf spot disease in sugar beet using deep learning algorithms. Physica A. 2019;535:122537.
Li Y, Wang H, Dang LM, Sadeghi-Niaraki A, Moon H. Crop pest recognition in natural scenes using convolutional neural networks. Comput Electron Agric. 2020;169:105174.
Zhong Y, Zhao Z. Research on deep learning in apple leaf disease recognition. Comput Electron Agric. 2020;168:105146.
Yang Y, Liu Z, Huang M, Zhu Q, Zhao X. Automatic detection of multi-type defects on potatoes using multispectral imaging combined with a deep learning mode. J Food Eng. 2023;336:111213.
Wang L, Sun J, Wu X, Shen J, Lu B, et al. Identification of crop diseases using improved convolutional neural networks. IET Comput Vision. 2020;14(7):538–45.
Arsenovic M, Karanovic M, Sladojevic S, Anderla A, Stefanovic D. Solving current limitations of deep learning-based approaches for plant disease detection. Symmetry. 2019;11(7):939.
Zhang S, Huang Z, Zhang C. Three-channel convolutional neural networks for vegetable leaf disease recognition. Cogn Syst Res. 2019;53:31–41.
Hu G, Wu H, Zhang Y, Wan M. A low shot learning method for tea leaf’s disease identification. Comput Electron Agric. 2019;163:104852.
Too EC, Yujian L, Njuki S, Yingchun L. A comparative study of fine-tuning deep learning models for plant disease identification. Comput Electron Agric. 2019;161:272–9.
Anwar Z, Masood S. Exploring Deep Ensemble Model for Insect and Pest Detection from Images. Proc Comput Sci. 2023;218:2328–37.
Altuntaş Y, Cömert Z, Kocamaz AF. Identification of haploid and diploid maize seeds using convolutional neural networks and a transfer learning approach. Comput Electron Agric. 2019;163:104874.
Ma R, Wang J, Zhao W, Guo H, Dai D, et al. Identification of maize seed varieties using MobileNetV2 with improved attention mechanism CBAM. Agriculture. 2023;13(1):11.
Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. Commun ACM. 2017;60(6):84–90.
Barbedo JG. Factors influencing the use of deep learning for plant disease recognition. Biosyst Eng. 2018;172:84–91.
Wallelign S, Polceanu M, and Buche C. Soybean plant disease identification using convolutional neural network. 13 International Florida Artificial Intelligence Research Society Conference (FLAIRS-31). 2018: 146–151, 2018.
Sapkal AT, Kulkarni UV. Comparative study of leaf disease diagnosis system using texture features and deep learning features. Int J Appl Eng Res. 2018;13(19):14334–40.
Szegedy C, Liu W, Jia Y, Sermanet P, Reed S et al. Going deeper with convolutions. Proceedings of 2015 IEEE Conference on Computer Vision and Pattern Recognition. 2015: 1–9.
Brahimi M, Boukhalfa K, Moussaoui A. Deep learning for tomato diseases: classification and symptoms visualization. Applied Artificial Intelligence. 2017;31(4):299–315.
Wang G, Sun Y, and Wang J. Automatic image-based plant disease severity estimation using deep learning. Comput Intell Neurosci. 2017;2017:2917536.
He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR). 2016: 770–778.
Amara J, Bouaziz B, and Algergawy A. A deep learningbased approach for banana leaf diseases classification. In B. Mitschang, N. Ritter, H. Schwarz, M. Klettke, A. Thor, O. Kopp, M. Wieland (Eds.), Datenbanksysteme für Business, Technologie und Web (BTW 2017). 2017: 79–88.
Srilatha D, Thillaiarasu N. Implementation of intrusion detection and prevention with deep learning in cloud computing. J Inform Technol Manag. 2023;15:1–8.
Sladojevic S, Arsenovic M, Anderla A, Culibrk D, and Stefanovic D. Deep neural networks-based recognition of plant diseases by leaf image classification. Comput Intell Neurosci. 2016;2016:3289801.
Fujita E, Kawasaki Y, Uga H, Kagiwada S, and Iyatomi H. Basic investigation on a robust and practical plant diagnostic system," Proceedings of 15th IEEE International Conference on Machine Learning and Applications (ICMLA), Anaheim, Canada. 2016: 989–992.
Huang G, Liu Z, Van Der Maaten L, and Weinberger KQ. Densely connected convolutional networks. Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). 2017: 4700–4708.
Zhang Y, Yang Y, Zhang J, Wang Y. Sensitivity study of multi-field information maps of typical landslides in mining areas based on transfer learning. Front Earth Sci. 2023;11:1105985.
Li P, Liu F, Jiao L, Li S, Li L, et al. Knowledge transduction for cross-domain few-shot learning. Pattern Recogn. 2023;141:109652.
Wu X, Fan X, Luo P, Choudhury SD, Tjahjadi T, et al. Laboratory to Field: Unsupervised Domain Adaptation for Plant Disease Recognition in the Wild. Plant Phenomics. 2023;5:00–38.
You H, Lu Y, Tang H. Plant disease classification and adversarial attack using SimAM-EfficientNet and GP-MI-FGSM. Sustainability. 2023;15(2):1233.
Singh V, Chug A, Singh AP. Classification of Beans Leaf Diseases using Fine Tuned CNN Model. Proc Comput Sci. 2023;218:348–56.
Tiwari S, Jain A, Sapra V, Koundal D, Alenezi F, et al. A smart decision support system to diagnose arrhythymia using ensembled ConvNet and ConvNet-LSTM model. Exp Syst Appl. 2023;213:118933.
Nie X, Wang L, Ding H, Xu M. Strawberry verticillium wilt detection network based on multi-task learning and attention. IEEE Access. 2019;7:170003–11.
Ramesh S, Vydeki D. Recognition and classification of paddy leaf diseases using optimized deep neural network with jaya algorithm. Inform Process Agric. 2020;7(2):249–60.
Shafik W, Tufail A, Liyanage CD, Apong RAAHM. Using a Novel Convolutional Neural Network for Plant Pests Detection and Disease Classification. J Sci Food Agric. 2023.
Zhang F, Xu X, Xiao Z, Wu J, Geng L, et al. Automated quality classification of colour fundus images based on a modified residual dense block network. Signal Image Video Process. 2020;14(1):215–23.
Jeevanantham R, Vignesh D, Abdul RA and Angeljulie J. Deep learning-based plant diseases monitoring and detection system. In 2023 International Conference on Sustainable Computing and Data Communication Systems (ICSCDS). 2023; 360-365, , Erode, India, IEEE.
Haridasan A, Thomas J, Raj ED. Deep learning system for paddy plant disease detection and classification. Environ Monit Assess. 2023;195(1):120.
Pahurkar AB and Deshmukh RM. SMoGW-based deep CNN: Plant disease detection and classification using SMoGW-deep CNN classifier. In Web Intelligence (No. Preprint, pp. 1-22). IOS Press.
Prathima S, Praveena NG, Sivachandar K, Nath SS, Sarala B. Generic Paddy Plant Disease Detector (GP2D2): an application of the Deep-CNN Model. Int J Electric Comput Eng Syst. 2023;14(6):647–56.
Sathiya V, Josephine MS, Jeyabalaraja V. Plant disease classification of basil and mint leaves using convolutional neural networks. Int J Intell Syst Appl Eng. 2023;11(2):153–63.
Code availability
Available upon request.
Funding
This research was fully funded by the School of Digital Science, Universiti Brunei Darussalam, and the Ministry of Education (MoE), Universiti Brunei Darussalam.
Author information
Authors and Affiliations
Contributions
Wasswa Shafik: Conceptualization, Methodology, Investigation (lead), Data Curation, Formal Analysis (lead), Writing – original draft, Software (lead). Ali Tufail: Funding Acquisition, Methodology, Resources, Supervision, Visualization, Writing – review and editing. Chandratilak De Silva Liyanage: Conceptualization (supporting); Supervision, Writing – review and editing (equal). Rosyzie Anna Awg Haji Mohd Apong: Supervision, Conceptualization (supporting); Writing – review and editing (equal). All authors read and approved the final manuscript.
Authors' information
Wasswa Shafik (Member, IEEE) received a Bachelor of Science degree in information technology engineering with a minor in mathematics from Ndejje University, Kampala, Uganda, in 2016 and a Master of Engineering degree in information technology engineering (MIT) from Yazd University, Yazd, Iran, in 2020. He is pursuing a Ph.D. in computer science with the School of Digital Science, Universiti Brunei Darussalam, Brunei Darussalam. He is also the Founder and a Principal Investigator of the Dig Connectivity Research Laboratory (DCRLab) after serving as a Research Associate at the Network Interconnectivity Research Laboratory at Yazd University. Prior to this, he worked as a Community Data Analyst at Population Services International (PSI-Uganda), Community Data Officer at Programme for Accessible Health Communication (PACE-Uganda), Research Assistant at the Socio-Economic Data Centre (SEDC-Uganda), Prime Minister’s Office, Kampala, Uganda, an Assistant Data Officer at TechnoServe, Kampala, IT Support at Thurayya Islam Media, Uganda, and Asmaah Charity Organization. He has more than 60 publications in renowned journals and conferences. His research interests include plant pathology, computer vision, AI-enabled IoT/IoMTs, IoT/IIoT/OT security, cyber security, and privacy.
Ali Tufail is currently working as a Senior Assistant Professor of Distributed and Cyber-Physical Systems at the School of Digital Science (SDS), Universiti Brunei Darussalam. He completed his Ph.D. in Information and Communication Engineering at Ajou University South Korea in 2011, Master of Science in Advanced Computing at the University of Bristol UK in 2006, and Bachelor's degree in Information Technology at the National University of Sciences and Technology Pakistan in 2005. Dr. Ali’s teaching and research specializations are in wireless sensor networks, the Internet of Things, and vehicular ad hoc networks. Dr. Ali has more than 10 years of teaching experience in countries such as Pakistan, South Korea, Turkey, and Saudi Arabia. He has 25+ publications in renowned journals and conferences. Dr. Ali is also serving as SDS Seminar Coordinator and Learning Technology Advisor.
Professor. Chandratilak De Silva Liyanage received a BSc Eng (Hons) degree from the University of Moratuwa Sri Lanka in 1985, an MPhil degree from The Open University of Sri Lanka in 1989, and MEng and PhD degrees from the University of Tokyo, Japan in 1992 and 1995, respectively. He was with the University of Tokyo, Japan, from 1989 to 1995. From April 1995 to March 1997, he pursued his postdoctoral research as a researcher at ATR (Advanced Telecommunication Research) Laboratories, Kyoto, Japan. In March 1997, he joined The National University of Singapore as a Lecturer, where he was an Assistant Professor till June 2003. He was with Massey University, New Zealand, from 2003 to 2007. Currently, he is the Professor of Engineering and the Deputy Dean of the Faculty of Integrated Technologies at the University of Brunei Darussalam. Liyanage has published over 160 technical papers in these areas in international conferences, journals, and Japanese national conventions and holds one Japanese national patent, which was successfully sold to Sony Corporation Japan for commercial utilization. He holds 1 US and 1 Brunei patent. Liyanage’s works have been cited as pioneering works in bimodal (audio and video signal-based) emotion recognition by many researchers. His papers so far have been cited more than 4500 times (according to scholar.google.com) with an h-index of 27. He received the Best Student Paper Award from SPIE (The International Society for Optical Engineering) for an outstanding paper contribution to the International Conference on Visual Communication and Image Processing (VCIP) in 1995. He also received the National University of Singapore Department of ECE Teaching commendation award in 2001 and 2002 consecutively. He is a senior member of IEEE USA and the interim chair of IEEE Brunei Darussalam Subsection. He was the General Chair of the 4th International Conference Computational Intelligence and Robotics and Autonomous Systems (CIRAS2007) held in New Zealand.
Rosyzie Anna Awg Haji Mohd Apong received her Ph.D. in Computer Science from Manchester University in 2018, her MSc in Multimedia and Internet Computing from Loughborough University in 2016, and her BSc in Computer Science from Strathclyde University in 2004. She is currently a lecturer at the School of Digital Science, Universiti Brunei Darussalam. Her research interests are Text Mining, the Internet of Things, and Information Retrieval. She has published and reviewed about ten papers.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Shafik, W., Tufail, A., De Silva Liyanage, C. et al. Using transfer learning-based plant disease classification and detection for sustainable agriculture. BMC Plant Biol 24, 136 (2024). https://doi.org/10.1186/s12870-024-04825-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-024-04825-y