Abstract
Background
Chrysanthemum is a popular ornamental plant worldwide. MYB (v-myb avian myeloblastosis viral oncogene homolog) transcription factors play an important role in everything from stress resistance to plant growth and development. However, the MYB family of chrysanthemums has not been the subject of a detailed bioinformatics and expression investigation.
Results
In this study, we examined 324 CnMYB transcription factors from Chrysanthemum nankingense genome data, which contained 122 Cn1R-MYB, 183 CnR2R3-MYB, 12 Cn3R-MYB, 2 Cn4R-MYB, and 5 atypical CnMYB. The protein motifs and classification of CnMYB transcription factors were analyzed. Among them, motifs 1, 2, 3, and 4 were found to encode the MYB DNA-binding domain in R2R3-MYB proteins, while in other-MYB proteins, the motifs 1, 2, 3, 4, 5, 6, 7, and 8 encode the MYB DNA-binding domain. Among all CnMYBs, 44 genes were selected due to the presence of CpG islands, while methylation is detected in three genes, including CnMYB9, CnMYB152, and CnMYB219. We analyzed the expression levels of each CnMYB gene in ray floret, disc floret, flower bud, leaf, stem, and root tissues. Based on phylogenetic analysis and gene expression analysis, three genes appeared likely to control cellulose and lignin synthesis in stem tissue, and 16 genes appeared likely to regulate flowering time, anther, pollen development, and flower color. Fifty-one candidate genes that may be involved in stress response were identified through phylogenetic, stress-responseve motif of promoter, and qRT-PCR analyses. According to genes expression levels under stress conditions, six CnMYB genes (CnMYB9, CnMYB172, CnMYB186, CnMYB199, CnMYB219, and CnMYB152) were identified as key stress-responsive genes.
Conclusions
This research provides useful information for further functional analysis of the CnMYB gene family in chrysanthemums, as well as offers candidate genes for further study of cellulose and lignin synthesis, flowering traits, salt and drought stress mechanism.
Similar content being viewed by others
Background
Transcription factors (TFs) bind to the DNA regulator sequence to regulate plant growth and development. They can also boost plant tolerance to abiotic and biotic stressors by orchestrating regulatory networks. The MYB family is a prominent TF family in eukaryotes that regulates a variety of biological processes [1]. The name of MYB TFs derives from their conserved N-terminal MYB DNA-binding domain (DBD) repeats (Rs), which usually contain 52 amino acid residues per repetition. The MYB domain comprises one to four imperfect amino acid repeats. Accordingly, the MYB protein family can be divided into four subclasses based on the number of MYB domains: 1R, R2R3-, 3R-, and 4R-MYB proteins [1].
The first MYB gene was discovered in maize in 1997 [2]. Since then, a considerably wider variety of plant species, such as Arabidopsis thaliana [3], Oryza sativa (rice) [4], Morella Rubra (Chinese bayberry) [5], Cajanus Cajan (pigeon pea) [6], etc. have shown evidence of the MYB family in genome-wide analyses. Previous studies have shown that the MYB family participates in diverse processes, including primary and secondary metabolism [1, 7,8,9,10,11], root hair development [12,13,14], flower development [15, 16], and responses to biotic and abiotic stresses [1, 17,18,19,20,21,22,23,24,25,26,27].
DNA methylation is a conserved epigenetic mark that is important for the development and stress responses in plants and many animals [28]. The CpG islands in plant genomes are more frequently methylated, and these variations in methylation status at promoters limit or increase the capacity of transcription factor (TF) proteins to bind to the DNA promoter, which in turn inhibits or increases the transcriptional activity of genes [28, 29]. Although multiple studies suggested that DNA methylation controlled anthocyanin accumulation and the MYB TFs’ role in the stress response [30, 31]. Studies on the effect of methylation in controlling MYB TF activities via CpG islands are scarce.
The productivity and quality of the commercially grown ornamental plant Chrysanthemum morifolium Ramat are considerably impacted by harsh environmental conditions [32]. The MYB TF family, which are essential transcription factors in the plant stress response system, should be crucial for the growth and development of chrysanthemums. Chrysanthemum, on the other hand, has a substantial number of hybrid polyploidy variants lacking high-quality reference genomes. Because there is currently a dearth of reliable information about the Chrysanthemum reference genome, only a portion of the MYB family of genes has been studied in Chrysanthemum [33]. It has become crucial to conduct genome-wide analysis of the MYB superfamily in chrysanthemum. Fortunately, a whole-genome fine mapping of C. nankingense was created using next-generation sequencing [34]. C. nankingense is one of the progenitors of Chrysanthemum×morifolium (Ramat) [32]. Due to its simple diploid genetic background, it is suitable to use as a model plant in genetic studies. Identification and incorporation of important genes into commercial varieties improve the quality and stress tolerance of the plant. Therefore, it is of great significance to identify the MYB family genes in C. nankingense.
In this study, 324 MYB genes were found in the genome of C. nankingense (http://www.amwayabrc.com). In addition, we looked into the MYB family genes’ evolutionary relationships, conserved protein domains, exon-intron architectures, cis-acting elements, and recognition of CpG islands. We further analyzed the expression patterns of these genes in different tissues based on RNA-Seq data. Finally, a phylogenetic tree that included 51 CnMYBs and 64 functional MYBs regulating flavonoid biosynthesis or defense responses from other species was generated in order to find additional CnMYBs involved in the regulation of flavonoid biosynthesis and defense responses in C. nankingense. The expression levels of selected 18 response genes and the methylation status of 3 genes of the CnMYB genes family in shoots under salt and drought stresses were then analyzed by quantitative real-time PCR and methylation-specific polymerase chain reaction (MSP). Together, these findings lay a strong framework for further research into the functions of the CnMYB gene family in C. nankingense under various environmental stresses.
Results
Identification and sequence features of MYB genes in Chrysanthemum nankingense
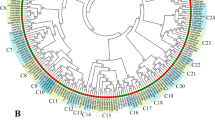
To identify CnMYB genes present in the C. nankingense genome, the MYB domain (PF00249) from the Pfam database was used as a query in the HMM search against the genome. A total of 324 deduced amino acid sequences that might contain MYB repeats or MYB-like repeats were obtained, named CnMYB1 to CnMYB324. All putative MYBs were further examined by the NCBI conserved Domain Search and SMART program for the presence of the MYB DNA-binding domains. Four different subfamilies were differentiated based on the number and location of domain repeats, including 1R-MYB (117), R2R3-MYB (183), 3R-MYB (12), and 4R-MYB (2) genes (Fig. 1, Table S1). A phylogenetic tree of CnMYBs was constructed by aligning the whole set of MYB protein sequences from C. nankingense (Fig. 1). Based on the genome data, five CnMYB contained five or six complete MYB domains, and therefore these five MYB members are classified as atypical MYBs (Table S1). These genes’ complete information includes gene name, gene ID, protein length, molecular weight (MW), isoelectric point (pI), and scaffold location is available in Table S1. The protein length varied, ranging from 70 amino acids to 1451 amino acids, the MW ranged from 8.03 to 162.55 kDa, and the pI varied from 4.08 to 11.51. Since the genome information of C.nankingense did not provide chromosome information, all 324 CnMYB genes were located in 289 scaffolds (Table S1).
CpG islands, which are rich in CG, are generally located near the promoters of genes [28, 29]. DNA methylation is higher in these regions in plant genomes [29]. CpG islands were detected in 44 members (Fig. 1, Table S2). Amongst them, ten genes contained CpG islands in the promoter region, 19 members were found to have it in the gene body, and 18 in the cross-promoter and gene region. Three members have two CpG islands, and the island length ranged from 305 to 965 bp (Table S2).
The classification, motif composition, and gene structure of the CnMYB gene family in C. nankingense
To further classify and understand the functions of the R2R3-CnMYB gene family members, phylogenetic analyses were carried out based on complete protein sequences of 183 R2R3-CnMYBs and 126 R2R3-AtMYBs. Based on the R2R3-AtMYBs classification principle [1], 183 R2R3-CnMYBs can be divided into 32 subfamilies (C1-C32) (Fig. 2). The C1-C25 subfamilies corresponded to S1-S25 in Arabidopsis. C1-C25 subfamilies included 117 R2R3-CnMYBs. There were seven specific clades (C26-C32) in C. nankingense that did not cluster with Arabidopsis, and none of the R2R3-CnMYBs belonged to the S6, S15, S19, and S23 Arabidopsis subfamilies. The largest clade was C27 with 15 R2R3-CnMYBs. Other CnMYBs (1R-MYB, 3R-MYB, 4R-MYB, and atypical MYBs) proteins could be divided into I-XVI subfamilies (Fig. S2).
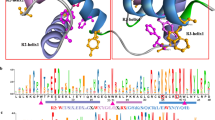
Ten conserved motifs were identified in the R2R3-CnMYBs proteins by MEME software, motif 1, motif 2, motif 3, and motif 4 were found to encode the MYB DNA-binding domain, while the other motifs didn’t have function annotation (Fig. 3A, Fig. S1). Each R2R3-CnMYB protein contained one to seven conserved motifs. Most R2R3-CnMYB proteins (83%) contain a motif group that has 1, 2, 3, and 4 motifs, with C26 (CnMYB116, CnMYB164, and CnMYB143), C27 (CnMYB60, CnMYB163, et al.,) and C32 (CnMYB103, CnMYB227, et al.,) subfamilies lacking this motif group (Fig. 3A). Other CnMYB proteins (1R-MYB, 3R-MYB, 4R-MYB, and atypical MYBs) have one to ten conserved motifs (Fig. S3). Most proteins contain two motifs, VIII, X, XII, XIV, and XVI had several motifs (Fig. S2A). The MYB DNA-binding domains were represented by motifs 1, 2, 3, 4, 5, 6, 7, and 8 (Fig. S3).
Conserved motif and gene structure analysis of Chrysanthemum nankingense R2R3-MYB genes. A Motif distribution of R2R3-MYB genes. B Gene structure of R2R3-MYB genes. The tree was constructed in MEGA 7.0 using the full-length amino acid sequences of 183 R2R3-MYBs in Chrysanthemum nankingense. The ten conserved motifs are shown in different colors and their specific sequence information is provided in Supplementary Fig. S1. The exon-intron structural diagram shows CDS, UTR, and introns as green boxes, yellow boxes, and black lines, respectively
As shown in Fig. 3B, the structure of R2R3-CnMYBs was also examined. The 183 R2R3-MYB genes contained different number of exons, varying from 1 to 13, with an average of 3.2. Most of the R2R3-MYB genes had three (109/183) exons and accounted for approximately 60% of R2R3-MYB gene family members, whereas 23% and of R2R3-MYB genes have less than three exons, while 18% have more than three exons. In addition, most of the R2R3-MYB genes clustered in C26 (CnMYB109, CnMYB236 et al.,) and C28 (CnMYB42, CnMYB313, et al.,) having more than three exons. As shown in Fig. S2B, the number of exons in other CnMYB genes (1R-MYB, 3R-MYB, 4R-MYB, and atypical MYBs) ranged from one to eighteen. In addition, combined with phylogenetic analysis, most genes clustered in the same group had the same or a similar number of exons (Fig. S2B).
Expression profiles of CnMYB genes in different tissues
The expression trends of CnMYB genes in different tissues, such as root, stem, leaf, flower bud, tubular flower, and tongue flower, were calculated by using FPKM values based on previously generated Illumina RNA-Seq data [34]. An expression heatmap of the CnMYB genes was generated (Fig. 4, Fig. S4). The R2R3-MYB genes expression results showed that 64 genes (FPKM> 0) had expression and 26 genes (FPKM = 0) were not expressed in any tissue (Table S3). The number of R2R3-MYB genes expressed in the root, stem, leaf, bud, tubular flowers, and tongue flowers was 62, 73, 61, 93, 72, and 55, respectively (Table S3), the expression level of these genes was over 1 (FPKM) (Table S3). R2R3-MYB genes expression were highest in the flower bud tissue and lowest in the tongue flowers (Fig. 4, Table S3).
Based on transcriptome data, 85 R2R3-MYB genes (FPKM> 0) were expressed in all three flower tissues. Among them, CnMYB219, CnMYB152, CnMYB242, CnMYB308, CnMYB81, CnMYB224, CnMYB127, and CnMYB296 genes were highly expressed in the three tissues (FPKM> 20) (Table S3). We also identified genes that were specifically expressed in the bud (23 genes, CnMYB167, CnMYB102, CnMYB212, CnMYB95, and CnMYB284 genes were highly expressed in this tissue (FPKM> 3)), tubular flower (10 genes, CnMYB175’ FPKM> 3 in this tissue), and tongue flower tissue (1 gene, CnMYB206’ FPKM = 0.09 in this tissue) (Table S3). Of these 183 R2R3-MYB genes, transcript levels of 46 (CnMYB219, CnMYB222, CnMYB152, et al.,) and 57 (CnMYB240, CnMYB222, CnMYB152, et al.,) genes were higher in the flower bud than in the tubular flower and tongue flower, respectively. However, transcript levels of 31 (CnMYB127, CnMYB308, CnMYB224, et al.,) and 14 genes (CnMYB127, CnMYB219, CnMYB21, et al.,) were lower in the flower bud than in the tubular flower and tongue flower (Table S3).
Regarding the remaining MYB genes (1R-MYB, 3R-MYB,4R-MYB, and atypical MYBs), tissues from the root, stem, leaf, flower bud, tubular flower, and tongue flower all showed detectable transcript levels for 64 (CnMYB15, CnMYB256, CnMYB151, CnMYB45, CnMYB217, CnMYB265, CnMYB26, CnMYB223, CnMYB61, CnMYB46, and CnMYB247) genes were highly expressed in the root tissue (FPKM> 20), 76 (26 genes such as CnMYB15, CnMYB181, and CnMYB126 were highly expressed in the stem tissue (FPKM> 20)), 71 (20 genes such as CnMYB277, CnMYB245, and CnMYB126 were highly expressed in the leaf tissue (FPKM> 20)), 85 (17 genes, CnMYB15, CnMYB45, and CnMYB61, et al., (FPKM> 20)), 70 (15 genes, CnMYB126, CnMYB245, and CnMYB181, et al., (FPKM> 20)), and 68 (15 genes, CnMYB126, CnMYB245, and CnMYB256, et al., (FPKM> 20)) out of the 141 genes, respectively (FPKM> 1). These MYB genes were expressed most in flower bud tissue and least in root tissue (Fig. S4, Table S4). An expression heatmap of other MYB genes was generated (Fig. S4). These CnMYB genes expression results showed that 71 genes (FPKM> 0) exhibited expression and 22 genes (FPKM = 0) were not expressed in any tissue (Table S4).
A total of 82 CnMYB genes (FPKM> 0) were expressed in all three flower tissues (Table S4). Among them, CnMYB15, CnMYB45, CnMYB61, CnMYB256, CnMYB217, CnMYB151, CnMYB265, CnMYB126, CnMYB181, and CnMYB26 genes were highly expressed in the three tissues (FPKM> 20) (Table S4). We also identified genes that were specifically expressed in the bud (11 genes, CnMYB40 and CnMYB89 genes were highly expressed in this tissue (FPKM> 3)), tubular flower (7 genes, none of the genes were highly expressed in this tissue), and tongue flower tissue (1 gene, it’s FPKM = 0.093) (Table S4). Of these 141 CnMYB genes, transcript levels of 26 and 27 genes (such as CnMYB15, CnMYB45, and CnMYB61, et al.,) were higher in the flower bud than in the tubular flower and tongue flower. However, transcript levels of 20 (such as CnMYB126, CnMYB245, and CnMYB181, et al.,) and 15 genes (CnMYB126, CnMYB245, and CnMYB256, et al.,) were lower in the flower bud than in the tubular flower and tongue flower (Table S4).
The expression type of all CnMYBs could be divided into three major patterns (Fig. 5, Table S5, S6, S7). The first trend (56 genes) had the highest expression in the stem (the FPKM values of CnMYB28, CnMYB139, CnMYB60, CnMYB90, CnMYB247, CnMYB76, CnMYB180, CnMYB38, CnMYB10, CnMYB207, CnMYB314, and CnMYB153 genes were greater than 2) and lowest in tongue flower tissues (the FPKM values of CnMYB28, CnMYB314, and CnMYB76 genes were less than − 2) (Fig. 5A, Table S5). The second trend (72 genes, CnMYB222, CnMYB240, CnMYB322, CnMYB63, CnMYB282, CnMYB167, CnMYB111, CnMYB321, and CnMYB212 genes’ FPKM> 2) had the highest expression in the bud (Fig. 5B, Table S6). The members of the third trend (196 genes) had approximately similar gene expression patterns in all tissues (Fig. 5C, Table S7).
Identification and expression analysis of CnMYBs in response to drought and salt stresses
To identify additional CnMYBs participating in the regulation of drought and salt stress in Chrysanthemum, a phylogenetic tree that includes 51 CnMYB members (candidate genes can respond to stress resistance) and 66 functional MYBs regulating flavonoids, flavanols, proanthocyanidins, anthocyanins biosynthesis, and response to stress resistance from other species was generated (Fig. 6). According to Fig. 6, three CnMYBs were selected as candidates, potentially regulating the biosynthesis of flavonoids, i.e., CnMYB127, CnMYB183, and CnMYB229, six for flavonols, i.e., CnMYB8, CnMYB21, CnMYB220, CnMYB39, CnMYB47, and CnMYB218, two for proanthocyanidins, i.e., CnMYB175 and CnMYB93, two for anthocyanins, i.e., CnMYB106 and CnMYB77, and 38 CnMYBs may be involved in abiotic stress responses in C. nankingense. CpG islands were detected in four members (Fig. 6).
To verify whether the candidate TFs had the ability to regulate abiotic stress responses in C. nankingense, the 1.5 kb sequence upstream of their start codons (ATG) of 51 selected CnMYB genes were submitted to the PlantCare online tool and searched for cis-acting elements. A diverse range of cis-acting elements was detected. Despite analyzing a huge number of cis-acting elements, we only focused on stress response elements according to our research objectives (Fig. 7, Table S8). These included anaerobic induction (ARE), drought stress (MYB, MYC, W-box, and MBS), salinity stress (TC-rich repeats), low temperature-responsive (LTR), abscisic acid-responsive (ABRE), and salicylic acid-responsive (TCA) cis-acting elements. Most of the CnMYB promoters contained ARE, ABRE, MYB, and MYC cis-acting elements. The proportion of ABRE was the largest amongst the cis-acting elements. Promoters of 51 stress-responsive candidate genes contain more or less stress response elements (Fig. 7, Table S8).
Predicted cis-acting elements related with stress response and hormones in 51 stress-resistant candidate CnMYB genes’ promoters. Promoter sequences (− 1500 bp) of CnMYBs were analyzed using PlantCARE. Different shapes and colors represent different elements and annotations of cis- acting elements are listed in Supplementary Table S9
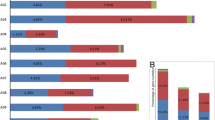
To investigate the function of these CnMYB genes in C. nankingense, the expression patterns of 18 candidate genes that contained ARE and ABRE two stress response elements were analyzed under drought and salt stresses. The expression levels of the 18 selected CnMYBs were detected by qRT-PCR at 0 h, 1 h, 3 h, 6 h, 12 h, and 24 h after these treatments. All genes of the 18 CnMYBs showed fluctuating changes in their transcript levels after the drought stress treatments (Fig. 8A). CnMYB9, CnMYB76, CnMYB109, CnMYB137, CnMYB157, CnMYB186, and CnMYB199 were significantly down-regulated by drought stress, however, except CnMYB186, others genes expression increased to the highest level at 24 h after treatment. The expression of CnMYB172 and CnMYB11 was significantly up-regulated after drought treatment. The expression of CnMYB55, CnMYB149, CnMYB158, and CnMYB127 was lower than CK at the initial stage of drought treatment but gradually increased with increasing stress time and reached a peak at a later stage, suggesting that these CnMYBs may be involved in responses to drought stress.
The expression of 19 CnMYBs was induced by abiotic stresses. The eight leaves seedlings were treated with 15% PEG6000 (A), 150 mM NaCl (B) for qRT-PCR. Samples were taken at 0, 1, 3, 6, 12, and 24 h after the stress treatment. CnActin gene was used as internal reference. The error bar is standard deviation (n = 3; *P < 0.05; **P < 0.01; Student’s t test)
All 18 CnMYBs showed fluctuating changes in their transcript levels after the salt stress treatments (Fig. 8B). CnMYB199, CnMYB186, and CnMYB172 were significantly down-regulated by salt stress. The expression of CnMYB11 and CnMYB137 was significantly up-regulated after salt treatment and reached its peak at 1 h. The expression of CnMYB9, CnMYB158, and CnMYB76 was higher than CK at the initial stage of salt treatment but gradually decreased with increasing stress time. The expression of CnMYB219, CnMYB55, CnMYB149, and CnMYB157 increased first, then decreased, and then increased with increasing stress time, suggesting that these CnMYBs may be involved in responses to salt stresses.
Based on the levels of expression under drought and salt stress, 18 genes were sorted into three categories (when the treatment time was zero, the expression of the CnMYB71 gene was set to one, as a control), among them, CnMYB9, CnMYB172, CnMYB199, CnMYB219, CnMYB186, and CnMYB152 genes were highly expressed, CnMYB11, CnMYB74, CnMYB157, CnMYB149, CnMYB109, and CnMYB127 genes were moderate, CnMYB71, CnMYB55, CnMYB226, CnMYB76, CnMYB137, and CnMYB158 genes have relatively low expression levels (Fig. 8). All 18 genes were regulated by drought and salt stress, but the genes with moderate and high expression levels will be the focus of future research.
CnMYB9, CnMYB152, and CnMYB219 have CpG islands in the gene body (Fig. 6), and the methylation status detected by MSP was analyzed through gel electrophoresis. The expression of CnMYB9, CnMYB152, and CnMYB219 was regulated under drought and salt stresses. Total genomic DNA of C. nankingense was extracted from the leaf of the treated seedlings with drought and salt stresses at the duration of was 0, 1, and 6 h. Gene expression varied greatly compared with CK in these tissues. The MSP results showed the presence of methylation in CnMYB9 and CnMYB152 in leave tissues at different stress times, but the CnMYB219 gene is demethylated at three stress times. So the methylation status of the three genes’ CpG islands did not change under drought and salt stress treatments in three stress times (Fig. S5).
Discussion
In all eukaryotes, the MYB gene family participates in a variety of biological activities, such as metabolism, plant development, and responses to biotic and abiotic stimuli [1]. Numerous species, including model plants [1, 3, 24], significant crops [2, 4, 12], and other plants have been studied in relation to the MYB family. Different plants had varying numbers of MYB family members, including 198 in Arabidopsis [1], 379 in soybeans [35], 129 in the Chines pear [36], 170 in grape [37], and 174 in Chinese bayberry [5]. Here, 324 CnMYB genes from the C. nankingense genome were discovered using bioinformatics research (Fig. 1, Table S1). As a result, there was no correlation between the number of MYB genes and genome size. Based on phylogenetic research, the R2R3-MYB gene family from C. nankingense was divided into 32 subgroups, most of which had conserved motif compositions and exon-intron organizations except for the C26 and C28 subgroups, which showed the need for species to adapt to their particular environments (Figs. 2, 3). This outcome is somewhat consistent with the earlier reports [38]. Other MYBs from C. nankingense were classified into 16 subgroups based on phylogenetic analysis, with a certain degree of divergent conserved motif compositions and exon-intron organizations (Fig. S2).
Based on transcriptome data, we analyzed the expression patterns of 324 CnMYB genes in six tissues and divided them into 3 trends according to different expression patterns (Fig. 5). The expression levels of 60% CnMYBs (196 genes) are similar across all tissues (Third trend) (Fig. 5C). Second trend has 72 genes (22%) and highly expressed in the bud tissues (Fig. 5B). 20% of CnMYB (50 genes) genes are highly expressed in the stem (Fig. 5A). This result indicated that CnMYBs might perform diversified functions during the C. nankingense lifespan.
Lodging is one of the most important agronomic traits that play an essential role in the quality and yield of crops [39, 40]. Cellulose and lignin contents of plant stems can affect the resistance of plants against lodging [39, 41]. A previous study reported that AtMYB46 and AtMYB83 are considered secondary main switches for controlling secondary cell wall biosynthesis [42] AtMYB46 and AtMYB83 can induce the expression of cellulose and lignin synthesis genes to promote the thickening of the secondary wall and their downstream MYB TFs (AtMYB58, AtMYB63, and AtMYB85) also play an important role in the regulation of secondary wall biosynthesis [43,44,45]. Moreover, there are also some MYB TFs, such as AtMYB52, AtMYB54, AtMYB69, and AtMYB103 may also be involved in the regulation of secondary cell walls [44]. Except, for positively regulating second cell wall synthesis genes of MYB transcription, AtMYB7, AtMYB4, and AtMYB32 may be negative regulators of lignin synthesis [46,47,48,49]. The combined transcriptomic data and phylogenetic tree analysis showed that CnMYB278, CnMYB226, CnMYB180, CnMYB76, CnMYB127, and CnMYB219 may be involved in the regulation of cellulose and lignin synthesis (Fig. 2, Table S7). Among them, the CnMYB278 was specifically expressed in the tubular flower tissue, and the CnMYB127 and CnMYB219 genes were highly expressed in all tissues. While CnMYB226, CnMYB180, and CnMYB76 genes were highly expressed in the stem and slightly expressed in other tissues. These results suggest that CnMYB226, CnMYB180, and CnMYB76 genes should be considered important candidate genes involved in the regulation of cellulose and lignin synthesis in stem tissue (Table S7).
MYB transcription factors family is one of the largest transcription factor families in plants. Some MYB TFs have effects on flowering time, anther, and pollen development [15, 16], while other MYB proteins have been found to participate in secondary metabolism processes, such as the biosynthesis of proanthocyanidins, anthocyanins, flavonols, and flavonoids in plants. These metabolites directly affect the color of flowers and fruits, so some MYB TFs are involved in flower color development [8, 50,51,52,53,54,55,56,57]. In this study, we identified a number of CnMYB genes that had higher expression levels in flower bud tissue are higher than in tubular flower and tongue flower tissues (Table S7). These results indicate that most CnMYB were involved in the flower bud development. The combined genes’ higher expression in flower tissues of transcriptomic data and phylogenetic tree analysis showed that CnMYB279, CnMYB288, CnMYB157, CnMYB9, CnMYB152, and CnMYB219 TFs may be involved in the regulation of flowering time, anther, and pollen development (Fig. S2). CnMYB175, CnMYB93, CnMYB229, CnMYB8, CnMYB106, CnMYB39, CnMYB21, CnMYB127, CnMYB47, and CnMYB183 TFs may participate in the regulation of flower color (Fig.6).
Numerous prior investigations [1, 11, 17, 58] have documented the crucial involvement of MYB TFs in the Arabidopsis stress response, particularly in response to abiotic stressors. Secondary Phenylpropanoid metabolites have been widely documented as defense metabolites for their role as antioxidants in protecting plants from abiotic stressors by scavenging reactive oxygen species [59]. In this study, we integrated stress response and phenylpropanoid secondary metabolite proteins with CnMYB proteins to build a phylogenetic tree (Fig. 6). We selected 51 CnMYB proteins that may be involved in abiotic stress response in C. nankingense based on their similarities.
Analysis of cis-elements on gene promoters is very important because it allows predicting the functional role of the genes [60, 61]. A large number of cis-acting regulatory elements were obtained from these 51 CnMYB genes promoter (Fig. 7), including MYC recognition site, WRKY recognition site, MYB recognition site, MYB binding site involved in drought-inducibility (MBS) [62], MYB binding site involved in the flavonoid biosynthetic genes regulation (MBSI), low-temperature responsiveness (LTR) [63], abscisic acid responsiveness (ABRE) [61, 64, 65], anaerobic induction site (ARE), MeJA responsiveness (TGACG-motif and CGTCA-motif), defense and stress responsiveness (TC-rich repeats) that are known to involve in various plant development and stress response functioning [66, 67]. The combined phylogenetic tree and stress-related cis-elements analysis would help us in selecting stress response candidate genes.
To further validate the stress-related candidate genes, we determined the expression patterns of 18 genes from 51 candidate CnMYB genes under drought and salt stresses by analyzing qRT-PCR. Eighteen CnMYB genes expressed differently at different time points, these patterns are similar to AtMYB genes in Arabidopsis under drought and salt stresses (Fig. 6). We focused on moderately and highly expressed genes. Among them, CnMYB127 clustered with Arabidopsis AtMYB4 and AtMYB7, indicating that CnMYB127 can negatively regulate abiotic stress [19, 26, 68]. CnMYB11, CnMYB186, CnMYB199, and CnMYB149, as AtMYB60 homologs; AtMYB88, and AtMYB124, as CnMYB109 homolog; CnMYB219, CnMYB9, and CnMYB152, as AtMYB77 homologs; AtMYB13, and AtMYB15, as CnMYB172 homologs; AtMYB33 as CnMYB157 homolog, can positively regulate some abiotic stress [29, 30, 69,70,71,72,73]. These 18 genes were randomly selected from 51 stress-resistant candidate genes and the stress-resistant functions of the other 33 genes need to be further verified, but the stress resistance of these 18 genes has demonstrated the accuracy of our predicted stress-resistant candidate genes.
There are many reports on MYB TFs’ regulation of abiotic stress in the plant [17, 30, 69, 70, 73], but its link with epigenetic control is rarely reported [30, 31, 74]. As CG or CpG is the most abundant source of DNA methylation, methylation-sensitive amplification polymorphism and methylation-specific PCR (MSP) are two methods that have been applied to methylated genes [75, 76]. Here we have identified 44 genes that may alter the epigenetic makeup of chrysanthemum by having high CpG content in the promoter and gene body regions (Fig. 1, Table S2). Four of the 51 stress response candidate genes had CpG islands, among them, CnMYB219, CnMYB9, and CnMYB152 were identified on the basis of methylation-specific PCR (MSP) results (Fig. 6, Fig. S5). These three genes can regulate gene expression and may play a role in drought and salt stresses, as their expression significantly changed under stress conditions (Fig. 8). We only examined three brief treatment intervals to identify methylation; it would be preferable to investigate the methylation status over a wider range of treatment times in the future. Our future research will focus on the regulation of flower development and drought and salt stress functional candidate genes.
Conclusions
In this study, 324 MYB genes were identified in the genome of C. nankingense. A comprehensive bioinformatics analyses were performed to investigate phylogenetic relationships, conserved motifs, gene structure, and promoter analysis. The present study investigated the expression patterns of these genes in different tissues by analyzing RNA-Seq data. CnMYB226, CnMYB180, and CnMYB76 genes were selected to further explore the regulation of cellulose and lignin synthesis in stem tissue. The regulation of flowering time, anthers development, pollen formation, and flower colors may be mediated by 16 MYB genes. We found 51 candidate genes by bioinformatics analysis that may be involved in stress resistance, and we also confirmed 18 of these candidate genes through qRT-PCR. According to gene expression levels under stress conditions, six CnMYB genes were identified as key stress-responsive genes. The information provided by these results may be helpful for further functional analysis of CnMYB gene to elucidate its development and abiotic stress mechanism in C. nankingense.
Methods
Plant growth conditions and treatments
The wild C. nankingense plants (the deposit number is NEAU0006698) were obtained from the Plant Germplasm Resources and Genetic Engineering, Henan University, Kaifeng, Henan Province, China. The plants were planted in the growth room of the Plant Germplasm Resources and Genetic Engineering, Henan University, and were maintained under the following conditions: 25 °C, with a 16 h light /8 h- dark cycle. The materials were propagated by cuttings and grown in pots. Seedlings with more than eight leaves were treated with two stress conditions: 15% PEG6000, and 150 mM NaCl. The duration of treated Seedlings with two stress conditions was 0, 1, 3, 6, 12, and 24 h, and then used for detection of gene transcript levels. A sample with 0 h duration served as the control. Each experiment was repeated three times.
Database searches, sequence retrieval, and gene identification
The C. nankingense genome and protein sequences were downloaded from the Chrysanthemum genome database (http://www.amwayabrc.com). The Hidden Markov Model (HMM) profile of the MYB DNA binding domain (PF00249) was downloaded from the Pfam protein family database (http://pfam.xfam.org/), then used it as a query (p < 0.001) for the identification of all putative CnMYB genes [77]. The SMART database (http://smart.embl-heidelberg.de/), the NCBI Conserved Domain database (http://www.ncbi.nlm.nih.gov/Stucture/cdd/wrpsb.cgi), and the Pfam database (http://pfam.janelia.org/) program were exploited to test for the presence of the MYB domain. The number of amino acids, open reading frame (ORF) length, molecular weight (MW) and isoelectric point (pI) of CnMYBs were obtained through the ExPASy (http://www.expasy.ch/tools/pi_tool.html) [78]. A. thaliana genome sequences were obtained from TAIR (http://www.Arabidopsis.org/) [79].
Phylogenetic analyses of CnMYBs
The protein sequences of CnMYB proteins were aligned by the ClustalW program and adjusted manually, the phylogenetic tree was constructed by the neighbor-joining method of MEGA 7.0 with 1000 bootstrap replicates [80]. All subsequent phylogenetic trees were constructed by the same method as above.
Gene structure and protein motifs analysis of the CnMYB gene family
The exon, intron and UTR of the CnMYB genes were determined by the Gene Structure Display Sever (GSDS 2.0) (http://gsds.cbi.pku.edu.cn/index.php) using the genomic sequences and coding regions, including intron numbers [81]. The multiple expectations for motif elicitation (MEME) online program (http://meme-suite.org) was used for the identification of conserved motifs in the CnMYB protein sequences, with the following parameters: maximum number of motifs 10, minimum motif width 6, and maximum motif width 200 [82]. The MEME results were also visualized by TBtools software [83].
CpG island analysis of the CnMYB gene family
“Methyl Primer Express V1.0” software (Methyl Primer Express Software v 1.0 Quick Reference Card (thermofisher.com)) was used for the CpG islands analysis of a 1500 bp upstream sequence and DNA (coding protein region). The conditions set for analysis are as follows: the maximum length of the CpG island was 2000 bp, the minimum length was 200 bp; the (C + G)/total bases ratio was 0.5; the criterion of CpG Observed/CpG Expected” was 0.6. The sequences were submitted to software to identify CpG islands or rich regions [84].
Prediction of cis-acting elements in promoter regions
PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) was used for the cis-acting analysis of a 1500 bp upstream sequence of the identified candidate CnMYB genes in the abiotic stress response. TBtools software was used to display the number of identified elements.
RNA-seq based expression analysis of MYB genes in six different tissues
We downloaded raw RNA-Seq data of leaf, stem, root, floral bud, disc florets, and ray florets from the chrysanthemum genome database (http://www.amwayabrc.com) [23]. A value for fragments per kilobase of transcript per million fragments mapped (FPKM) was calculated for each gene, and the log2 (Fold Change) transformed values for each CnMYB family gene were used to generate a heatmap. A heatmap was generated and visualized using the TBTOOLS software [83], the color scale shown represents log2 transformed FPKM values. R software (https://www.r-project.org) was used for the clustering analysis.
Quantitative real-time polymerase chain reaction (qRT-PCR) analysis
Total RNA was extracted from leaves using the RNAprep Pure Plant Plus Kit (spin column) (Takara, Dalian, China) based on specifications. Approximately 1 μg total RNA was reverse-transcribed into cDNA using the “StarScript ǁ First-stand cDNA Synthesis Mix with gDNA Remover”. The qRT-PCR was performed on using a Roche LightCycler 480II detection system, using Hiscript II one-step RT-PCR SYBR Green kit (Vazyme, Nanjing, China). The primers used in this study are listed in Supplementary Table 1. Each sample was conducted in three biological replications. CnActin (KF305683) was used as the reference gene. The relative expression level of each gene was calculated as 2-ΔΔCT equation [85].
Methylation-specific polymerase chain reaction (MSP)
Total DNA was extracted from leaves using “Plant Genomic DNA Kit” (TOLOBIO) based on specifications. The integrity and concentration of the DNA was verified by 1% agarose gel electrophoresis and NanoDrop™ 2000/2000c (ThermoFisher Scientific, USA). Total DNA was treated with sodium bisulfites using “DNA Bisulfite Conversion Kit” (Tiangen) based on specifications. Sodium bisulfites treated DNA was used as the template for methylation-specific PCR. The methylated and unmethylated primers of CpG islands were designed using “Methyl Primer Express Version 1.0” software (Table S9). The PCR product length was between 100 and 175 bp. Three biological replicates and 35 cycles for each reaction were performed [84].
Availability of data and materials
The C. nankingense genome and protein sequences were downloaded from the Chrysanthemum genome database (http://www.amwayabrc.com). Other data have been submitted as supplementary materials. All data analyzed or generated of this study are available from the first or corresponding author on reasonable request.
References
Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010;15(10):573–81.
Paz-Ares J, Ghosal D, Wienand U, Peterson PA, Saedler H. The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J. 1987;6(12):3553–8.
Stracke R, Werber M, Weisshaar B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol. 2001;4(5):447–56.
Jia L, Clegg MT, Jiang T. Evolutionary dynamics of the DNA-binding domains in putative R2R3-MYB genes Identified from Rice subspecies indica and japonica genomes. Plant Physiol. 2004;134(2):575–85.
Cao Y, Jia H, Xing M, Jin R, Grierson D, Gao Z, et al. Genome-wide analysis of MYB gene family in Chinese bayberry (Morella rubra) and identification of members regulating flavonoid biosynthesis. Front Plant Sci. 2021;12:691384.
Yang J, Zhang S, Li H, Wang L, Liu Y, Niu L, et al. Genome-wide analysis and characterization of R2R3-MYB family in pigeon pea (Cajanus cajan) and their functional identification in phenylpropanoids biosynthesis. Planta. 2021;254(4):64.
Yan H, Pei X, Zhang H, Li X, Zhang X, Zhao M, et al. MYB-mediated regulation of anthocyanin biosynthesis. Int J Mol Sci. 2021;22(6):3103.
Ma D, Constabel CP. MYB repressors as regulators of Phenylpropanoid metabolism in plants. Trends Plant Sci. 2019;24(3):275–89.
Liu J, Osbourn A, Ma P. MYB transcription factors as regulators of Phenylpropanoid metabolism in plants. Mol Plant. 2015;8(5):689–708.
Cao Y, Li K, Li Y, Zhao X, Wang L. MYB transcription factors as regulators of secondary metabolism in plants. Biology. 2020;9(3):61.
Roy S. Function of MYB domain transcription factors in abiotic stress and epigenetic control of stress response in plant genome. Plant Signal Behav. 2016;11(1):e1117723.
Li P, Wen J, Chen P, Guo P, Ke Y, Wang M, et al. MYB superfamily in Brassica napus: evidence for hormone-mediated expression profiles, large expansion, and functions in root hair development. Biomolecules. 2020;10(6):875.
Albert NW, Lewis DH, Zhang H, Schwinn KE, Jameson PE, Davies KM. Members of an R2R3-MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. Plant J. 2011;65(5):771–84.
Lee MM, Schiefelbein J. WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell. 1999;99(5):473–83.
Zhang Y, Cao G, Qu LJ, Gu H. Characterization of Arabidopsis MYB transcription factor gene AtMYB17 and its possible regulation by LEAFY and AGL15. J Genet Genomics. 2009;36(2):99–107.
Punwani JA, Rabiger DS, Lloyd A, Drews GN. The MYB98 subcircuit of the synergid gene regulatory network includes genes directly and indirectly regulated by MYB98. Plant J. 2008;55(3):406–14.
Wang X, Niu Y, Zheng Y. Multiple functions of MYB transcription factors in abiotic stress responses. Int J Mol Sci. 2021;22(11):6125.
Zhao J, Zhang W, Zhao Y, Gong X, Guo L, Zhu G, et al. SAD2, an importin -like protein, is required for UV-B response in Arabidopsis by mediating MYB4 nuclear trafficking. Plant Cell. 2007;19(11):3805–18.
Fornalé S, Lopez E, Salazar-Henao JE, Fernández-Nohales P, Rigau J, Caparros-Ruiz D. AtMYB7, a new player in the regulation of UV-sunscreens in Arabidopsis thaliana. Plant Cell Physiol. 2014;55(3):507–16.
Kim JH, Hyun WY, Nguyen HN, Jeong CY, Xiong L, Hong SW, et al. AtMyb7, a subgroup 4 R2R3 Myb, negatively regulates ABA-induced inhibition of seed germination by blocking the expression of the bZIP transcription factor ABI5. Plant Cell Environ. 2015;38(3):559–71.
Qian C, Chen Z, Liu Q, Mao W, Chen Y, Tian W, et al. Coordinated transcriptional regulation by the UV-B photoreceptor and multiple transcription factors for plant UV-B responses. Mol Plant. 2020;13(5):777–92.
Nakabayashi R, Yonekura-Sakakibara K, Urano K, Suzuki M, Yamada Y, Nishizawa T, et al. Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 2014;77(3):367–79.
Mehrtens F, Kranz H, Bednarek P, Weisshaar B. The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol. 2005;138(2):1083–96.
Wang F, Kong W, Wong G, Fu L, Peng R, Li Z, et al. AtMYB12 regulates flavonoids accumulation and abiotic stress tolerance in transgenic Arabidopsis thaliana. Mol Gen Genomics. 2016;291(4):1545–59.
Teng S, Keurentjes J, Bentsink L, Koornneef M, Smeekens S. Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol. 2005;139(4):1840–52.
Cominelli E, Sala T, Calvi D, Gusmaroli G, Tonelli C. Over-expression of the Arabidopsis AtMYB41 gene alters cell expansion and leaf surface permeability. Plant J. 2008;53(1):53–64.
Seo PJ, Lee SB, Suh MC, Park MJ, Go YS, Park CM. The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell. 2011;23(3):1138–52.
Anastasiadi D, Venney CJ, Bernatchez L, Wellenreuther M. Epigenetic inheritance and reproductive mode in plants and animals. Trends Ecol Evol. 2021;36(12):1124–40.
Illingworth RS, Bird AP. CpG islands—‘a rough guide’. FEBS Lett. 2009;583(11):1713–20.
Cominelli E, Galbiati M, Vavasseur A, Conti L, Sala T, Vuylsteke M, et al. A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr Biol. 2005;15(13):1196–200.
Xu R, Wang Y, Zheng H, Lu W, Wu C, Huang J, et al. Salt-induced transcription factor MYB74 is regulated by the RNA-directed DNA methylation pathway in Arabidopsis. J Exp Bot. 2015;66(19):5997–6008.
Dai S, Wang W, Huang J. Advances of reseraches on phylogeny of Dendranthema and origin of chrysanthemum. J Northeast For Univ. 2002;24(5/6):230–4.
Xiang L, Liu X, Li H, Yin X, Grierson D, Li F, et al. CmMYB#7, an R3 MYB transcription factor, acts as a negative regulator of anthocyanin biosynthesis in chrysanthemum. J Exp Bot. 2019;70(12):3111–23.
Song C, Liu Y, Song A, Dong G, Zhao H, Sun W, et al. The Chrysanthemum nankingense genome provides insights into the evolution and diversification of Chrysanthemum flowers and medicinal traits. Mol Plant. 2018;11(12):1482–91.
Du H, Yang SS, Liang Z, Feng BR, Liu L, Huang YB, et al. Genome-wide analysis of the MYB transcription factor superfamily in soybean. BMC Plant Biol. 2012;12:106.
Cao Y, Han Y, Li D, Lin Y, Cai Y. MYB transcription factors in Chinese pear (Pyrus bretschneideri Rehd.): genome-wide identification, classification, and expression profiling during fruit development. Front. Plant Sci. 2016;7:577.
Wilkins O, Nahal H, Foong J, Provart NJ, Campbell MM. Expansion and diversification of the Populus R2R3-MYB family of transcription factors. Plant Physiol. 2009;149(2):981–93.
Liu J, Wang J, Wang M, Zhao J, Zheng Y, Zhang T, et al. Genome-wide analysis of the R2R3-MYB gene family in Fragaria × ananassa and its function identification during anthocyanins biosynthesis in pink-flowered strawberry. Front Plant Sci. 2021;12:702160.
Chen H, Fang R, Deng R, Li J. The OsmiRNA166b-OsHox32 pair regulates mechanical strength of rice plants by modulating cell wall biosynthesis. Plant Biotechnol J. 2021;19(7):1468–80.
Butrón A, Malvar RA, Revilla P, Soengas P, Ordás A, Geiger HH. Rind puncture resistance in maize: inheritance and relationship with resistance to pink stem borer attack. Plant Breed. 2002;121(5):378–82.
Zhao W, Ding L, Liu J, Zhang X, Li S, Zhao K, et al. Regulation of lignin biosynthesis by an atypical bHLH protein CmHLB in Chrysanthemum. J Exp Bot. 2022;73(8):2403–19.
Zhong R, Ye ZH. Secondary cell walls: biosynthesis, patterned deposition and transcriptional regulation. Plant Cell Physiol. 2015;56(2):195–214.
McCarthy RL, Zhong R, Ye ZH. MYB83 is a direct target of SND1 and acts redundantly with MYB46 in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell Physiol. 2009;50(11):1950–64.
Zhong R, Lee C, Zhou J, McCarthy RL, Ye ZH. A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell. 2008;20(10):2763–82.
Zhong R, Richardson EA, Ye ZH. The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell. 2007;19(9):2776–92.
Sonbol FM, Fornalé S, Capellades M, Encina A, Touriño S, Torres JL, et al. The maize ZmMYB42 represses the phenylpropanoid pathway and affects the cell wall structure, composition and degradability in Arabidopsis thaliana. Plant Mol Biol. 2009;70(3):283–96.
Fornalé S, Shi X, Chai C, Encina A, Irar S, Capellades M, et al. ZmMYB31 directly represses maize lignin genes and redirects the phenylpropanoid metabolic flux. Plant J. 2010;64(4):633–44.
Legay S, Sivadon P, Blervacq AS, Pavy N, Baghdady A, Tremblay L, et al. EgMYB1, an R2R3 MYB transcription factor from eucalyptus negatively regulates secondary cell wall formation in Arabidopsis and poplar. New Phytol. 2010;188(3):774–86.
Shen H, He X, Poovaiah CR, Wuddineh WA, Ma J, Mann DGJ, et al. Functional characterization of the switchgrass (Panicum virgatum) R2R3-MYB transcription factor PvMYB4 for improvement of lignocellulosic feedstocks. New Phytol. 2012;193(1):121–36.
Falcone Ferreyra ML, Rius SP, Casati P. Flavonoids: biosynthesis, biological functions, and biotechnological applications. Front Plant Sci. 2012;3:222.
Mao Z, Jiang H, Wang S, Wang Y, Yu L, Zou Q, et al. The MdHY5-MdWRKY41-MdMYB transcription factor cascade regulates the anthocyanin and proanthocyanidin biosynthesis in red-fleshed apple. Plant Sci. 2021;306:110848.
Schaart JG, Dubos C, Romero De La Fuente I, van Houwelingen A, de Vos RCH, Jonker HH, et al. Identification and characterization of MYB-bHLH-WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry (Fragaria × ananassa) fruits. New Phytol. 2013;197(2):454–67.
Li SF, Milliken ON, Pham H, Seyit R, Napoli R, Preston J, et al. The Arabidopsis MYB5 transcription factor regulates mucilage synthesis, seed coat development, and trichome morphogenesis. Plant Cell. 2009;21(1):72–89.
Muñoz-Gómez S, Suárez-Baron H, Alzate JF, González F, Pabón-Mora N. Evolution of the subgroup 6 R2R3-MYB genes and their contribution to floral color in the Perianth-bearing Piperales. Front Plant Sci. 2021;12:633227.
Deluc L, Bogs J, Walker AR, Ferrier T, Decendit A, Merillon JM, et al. The transcription factor VvMYB5b contributes to the regulation of anthocyanin and proanthocyanidin biosynthesis in developing grape berries. Plant Physiol. 2008;147(4):2041–53.
Zhang X, He Y, Li L, Liu H, Hong G. Involvement of the R2R3-MYB transcription factor MYB21 and its homologs in regulating flavonol accumulation in Arabidopsis stamen. J Exp Bot. 2021;72(12):4319–32.
Nakatsuka T, Saito M, Yamada E, Fujita K, Kakizaki Y, Nishihara M. Isolation and characterization of GtMYBP3 and GtMYBP4, orthologues of R2R3-MYB transcription factors that regulate early flavonoid biosynthesis, in gentian flowers. J Exp Bot. 2012;63(18):6505–17.
Li J, Han G, Sun C, Sui N. Research advances of MYB transcription factors in plant stress resistance and breeding. Plant Signal Behav. 2019;14(8):1613131.
Nakabayashi R, Saito K. Integrated metabolomics for abiotic stress responses in plants. Curr Opin Plant Biol. 2015;24:10–6.
Biłas R, Szafran K, Hnatuszko-Konka K, Kononowicz AK. Cis-regulatory elements used to control gene expression in plants. Plant Cell Tissue Organ Cult. 2016;127(2):269–87.
Kaur G, Pati PK. Analysis of cis-acting regulatory elements of respiratory burst oxidase homolog (Rboh) gene families in Arabidopsis and rice provides clues for their diverse functions. Comput Biol Chem. 2016;62:104–18.
Prabu G, Theertha PD. Functional characterization of sugarcane MYB transcription factor gene promoter (PScMYBAS1) in response to abiotic stresses and hormones. Plant Cell Rep. 2012;31(4):661–9.
Mittal D, Chakrabarti S, Sarkar A, Singh A, Grover A. Heat shock factor gene family in rice: genomic organization and transcript expression profiling in response to high temperature, low temperature and oxidative stresses. Plant Physiol Biochem. 2009;47(9):785–95.
Bhuria M, Goel P, Kumar S, Singh AK. The promoter of AtUSP is co-regulated by phytohormones and abiotic stresses in Arabidopsis thaliana. Front Plant Sci. 2016;7:1957.
Abu El-Heba GA, Hussein GM, Fahmy IF, Abdou SM, Faisal A, Taha O, et al. Impact of cis-acting elements’ frequency in transcription activity in dicot and monocot plants. 3 Biotech. 2015;5(6):1007–19.
Cui D-L, Meng J-Y, Ren X-Y, Yue J-J, Fu H-Y, Huang M-T, et al. Genome-wide identification and characterization of DCL, AGO and RDR gene families in Saccharum spontaneum. Sci Rep. 2020;10(1):1–17.
Zuo C, Liu H, Lv Q, Chen Z, Tian Y, Mao J, et al. Genome-wide analysis of the apple (Malus domestica) cysteine-rich receptor-like kinase (CRK) family: annotation, genomic organization, and expression profiles in response to fungal infection. Plant Mol Biol Rep. 2020;38(1):14–24.
Kim JH, Hyun WY, Nguyen HN, Jeong CY, Xiong L, Hong SW, et al. AtM yb7, a subgroup 4 R 2 R 3 M yb, negatively regulates ABA-induced inhibition of seed germination by blocking the expression of the bZIP transcription factor ABI 5. Plant Cell Environ. 2015;38(3):559–71.
Xie Z, Lee E, Lucas JR, Morohashi K, Li D, Murray JA, et al. Regulation of cell proliferation in the stomatal lineage by the Arabidopsis MYB FOUR LIPS via direct targeting of core cell cycle genes. Plant Cell. 2010;22(7):2306–21.
Tran L-SP, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, et al. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell. 2004;16(9):2481–98.
Yang Y, Zhang L, Chen P, Liang T, Li X, Liu H. UV-B photoreceptor UVR8 interacts with MYB73/MYB77 to regulate auxin responses and lateral root development. EMBO J. 2020;39(2):e101928.
Agarwal M, Hao Y, Kapoor A, Dong C-H, Fujii H, Zheng X, et al. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J Biol Chem. 2006;281(49):37636–45.
Reyes JL, Chua NH. ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J. 2007;49(4):592–606.
Song Y, Ji D, Li S, Wang P, Li Q, Xiang F. The dynamic changes of DNA methylation and histone modifications of salt responsive transcription factor genes in soybean. PLoS One. 2012;7(7):e41274.
Coronel CJ, González AI, Ruiz ML, Polanco C. Analysis of somaclonal variation in transgenic and regenerated plants of Arabidopsis thaliana using methylation related metAFLP and TMD markers. Plant Cell Rep. 2018;37(1):137–52.
Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci. 1996;93(18):9821–6.
Qing J, Dawei W, Jun Z, Yulan X, Bingqi S, Fan Z. Genome-wide characterization and expression analyses of the MYB superfamily genes during developmental stages in Chinese jujube. PeerJ. 2019;7:e6353.
Wang YJ, Zhang Y, Fan CJ, Wei YC, Meng JX, Li Z, et al. Genome-wide analysis of MYB transcription factors and their responses to salt stress in Casuarina equisetifolia. BMC Plant Biol. 2021;21:328.
Won SY, Jung JA, Kim JS. Genome-wide analysis of the MADS-box gene family in Chrysanthemum. Comput Biol Chem. 2021;90:107424.
Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–4.
Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31(8):1296–7.
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37(Web Server issue):W202–8.
Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020;13(8):1194–202.
Ayoub Khan M, Dongru K, Yifei W, Ying W, Penghui A, Zicheng W. Characterization of WRKY gene family in whole-genome and exploration of flowering improvement genes in Chrysanthemum lavandulifolium. Front Plant Sci. 2022;13:861193.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25(4):402–8.
Acknowledgements
We are thankful to funding agencies for funding.
Funding
This work was supported by the National Key Research and Development Program (2020YFD1000401), National Natural Science Foundation of China (32201614), Project of Science and Technology Department of KaiFeng (2202001), Project of Science and Technology Department of KaiFeng (2019009 ZK), and Project of Innovation and Entrepreneurship Training program for college students in Henan Province (202213501007).
Author information
Authors and Affiliations
Contributions
APH finished refining and writing the manuscript. APH, XJD, SZY, WKX, and KDR performed the experiments. LYR, LT, LZA, and ZWQ analyzed the RNA-Seq data. WZC and Muhammad Ayoub Khan refined and revised the manuscript. The author(s) read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
We declare that the C. nankingense (the deposit number is NEAU0006698) used in this study comply with relevant institutional, national, and international guidelines and legislation. And we all comply with the IUCN Policy Statement on Research Involving Species at Risk of Extinction and the Convention on the Trade in Endangered Species of Wild Fauna and Flora. All the authors of this article agree with this statement.
In addition, we dispose of experimental plants and materials in strict accordance with relevant regulations to ensure that they will not cause any impact on the surrounding ecological environment and meet the requirements of completeness.
Consent for publication
Not applicable.
Competing interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1:
Table S1. The isoelectric point, protein length, molecular weight, Exon numbers, scaffold No, and MYB-domain type of the members of CnMYB gene family. CnMYB genes were named according to the order of scaffold No. Table S2. Details of each of the 41 genes with CpG island: the 324 CnMYB genes were analyzed for CG-rich regions using “Methyl Primer Express V1.0” software. Table S3. List of R2R3-CnMYB genes expression in different tissues. Table S4. List of CnMYB (1R-MYB, 3R-MYB,4R-MYB, and atypical MYBs) genes expression in different tissues. Table S5. List of First trend’ CnMYB genes expression in different tissues. Table S6. List of Second trend’ CnMYB genes expression in different tissues. Table S7. List of Third trend’ CnMYB genes expression in different tissues. Table S8. List of main motif numbers in 51 stress-resistant candidate genes’ promoters. Table S9. List of the primers used for qRT-PCR and MSP alaysis in this study.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Ai, P., Xue, J., Shi, Z. et al. Genome-wide characterization and expression analysis of MYB transcription factors in Chrysanthemum nankingense. BMC Plant Biol 23, 140 (2023). https://doi.org/10.1186/s12870-023-04137-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-023-04137-7