Abstract
Background
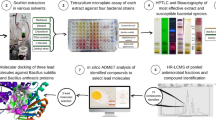
Antimicrobial resistance became the leading cause of death globally, resulting in an urgent need for the discovery of new, safe, and efficient antibacterial agents. Compounds derived from plants can provide an essential source of new types of antibiotics. A. indica (neem) plant is rich in antimicrobial phytoconstituents. Here, we used the sensitive and reliable gas chromatography-mass spectrometry (GC–MS) approach, for the quantitative and quantitative determination of bioactive constituents in methanolic extract of neem leaves grown in Sudan. Subsequently, antibacterial activity, pharmacokinetic and toxicological properties were utilized using in silico tools.
Results
The methanolic extract of neem leaves was found to have antibacterial activity against all pathogenic and reference strains. The lowest concentration reported with bacterial activity was 3.125%, which showed zones of inhibition of more than 10 mm on P. aeruginosa, K. pneumoniae, Citrobacter spp., and E. coli, and 8 mm on Proteus spp., E. faecalis, S. epidermidis, and the pathogenic S. aureus. GC–MS analysis revealed the presence of 30 chemical compounds, including fatty acids (11), hydrocarbons (9), pyridine derivatives (2), aldehydes (2), phenol group (1), aromatic substances (1), coumarins (1), and monoterpenes (1). In silico and in vitro tools revealed that.beta.d-Mannofuranoside, O-geranyl was the most active compound on different bacterial proteins. It showed the best docking energy (-8 kcal/mol) and best stability with different bacterial essential proteins during molecular dynamic (MD) simulation. It also had a good minimum inhibitory concentration (MIC) (32 μg/ml and 64 μg/ml) against S. aureus (ATCC 25,923) and E. coli (ATCC 25,922) respectively.
Conclusion
The methanolic extract of A. indica leaves possessed strong antibacterial activity against different types of bacteria. Beta.d-Mannofuranoside, O-geranyl was the most active compound and it passed 5 rules of drug-likeness properties. It could therefore be further processed for animal testing and clinical trials for its possible use as an antibacterial agent with commercial values.
Similar content being viewed by others
Background
Medicinal plants are known to have a wide range of bioactive compounds that have antimicrobial, antifungal, anticancer, anti-inflammatory, and antioxidant activities [1,2,3]. Many researchers have documented the potent activity of plants’ bioactive compounds on drug-sensitive and resistant bacteria [4,5,6]. Although plants contain a very large number of bioactive compounds, few have been discovered [1]. The development of extraction methods and the use of molecular spectroscopic techniques such as GC–MS and fourier-transform infrared (FTIR) has led to the discovery and characterization of new plant bioactive compounds [1, 7,8,9]. Recently, in silico tools have emerged as promising, time and cost-saving approaches for drug discovery [10].
Azadirachta indica (A. indica) is one of the Meliaceae family known as neem. It has been used in traditional medicine since ancient times to treat a range of human diseases [11]. The leaves, seeds, and roots of neem contain antibacterial and antifungal agents [12, 13]. This biological activity of neem stems from many bioactive compounds that are structurally and chemically diverse, with more than 140 compounds found in different parts of the plant [14]. Several types of biological compounds are extracted from neem, including ketones, carotenoids, flavonoids, steroids, and phenolic compounds [15]. The antibacterial activity of A. indica leaves extract has been documented on different bacterial species including E. coli, Staphylococcus species, Streptococcus species, and Pseudomonas species [16,17,18].
Antimicrobial resistance is one of the major problems facing global health today [19], and it is a major source of morbidity and mortality globally [20]. Nowadays it is becoming the leading cause of death globally [21]. A large number of bacteria have acquired and developed antimicrobial resistance mechanisms [20], which constitutes a burden on the global health system with the increasing financial cost [22]. With the increase in drug resistance, there are very few alternatives for patients, and as a result, the number of deaths associated with it has increased [23]. In America, there are 23,000 deaths annually related to drug resistance [20]. The emergence of infectious diseases and the development of antibiotic resistance in bacteria resulting in decreased action or failure of existing antibacterial agents [24], has resulted in an urgent need for the discovery of new, safe, and efficient antibacterial agents [25]. Compounds derived from plants can provide an essential source of new types of antibiotics. There are many types of phytochemicals of plant extract that can exert potential activity on sensitive and multidrug-resistant bacteria [24, 26].
Although a number of studies have reported various neem bioactive compounds, there hasn't been much focus on.beta.d-Mannofuranoside, O-geranyl. Scanty information is available on this compound. In a study conducted in India, the authors documented the presence of this compound in mangroves associates crude extract with antimicrobial activity [27], and in another study conducted by Iga et al., they used the isomers of synthetic D-mannofuranoside as antiallergic and anti-inflammatory agents [28]. The present study focused on the extraction, GC–MS analysis, and investigation of the antimicrobial activity of crude neem methanolic extract on drug-resistant and sensitive bacteria. Subsequently, the molecular docking and MD simulation studies were explored for the evaluation of the activity of GC–MS-identified compounds on different bacterial essential proteins. Accordingly, in vitro study was explored for the analysis of the antibacterial activity of the pure.beta.d-Mannofuranoside, O-geranyl compound.
Results
Bacterial isolates
From 130 urine samples, 100 bacterial isolates were identified. From the 100 bacterial isolates, 90 were Gram-negative rods, and 10 were Gram-positive cocci. Escherichia coli represents the majority (70%) of isolates, followed by Klebsiella pneumoniae (9%), Enterococcus faecalis (8%), Pseudomonas aeruginosa (5%), Proteus spp. (5%), Citrobacter spp. (1%), Staphylococcus aureus (1%), and Staphylococcus epidermidis (1%).
Antibacterial Susceptibility tests
The antibacterial susceptibility tests showed that 81.4% of E. coli isolates were resistant to ciprofloxacin, 75.7% to ceftazidime, 71.4% to cotrimoxazole, 62.9% to gentamicin, and 10% to imipenem. P. aeruginosa showed resistance to ceftazidime (80%), ciprofloxacin (40%), gentamicin (40%), cotrimoxazole (40%), imipenem (0%). Proteus species showed high resistance to ceftazidime (60%), cotrimoxazole (60%) and imipenem (40%); a low resistance rate was also observed in ciprofloxacin (20%) and gentamicin (20%). K. pneumoniae was highly resistant to ceftazidime (66.7%), and cotrimoxazole (55.6%), the resistance rate to ciprofloxacin, gentamicin, and imipenem was 22.2%. E. faecalis was highly resistant to ceftazidime (50%), ciprofloxacin (37.5%), gentamicin (37.5%), imipenem (37.5%), and cotrimoxazole (25%) (Table 1).
Antimicrobial activity of A. indica
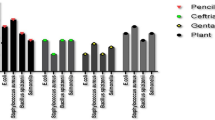
In this study, methanolic extract of A. indica showed antimicrobial activity against strains of S. aureus, P. aeruginosa, E. coli, Proteus spp., S. epidermidis, Citrobacter spp., K. pneumoniae, E. faecalis, S. aureus ATCC 25,923, and E. coli ATCC 25,922. A low (6.25%) concentration of the extract showed activity on K. pneumoniae, Citrobacter spp., P. aeruginosa, and control strains, while 12.5% concentration was active on E. faecalis, Proteus spp., S. epidermidis. The concentration of 3.125% moderately inhibits the bacteria, with the best activity (13 ± 1.8) recorded on P. aeruginosa. The concentration of 1.5% was not active on all bacterial strains (Table 2) (Supplementary figures S2 and S3).
Gas chromatography results
GC–MS analysis of neem leaves methanolic extract revealed 30 peaks, corresponding to 30 phytochemical compounds as shown in Table 3, including fatty acids (11), hydrocarbons (9), pyridine derivatives (2) and aldehydes (2), phenol group (1), aromatic substances (1), coumarins (1), and monoterpenes (1). The 1,5-Anhydro-2-deoxy-L-arabino-hex-1-enitol was the most predominant compound with a peak area percentage of 15.6.
Molecular docking
The validation of docking protocols revealed the same orientation of redocked inhibitors with experimentally determined positions, the Root Mean Square Deviation (RMSD) between superimposed SB-239629, 07 N, and cefepime were 1.2, 0.66, and 2 Å, and docking energy -11.6, -6.8, and -8.25 kcal/mol respectively (Fig. 1).
3D interaction of superimposed crystal structure of protein inhibitors and that after redocking the same inhibitors with Maestro. The dashed lines indicate dashed lines Pi-cation (green), H-bonds (blue), bad contact (brown), and ugly contact (red). Active site residues (labeled in brown) showed interaction with reference co-crystallized ligands (white) and re-docked the same ligands (violet). A. 1JIJ and SB-239629 inhibitor. B. 3TTZ and 07 N inhibitor and C. 5OJ0 and cefepime
The molecular docking of GC–MS-identified compounds in neem showed variable activities on different bacterial proteins. The.beta.d-Mannofuranoside, O-geranyl showed best docking energy on the four selected bacterial proteins, -8, 8.3, -8, and -9.7 kcal/mol on tyrosyl-tRNA synthetase (PDB ID: 1JIJ), DNA gyrase (PDB ID: 3TTZ), Penicillin-Binding Protein 2X (PBP2X) (PDB ID: 5OJ0), and penicillin-binding protein 4 (PBP4) (PDB ID: 1TVF) respectively (Figs. 2 and 3).
3D structures of the interaction of.beta.d-Mannofuranoside, O-geranyl (green) at protein binding sites. The protein backbones are colored blue, residues associated with the interaction are labeled in black, H-bonds (green), bad interactions (brown). A. tyrosyl-tRNA synthetase (PDB ID: 1JIJ). B. DNA gyrase (PDB ID: 3TTZ). C. Penicillin-Binding Protein 2X (PBP2X) (PDB ID: 5OJ0), and D. Penicillin-binding protein 4 (PBP4) (PDB ID: 1TVF)
2D interaction of.beta.d-Mannofuranoside, O-geranyl (black) and proteins binding site residues during molecular docking. H-bonds (violet), pi-cation (red), active site residues are colored red (negatively charged), deep blue (positively charged), green (hydrophobic), pale yellow (glycin), and pale blue (polar) A. tyrosyl-tRNA synthetase (PDB ID: 1JIJ) B. DNA gyrase (PDB ID: 3TTZ) C. Penicillin-Binding Protein 2X (PBP2X) (PDB ID: 5OJ0), and D. Penicillin-binding protein 4 (PBP4) (PDB ID: 1TVF)
Molecular Dynamics simulation
For predicting the interaction stability of ligand and protein complexes, a 50 ns simulation run was performed for the complex of.beta.d-Mannofuranoside, O-geranyl and tyrosyl-tRNA synthetase (PDB ID: 1JIJ), which showed the highest stability during the whole simulation time. The ligand was aligned with protein from the first 5 ns simulation until the end (50 ns) (Fig. 4A). The strongest molecular interaction between ligand and O-geranyl and tyrosyl-tRNA synthetase amino residues was found with Asp117 and Tyr36 (Fig. 5A). Whereas in Fig. 4B, which shows the interaction of.beta.d-Mannofuranoside, O-geranyl and DNA gyrase (PDB ID: 3TTZ), the ligand diverged within less than 3 Å, and with small fluctuation from 25-30 ns simulation time. The Glu50 of DNA gyrase showed 70% stable interaction during 50 ns MD simulation (Fig. 5B). The RMSD plot in Fig. 4C indicates the 50 ns trajectory of the complex of.beta.d-Mannofuranoside, O-geranyl and PBP2X (PDB ID: 5OJ0) revealed the high stability of the complex in which the ligand and protein aligned throughout the whole simulation time. The Phe450, Lys340, and Asn397 of the PBP2X had the most stable interaction during the simulation time (Fig. 5C). The complex of PBP4 (PDB ID: 1TVF) and.beta.d-Mannofuranoside, O-geranyl was stable within 1.2 Å, from 2.4–3.6 Å, this stability occurred after 15 ns simulation time (Fig. 4D). Three residues (Glu83, Ser116, and Ser262) of PBP4 protein showed more than 70% stable interaction with the ligand during the simulation period (Fig. 5D).
RMSD analysis of MD simulation trajectories generated from the interaction of.beta.d-Mannofuranoside, O-geranyl and backbones of proteins: A. tyrosyl-tRNA synthetase (PDB ID: 1JIJ) B. DNA gyrase (PDB ID: 3TTZ) C. Penicillin-Binding Protein 2X (PBP2X) (PDB ID: 5OJ0), and D. Penicillin-binding protein 4 (PBP4) (PDB ID: 1TVF), during 50 ns simulation time. The ligand is shown in red, and alpha carbon atoms of protein backbones are shown in blue color
2D interaction of.beta.d-Mannofuranoside, O-geranyl (colored black in the center), and the proteins binding sites residues, showing the percentage of H-bonds (violet) during 50 ns of MD simulation time. Active site residues are colored red (negatively charged), deep blue (positively charged), green (hydrophobic), pale yellow (glycin), pale blue (polar), and water molecules (gray). A. tyrosyl-tRNA synthetase (PDB ID: 1JIJ) B. DNA gyrase (PDB ID: 3TTZ) C. Penicillin-Binding Protein 2X (PBP2X) (PDB ID: 5OJ0), and D. Penicillin-binding protein 4 (PBP4) (PDB ID: 1TVF)
Drug likeness, ADME, and Toxicity prediction
In silico tool is used for prediction of.beta.d-Mannofuranoside, O-geranyl drug-likeness, absorption and distribution, Ames test, and carcinogenicity. As shown in Table 4, the compound showed some side effects with remarkable potential to be developed as an antibacterial agent.
Minimum Inhibitory Concentration (MIC)
The MIC revealed the.beta.d-Mannofuranoside, O-geranyl compound had good activity (MIC 32 μg/ml and 64 μg/ml) against S. aureus (ATCC 25,923) and E. coli (ATCC 25,922) respectively (Supplementary figure S4).
Discussion
Plants are now one of the most important sources for finding a new bioactive molecule to treat human diseases caused by pathogenic bacteria [1, 7,8,9] since more than 80% of people worldwide depend on herbal medicine for their basic healthcare requirements [29]. Neem trees have received worldwide attention as holy trees with remarkable therapeutic benefits such as immunomodulatory, anticancer, antibacterial, and hepatoprotective properties [30]. Neem activity could be due to the presence of a wide array of bioactive phytoconstituents, which include fatty acids, flavonoids, carbohydrates, anthocyanin, cardiac glycosides, phenols, and alkaloids [31]. The present analysis indicates the presence of different phytochemicals in neem extract, including fatty acids, pyridine derivatives, coumarins, and hydrocarbons. Fatty acids have a variety of health benefits and are commonly employed in the pharma industry, and they also have strong antioxidant properties [32]. The coumarin (benzoofuran, 2, 3-dihydro-) was reported to be anti-inflammatory [33].
Neem extract is rich in antimicrobial phytoconstituents such as alkaloids, glycosides flavonoids, phenolic compounds, steroids, triterpenoids, carotenoids, and tetra-triterpenoids azadirachtin [34]. In this study, the GC–MS analysis revealed the presence of 30 phytochemicals in the methanolic extract of neem leaves, the top detected phytochemical compounds were reported previously with antibacterial activity: 1,5-Anhydro-2-deoxy-L-arabino-hex-1-enitol [35], 3,7,11,15-Tetramethyl-2-hexadecane-1-ol [36] 1,3-Propanediol,2-(hydroxymethyl)-2-nitro- [37], n-Hexadecanoic acid [38],.beta. d-Mannofuranoside, O-geranyl [27], and phytol [39].
The methanolic extract of neem leaves used in this study had shown a potent antibacterial activity on different bacteria types. The antibacterial activity of crude neem extract was reported previously in many studies worldwide [40,41,42]. Methanol can extract a broad polarity range of compounds with antimicrobial activity [43, 43]. In this study, the activity of neem extract was observed on different uropathogens including S. aureus, E. coli, K. pneumoniae, Citrobacter spp., E. faecalis, P. aeruginosa, Proteus spp., and S. epidermidis. The activity of neem leaves ethanolic extract on urine isolates was documented in Sudan [18, 45], Pakistan [46], and India [47]. Additionally, Okemo et al. [48] and Pokhrel et al. [40] reported that the crude extract of the neem plant was very effective against S. aureus and E. coli. Our results are different from those obtained by Francine and his colleagues [49] in Rwanda. They reported the activity of neem methanolic extract only on S. aureus but not on E. coli. This variation could be due to differences in neem plants' active constituents and due to differences in environment, genetic factors, and climates [50, 51].
The lowest concentration of neem extract reported with bacterial activity was 3.125%, which showed zones of inhibition of more than 10 mm on P. aeruginosa, K. pneumoniae, Citrobacter spp., and E. coli, and 8 mm on Proteus spp. and Staphylococcus spp. These findings are better than Faujdar et al. [47], who also used the same concentrations of neem methanolic extract as in our study, and reported at 6.25 mg/dl concentration a 7 mm zone of inhibition on E. coli and Proteus spp, and 0 mm on P. aeruginosa. Our finding is in concordance with a previous study conducted in Sudan, in which the 6.25 mg/dl concentration was more active on P. aeruginosa, and K. pneumoniae [45]. This could be due to the presence of the same phytoconstituents in neem plants grown in our environment [18, 50, 51].
Our study showed a high activity of neem extract on bacteria resistant to B-lactam, quinolones, and aminoglycosides, which is consistent with previous findings [45, 47]. Although neem was active on pathogenic E. coli and S. aureus, we noticed better activity on the control strains of E. coli and S. aureus than on the pathogenic ones. This could be due to the presence of resistance mechanisms in pathogenic bacteria such as efflux pumps [52, 53].
The molecular docking study showed that.beta.d-Mannofuranoside, O-geranyl had potent activity on essential bacterial proteins. The.beta.d-Mannofuranoside, O-geranyl showed hydrogen bonds with residues that are closely interacted with active sites [54]. The Tyr36, Asp40, Tyr170, and Asp177 of S. aureus tyrosyl-tRNA synthetase protein was documented to also have hydrogen bonds with the known co-crystallized protein inhibitor (SB-239629). This activity is concise with our in vitro study and with another in vitro study, in which the authors identified mangrove associates extract with good antibacterial activity, the GC–MS analysis of this extract revealed the presence of a high concentration of.beta.d-Mannofuranoside, O-geranyl [27].
We further evaluated the docking complexes' stability using molecular dynamic simulation, which showed the stability of these complexes during simulation time (50 ns). The ligand was aligned with protein backbones, with a fluctuation of less than 3 Å at most of the simulation period. Usually, changes of the order of 1–3 Å are perfectly acceptable [55]. During the simulation, time residues (Lys340, Trp374, Asn397, and Gln452) of penicillin-binding protein 2X (PDB ID: 5OJ0) showed interaction with protein inhibitor (cefepime) showed a stable interaction with.beta. d-Mannofuranoside, O-geranyl, indicates stability is maintained [56]. On the other hand, the active compounds of neem (.beta.d-Mannofuranoside, O-geranyl) showed a stable interaction with S. aureus penicillin-binding protein 4 (PDB ID: 1TVF); the compound formed a stable water bridge with the catalytic residue of the SXXK motif of the penicillin-binding protein 4 [57].
Conclusion
Methanolic extract of A. indica leaves revealed a potent antimicrobial activity to different types of gram-negative, gram-positive, and control strains. In vitro and in silico experiments revealed the.beta. d-Mannofuranoside, O-geranyl is the most active compound on control strains and different bacterial essential proteins. Using in silico ADME/T prediction, the compound passed 5 rules of drug-likeness properties, so it could be further processed for animal testing and clinical trials for its possible use as an antibacterial agent with commercial values. Moreover, the GC–MS analysis of neem extract revealed the presence of a large number of bioactive components, and the most common were fatty acids (11), hydrocarbons (9), pyridine derivatives (2), and aldehydes (2), which played different biological activities in addition to their nutritional benefits. Based on these findings, the neem plant could help us produce safe and effective medications for a variety of diseases. More in-depth research into these identified phytochemicals will aid pharmaceutical explorations.
Materials and methods
Collection and identification of bacterial isolates
A total of 130 urine samples indicated for urine culture and sensitivity testing were collected randomly from patients at East Nile Hospital and Ribat University Hospital in Khartoum state, from January to April 2017. The urine samples were cultured on Cystine Lactose Electrolyte Deficient (CLED) agar media (Hi-Media laboratories PV + Ltd, India), and the clinical isolates were identified using conventional biochemical tests [58].
Antibacterial susceptibility testing
The Kirby-Bauer disk diffusion method [59] was used to test isolated organisms against various antibiotics including ceftazidime (30 mcg), imipenem (10 mcg), gentamicin (10 mcg), cotrimoxazole (25 mcg), and ciprofloxacin (10 mcg) (Hi-Media labs PV + Ltd, India) (5mcg). Results were interpreted according to the Clinical and Laboratory Standard Institute (CLSI) document M100 [59]. The following strains were used for quality control: S. aureus (ATCC 25,923) and E. coli (ATCC 25,922), to assess the media and antimicrobials disk efficiency.
Plant collection and extraction
Fresh leaves of wild neem were collected from Algazira (Alkamleen city) in central Sudan in March 2017. Leaves were collected from the same tree into clean, dry, labeled plastic bags. Samples were kept frozen at -80 °C until the time of their use [60]. A taxonomist authenticated the plant at Medicinal and Aromatic Plants and Traditional Medicine Research Institute (MAPRI) National Center for Research, Khartoum, Sudan. At MAPRI herbarium, a voucher sample (No. MAP/2017/4) was deposited. The collected leaves were washed and rinsed to remove dust and other impurities. They were then air-dried and then a total of 50 g of leaves were grounded using a mortar and pestle (Supplementary figure S1), 80% methanol was then used to soak the leaves for three days with daily filtration and evaporation. Then by using a rotary evaporator apparatus under reduced pressure, the solvent was evaporated to dryness [61].
Antibacterial activity of neem extract
The antibacterial activity of neem leaves was tested using the agar well diffusion method on Muller Hinton Agar (MHA) medium against the isolated bacteria and control strains (S. aureus (ATCC 25,923) and E. coli (ATCC 25,922)). Three colonies with similar features were dissolved in 1 mL normal saline and turbidity adjusted to 0.5 McFarland. The isolates were then streaked on the surface of the MHA plate with a sterile swab. Using a cork borer, 6 mm wells were created aseptically on MHA. At sterile conditions, 100 µl of each 50, 25, 12.5, 6.25, 3.125, and 1.5% concentrations of neem extract were poured into media wells [62]. The plates were placed refrigerated for 1 h to allow for extract diffusion before being incubated at 37 °C for 24 h. Methanol alone was used as a negative control. The zone of inhibition was measured (in mm), and the mean was calculated [63]. Three replicates were carried out for the activity of extracted neem against tested organisms. Then the data were presented as mean and standard deviation.
Phytochemical screening of A. indica (neem) extract
The GM-MS method was used to conduct a qualitative and quantitative characterization of neem extract, using the model (GC–MS-QP2010-Ultra) from Shimadzu Company, Japan, with a capillary column Rtx®-5MS column (30 m, 0.25 mm, 0.25 µm) [64]. The split mode was used for sample injection, and operated in electron ionization (EI) mode at 70 eV, inflow rate of 1.69 ml/min. Helium gas was used as carrier gas. The injector temperature was set at 300 °C, the temperature of the ion source was 200 °C, and 250 °C was used as interface temperature. The oven temperature program was as follows: the initial temperature at 50 °C rising at 7 °C /min to 180 °C, then the rate changed 10 °C/min reaching the final temperature at 280 °C with 2 min as hold time. In a total 22 min run, the sample was analyzed by the scan mode in a range of 40 to 500 m/z charges to ratio. The neem extract's components were identified by comparing the retention times and mass fragmentation patents with the National Institute of Standards and Technology (NIST) library, and then the results were recorded [65, 66].
In silico analysis
Molecular docking
Proteins selection and preparation
The crystal structures of four essential bacterial proteins were obtained from RCSB PDB database [67] according to their essential role in bacterial cell wall synthesis and protein production in most of our studied isolates, and according to published data [68,69,70,71]. These proteins were Staphylococcus aureus tyrosyl-tRNA synthetase (PDB ID: 1JIJ), DNA gyrase (PDB ID: 3TTZ), Penicillin-Binding Protein 2X (PBP2X) from Streptococcus pneumoniae (PDB ID: 5OJ0), and penicillin-binding protein 4 (PBP4) from Staphylococcus aureus (PDB ID: 1TVF). The proteins’ 3D structures were prepared with the Protein Preparation Wizard in Maestro using the default setting. For validation of the docking method, the co-crystalized ligands, SB-239629, 07 N, and cefepime, with their respective structures (1JIJ, 3TTZ, and 5OJ0, respectively), were redocked again using Maestro software [72].
Ligands preparation
The structures of compounds identified by GC–MS (Table 3) were obtained from NCBI PubChem and ChemSpider databases. The ligand's energy was minimized using LigPrep (Schrodinger software, version 2020–3).
Molecular docking
Proteins’ active sites were predicted using the Receptor Grid Generation module in Schrodinger. The grids were specified around the co-crystalized ligands or using the SiteMap module to predict S. aureus (PDB ID: 1TVF) protein according to published data [69]. The prepared molecules were docked on protein active sites using extra precision (XP) docking of Schrödinger Maestro software [72]. Ligands were set flexible while proteins were set rigid.
Molecular Dynamic (MD) simulation
Desmond package in the Schrödinger Maestro software [72] was used for MD simulation. The complexes with the best interaction and docking energy were first solvated into the TIP3P water model, an orthorhombic box with boundary 10 Å beyond any of the complex's atoms. Charges were neutralized and OPLS3e force field was used. The particle mesh Ewald method was used for the calculation of long-range electrostatic interactions [73] and cutoff of 12 Å. The molecular dynamic simulation was done in the NPT ensemble at a temperature of 300 K and 1.013 bar pressure over 50 ns and 30 ps for trajectory and 100 ps relaxation time. The trajectories were recorded in 50 ps intervals. After job completion, Root Mean Square Deviation (RMSD) and Root Mean Square Fluctuation (RMSF) were used to examine complexes' stability.
Drug-likeness, ADME & Toxicity Prediction Studies
The ADME profiles and toxicity analysis were carried out using PreADMET (http://preadmet.bmdrc.org).
Minimum Inhibitory Concentration (MIC)
Antibacterial activity of the most stable ligand (.beta.d-Mannofuranoside, O-geranyl) from the in silico study was evaluated using in vitro method. The compound was purchased from Apollo Scientific (UK), the compound ID: MolPort-019–937-357, purity 95%, and the Molecular weight was 316.394. The MIC of the compound was evaluated against S. aureus (ATCC 25,923) and E. coli (ATCC 25,922) using the microtitre broth dilution method [74]. A twofold serial dilution of the compound was prepared in broth media (Muller-Hinton), using 96-well microplates flat-bottom plates. One 100 μL of culture media containing bacterial growth adjusted to 5–105 CFU/ml was poured into each well. The MIC of the compound was determined at a concentration ranging from 0.5 to 256 μg/ml [75].
Availability of data and materials
All data are included in the manuscript, any additional information needed contact the corresponding author.
Abbreviations
- ATCC:
-
American Type Culture Collection
- WHO:
-
World Health Organization
- GC-MS:
-
Gas chromatography-mass spectrometry
- CID:
-
Compound ID
- HIA:
-
Human intestinal absorption
- CMC:
-
Comprehensive medicinal chemistry
- MDDR:
-
MDL Drug Data Report
- WDI:
-
World Drug Index
- MDCK:
-
Madin-Darby Canine Kidney Cell Permeability
- RMSD:
-
Root Mean Square Deviation
- RCSB PDB:
-
Structural Bioinformatics Protein Data Bank
- ADME/T:
-
Absorption, Distribution, Metabolism, Excretion and Toxicity
References
Konappa N, Udayashankar AC, Krishnamurthy S, Pradeep CK, Chowdappa S, Jogaiah S. GC–MS analysis of phytoconstituents from Amomum nilgiricum and molecular docking interactions of bioactive serverogenin acetate with target proteins. Sci Rep. 2020;10(1):1–23.
Jogaiah S, Shetty HS. Ito S-i, Tran L-SP: Enhancement of downy mildew disease resistance in pearl millet by the G_app7 bioactive compound produced by Ganoderma applanatum. Plant Physiol Biochem. 2016;105:109–17.
Jagannath S, Konappa N, Lokesh A, Dasegowda T, Udayashankar AC, Chowdappa S, Cheluviah M, Satapute P, Jogaiah S. Bioactive compounds guided diversity of endophytic fungi from Baliospermum montanum and their potential extracellular enzymes. Anal Biochem. 2021;614: 114024.
Shakeri A, Sharifi MJ, Fazly Bazzaz BS, Emami A, Soheili V, Sahebkar A, Asili J. Bioautography detection of antimicrobial compounds from the essential oil of salvia Pachystachys. Curr Bioact Compd. 2018;14(1):80–5.
Bazzaz BSF, Khameneh B, Ostad MRZ, Hosseinzadeh H. In vitro evaluation of antibacterial activity of verbascoside, lemon verbena extract and caffeine in combination with gentamicin against drug-resistant Staphylococcus aureus and Escherichia coli clinical isolates. Avicenna journal of phytomedicine. 2018;8(3):246.
Cowan MM. Plant products as antimicrobial agents. Clin Microbiol Rev. 1999;12(4):564–82.
Vanitha V, Vijayakumar S, Nilavukkarasi M, Punitha V, Vidhya E, Praseetha P. Heneicosane—A novel microbicidal bioactive alkane identified from Plumbago zeylanica L. Ind Crops Prod. 2020;154: 112748.
Abubaker M, Adamd I, Mohammed A, Liang T, Zhang J. Gas Chromatography-Mass Spectrum andFourier-transform infrared spectroscopy analysis of Fixed Oil from Sudanese Ziziphus spina Christi Fruits Pulp. Progress in Chemical and Biochemical Research. 2020;4(3):278–94.
Ralte L, Khiangte L, Thangjam NM, Kumar A, Singh YT. GC–MS and molecular docking analyses of phytochemicals from the underutilized plant, Parkia timoriana revealed candidate anti-cancerous and anti-inflammatory agents. Sci Rep. 2022;12(1):1–21.
Abishad P, Niveditha P, Unni V, Vergis J, Kurkure NV, Chaudhari S, Rawool DB, Barbuddhe SB. In silico molecular docking and in vitro antimicrobial efficacy of phytochemicals against multi-drug-resistant enteroaggregative Escherichia coli and non-typhoidal Salmonella spp. Gut pathogens. 2021;13(1):1–11.
Joshi M, Purwar R, Ali SW, Rajendran S. Antimicrobial textiles for health and hygiene applications based on eco-friendly natural products. In: Medical and Healthcare Textiles. Elsevier. 2010:84–92.
Alzohairy MA. Therapeutics Role of Azadirachta indica (Neem) and Their Active Constituents in Diseases Prevention and Treatment. Evidence-based complementary and alternative medicine : eCAM. 2016;2016:7382506.
Ospina Salazar DI, Hoyos Sanchez RA, Orozco Sanchez F, Arango Arteaga M, Gomez Londono LF. Antifungal activity of neem (Azadirachta indica: Meliaceae) extracts against dermatophytes. Acta Biológica Colombiana. 2015;20(3):181–92.
Mahmoud D, Hassanein N, Youssef K, Abou Zeid M. Antifungal activity of different neem leaf extracts and the nimonol against some important human pathogens. Braz J Microbiol. 2011;42(3):1007–16.
El-Hawary SS, El-Tantawy ME, Rabeh MA, Badr WK. DNA fingerprinting and botanical study of Azadirachta indica A. Juss.(neem) family Meliaceae. Beni-Suef University Journal of Basic and Applied Sciences. 2013;2(1):1–13.
Ali E, Islam MS, Hossen MI, Khatun MM, Islam MA. Extract of neem (Azadirachta indica) leaf exhibits bactericidal effect against multidrug resistant pathogenic bacteria of poultry. Veterinary medicine and science. 2021;7(5):1921–7.
Kumar PS, Debasis M, Goutam G, Panda CS. Biological action and medicinal properties of various constituent of Azadirachta indica (Meliaceae): an overview. Ann Biol Res. 2010;1(3):24–34.
Jessinta S, Azhari H, Saiful N, Abdurahman H. Impact of geographic variation on physicochemical properties of neem (Azadirachta indica) seed oil. Int J Pharm Sci Res. 2014;5(10):4406–13.
Dadgostar P. Antimicrobial Resistance: Implications and Costs. Infection and drug resistance. 2019;12:3903–10.
Reygaert WC. An overview of the antimicrobial resistance mechanisms of bacteria. AIMS microbiology. 2018;4(3):482–501.
Murray CJ, Ikuta KS, Sharara F, Swetschinski L, Aguilar GR, Gray A, Han C, Bisignano C, Rao P, Wool E. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399(10325):629–55.
Santajit S, Indrawattana N. Mechanisms of antimicrobial resistance in ESKAPE pathogens. BioMed research international 2016, 2016, 2475067.
World Health Organization. New report calls for urgent action to avert antimicrobial resistance crisis. Joint News Release. 29 April 2019. Available online: https://www.who.int/news-room/detail/29-04-2019-new-report-calls-for-urgent-action-to-avert-antimicrobial-resistance-crisis (accessed on 20 April 2022).
Khameneh B, Iranshahy M, Soheili V, Bazzaz BSF. Review on plant antimicrobials: a mechanistic viewpoint. Antimicrob Resist Infect Control. 2019;8(1):1–28.
Aslam B, Wang W, Arshad MI, Khurshid M, Muzammil S, Rasool MH, Nisar MA, Alvi RF, Aslam MA, Qamar MU, et al. Antibiotic resistance: a rundown of a global crisis. Infection and drug resistance. 2018;11:1645–58.
Subramani R, Narayanasamy M, Feussner KD. Plant-derived antimicrobials to fight against multi-drug-resistant human pathogens. 3 Biotech. 2017;7(3):172.
Manilal A, Merdekios B, Velappan JK, Paul JPV, Idhayadhulla A, Muthukumar C, Melkie M. An in vitro efficacy validation of mangrove associates. J Coast Life Med. 2014;2(7):560–5.
Iga S, Iga A, Nicolescu A, Iga DP. Synthesis of D, L-α-Tocopheryl-α-D-mannopyranoside, a Potential Antiallergic and Antiinflammatory Compound, and its-α-D-mannofuranoside Isomer. Rev Chim. 2010;61(5):475–8.
Kebede T, Gadisa E, Tufa A. Antimicrobial activities evaluation and phytochemical screening of some selected medicinal plants: A possible alternative in the treatment of multidrug-resistant microbes. PLoS ONE. 2021;16(3): e0249253.
Al-Sarraj FM. A Review on the impacts of Azadirachta indica on Multi-drug Resistant Extended Spectrum Beta Lactamase-positive of Escherichia coli and Klebsiella pneumonia. Advancements in Life Sciences. 2021;8(3):228–32.
Latif MJ, Hassan SM, Mughal SS, Aslam A, Munir M, Shabbir N, Mushtaq M, Perveiz S. Therapeutic Potential of Azadirachta indica (Neem) and Their Active Phytoconstituents against Diseases Prevention. 2020.
Dinagaran S, Sridhar S, Eganathan P. Chemical composition and antioxidant activities of black seed oil (Nigella sativa L.). International Journal of Pharmaceutical Sciences and Research. 2016;7(11):4473.
Correa MF, Barbosa AJ, Teixeira LB, Duarte DA, Simoes SC, Parreiras-e-Silva LT, Balbino AM, Landgraf RG, Bouvier M, Costa-Neto CM. Pharmacological Characterization of 5-Substituted 1-[(2, 3-dihydro-1-benzofuran-2-yl) methyl] piperazines: Novel Antagonists for the Histamine H3 and H4 Receptors with Anti-inflammatory Potential. Front Pharmacol. 2017;8:825.
Koona S, Budida S. Antibacterial Potential of the Extracts of the Leaves of Azadirachta indica Linn. Notulae Scientia Biologicae. 2011;3(1):65–9.
Setiawansyah A, Hakim A, Wirasisya DG. EVALUASI DAN IDENTIFIKASI GOLONGAN SENYAWA POTENSIAL ANTIBAKTERI PADA DAUN DAN KULIT BATANG MIMBA (Azhadirachta indica A. Juss) TERHADAP Escherichia coli. Jurnal Tumbuhan Obat Indonesia. 2018;11(2):40–8.
Nithya M, Ragavendran C, Natarajan D. Antibacterial and free radical scavenging activity of a medicinal plant Solanum xanthocarpum. Int J Food Prop. 2018;21(1):313–27.
Amponin DE, Przybek-Skrzypecka J, Zyablitskaya M, Takaoka A, Suh LH, Nagasaki T, Trokel SL, Paik DC. Ex vivo anti-microbial efficacy of various formaldehyde releasers against antibiotic resistant and antibiotic sensitive microorganisms involved in infectious keratitis. BMC Ophthalmol. 2020;20(1):1–10.
Aparna V, Dileep KV, Mandal PK, Karthe P, Sadasivan C, Haridas M. Anti-inflammatory property of n-hexadecanoic acid: structural evidence and kinetic assessment. Chem Biol Drug Des. 2012;80(3):434–9.
Lee W, Woo E-R, Lee DG. Phytol has antibacterial property by inducing oxidative stress response in Pseudomonas aeruginosa. Free Radical Res. 2016;50(12):1309–18.
Pokhrel B, Rijal S, Raut S, Pandeya A. Investigations of antioxidant and antibacterial activity of leaf extracts of Azadirachta indica. Afr J Biotech. 2015;14(46):3159–63.
Tirumalasetty J, Anuradha B, Praveena A. Antimicrobial activity of methanolic extracts of Azadirachta indica, Rosmarinus officinalis and Lagenaria siceraria leaves on some important pathogenic organisms. J Chem Pharm Res. 2014;6:766–70.
Mistry KS, Sanghvi Z, Parmar G, Shah S. The antimicrobial activity of Azadirachta indica, Mimusops elengi, Tinospora cardifolia, Ocimum sanctum and 2% chlorhexidine gluconate on common endodontic pathogens: An in vitro study. European journal of dentistry. 2014;8(2):172.
Dzulkarnain S, bin Abdul Rahim I. Antimicrobial activity of methanolic Neem extract on wound infection bacteria. In: International Conference on Biological, Chemical and Environmental Sciences: 2014;4:72–5.
Parekh J, Jadeja D, Chanda S. Efficacy of aqueous and methanol extracts of some medicinal plants for potential antibacterial activity. Turk J Biol. 2006;29(4):203–10.
Mohammed HA, Al Fadhil AO. Antibacterial activity of Azadirachta indica (Neem) leaf extract against bacterial pathogens in Sudan. Afr J Med Sci. 2017;3:246–2512.
Aziz F, Taqdees M, Ifrah I, Sayyada G. Phytochemical screening and antibacterial activity of neem extracts on uropathogens. Pure Appl Biol. 2020;9(1):148–53.
Faujdar SS, Bisht D, Sharma A. Antibacterial potential of neem (Azadirachta indica) against uropathogens producing beta-lactamase enzymes: A clue to future antibacterial agent? Biomedical and Biotechnology Research Journal (BBRJ). 2020;4(3):232.
Okemo P, Mwatha W, Chhabra S, Fabry W. The kill kinetics of Azadirachta indica A. Juss.(Meliaceae) extracts on Staphylococcus aureus, Escherichia coli, Pseudomonas aeruginosa and Candida albicans. African Journal of Science and Technology. 2001;2(2):113–8.
Francine U, Jeannette U, Pierre RJ. Assessment of antibacterial activity of neem plant (Azadirachta indica) on Staphylococcus aureus and Escherichia coli. J Med Plants Stud. 2015;3(4):85–91.
Eid A, Jaradat N, Elmarzugi N. A Review of chemical constituents and traditional usage of Neem plant (Azadirachta Indica). Palestinian Medical and Pharmaceutical Journal. 2017;2(2):75–81.
Uchegbu M, Okoli I, Esonu B, Iloeje M. The grovving importance of neem (Azadirachta indica A. Juss) in agriculture, industry, medicine and Eenvironment: A review. Research Journal of Medicinal Plant. 2011;5(3):230–45.
Lynch AS. Efflux systems in bacterial pathogens: an opportunity for therapeutic intervention? An industry view Biochemical pharmacology. 2006;71(7):949–56.
Chetri S, Bhowmik D, Paul D, Pandey P, Chanda DD, Chakravarty A, Bora D, Bhattacharjee A. AcrAB-TolC efflux pump system plays a role in carbapenem non-susceptibility in Escherichia coli. BMC Microbiol. 2019;19(1):210.
Beg M, Ansari S, Athar F. Molecular docking studies of Calotropis gigantea phytoconstituents against Staphylococcus aureus tyrosyl-tRNA synthetase protein. J Bacteriol Mycol Open Access. 2020;8(3):78–91.
Sangai NP, Patel CN, Pandya HA. Ameliorative effects of quercetin against bisphenol A-caused oxidative stress in human erythrocytes: an in vitro and in silico study. Toxicology research. 2018;7(6):1091–9.
Sadiq S, Rana NF, Zahid MA, Zargaham MK, Tanweer T, Batool A, Naeem A, Nawaz A, Muneer Z, Siddiqi AR. Virtual Screening of FDA-Approved Drugs against LasR of Pseudomonas aeruginosa for Antibiofilm Potential. Molecules. 2020;25(16):3723.
Navratna V, Nadig S, Sood V, Prasad K, Arakere G, Gopal B. Molecular basis for the role of Staphylococcus aureus penicillin binding protein 4 in antimicrobial resistance. J Bacteriol. 2010;192(1):134–44.
Rave A, Kuss A, Peil G, Ladeira S, Villarreal J, Nascente P. Biochemical identification techniques and antibiotic susceptibility profile of lipolytic ambiental bacteria from effluents. Braz J Biol. 2019;79(4):555–65.
CLSI: Performance standards for antimicrobial susceptibility testing. Approved Standard M100 2016;35(3):16–38.
Ney FP, Malco DCL, Senoro DB, Catajay-Mani M. The bio-mechanical properties of coco wood applied with Neem extracts: a potential preservative for sustainable building in Marinduque. Philippines Sustainable Environment Research. 2019;29(1):1–13.
Gennaro L. Extraction technology for medicinal and aromatic Plant. United Nations Industrial Development Organization and the International Centre for Science and High Technology, Trieste, Italy P, 2008, 23.
Costa C, Bevilaqua C, Camurça-Vasconcelos A, Maciel M, Morais S, Castro C, Braga R, Oliveira L. In vitro ovicidal and larvicidal activity of Azadirachta indica extracts on Haemonchus contortus. Small Rumin Res. 2008;74(1–3):284–7.
RAI AK, Joshi R. Evaluation of antimicrobial properties of fruit extracts of Terminalia chebula against dental caries pathogens. 2009.
Kumar A, Gill JPS, Bedi JS, Kumar A. Pesticide residues in Indian raw honeys, an indicator of environmental pollution. Environ Sci Pollut Res. 2018;25(34):34005–16.
A Adam A, A. Ahmed S, A Mohamed T, A Azrag R, E Mustfa S, AA Hamdi O. Evaluation of repellent activities of the essential oil of Ocimum basilicum against Anopheles mosquito and formulation of mosquito repellent cream. Biomedical Research and Clinical Practice. 2019;4:2–5.
Kalt F, Cock I. Gas chromatography-mass spectroscopy analysis of bioactive Petalostigma extracts: Toxicity, antibacterial and antiviral activities. Pharmacogn Mag. 2014;10(Suppl 1):S37.
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 2000;28(1):235–42.
Pisano MB, Kumar A, Medda R, Gatto G, Pal R, Fais A, Era B, Cosentino S, Uriarte E, Santana L. Antibacterial activity and molecular docking studies of a selected series of hydroxy-3-arylcoumarins. Molecules. 2019;24(15):2815.
Kulanthaivel L, Jeyaraman J, Biswas A, Subbaraj GK, Santhoshkumar S. Identification of potential inhibitors for Penicillinbinding protein (PBP) from Staphylococcus aureus. Bioinformation. 2018;14(9):471.
Varkey DR, Kingsley DJ, Abraham J. Antimicrobial Activity and In-Silico Analysis of 3, 5, 6-Trichloro-2-Pyridinol. RESEARCH JOURNAL OF PHARMACEUTICAL BIOLOGICAL AND CHEMICAL SCIENCES. 2015;6(4):272–84.
Rahmania TA, Ritmaleni R, Setyowati EP. In silico and in vitro assay of Hexagamavunon-6 analogs, Dibenzilyden-N-Methyl-4-piperidone as antibacterial agents. Journal of Applied Pharmaceutical Science. 2020;10(03):039–43.
Release S: 3: Maestro-Desmond Interoperability Tools, Schrödinger, New York, NY, USA, 2020.
Toukmaji AY, Board JA Jr. Ewald summation techniques in perspective: a survey. Comput Phys Commun. 1996;95(2–3):73–92.
Mogana R, Adhikari A, Tzar M, Ramliza R, Wiart C. Antibacterial activities of the extracts, fractions and isolated compounds from Canarium patentinervium Miq. against bacterial clinical isolates. BMC complementary medicine and therapies. 2020;20(1):1–11.
Kouidhi B, Zmantar T, Jrah H, Souiden Y, Chaieb K, Mahdouani K, Bakhrouf A. Antibacterial and resistance-modifying activities of thymoquinone against oral pathogens. Ann Clin Microbiol Antimicrob. 2011;10(1):1–7.
Acknowledgements
Not applicable
Funding
This research work was funded by institutional fund projects under the grant (IFPHI-344–130-2020). Therefore, the authors gratefully acknowledge technical and financial support from the ministry of education and King Abdulaziz University, DSR, Jeddah, Saudi Arabia.
Author information
Authors and Affiliations
Contributions
HNA, and NFY contribute to data collection experimental design, conceptualization, and manuscript writing, SH, and IK wrote the manuscript, critically reviewed the final version of the manuscript and project supervision, HNA did the bioinformatics analysis. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Review Board of Deanship of Scientific Research, Sudan University of Science and Technology (No: DSR-IEC12-02–17). Verbal consent was obtained from participants (in the case of neonates, parental consent was obtained). Written consent was waived by the ethical committee of Sudan University of Science and Technology, meeting no. (SUST/DSR/1EC/EA12/17) date (12/02/2017) because we are using the remaining bio-samples without personal data. A taxonomist authenticated the plant at Medicinal and Aromatic Plants and Traditional Medicine Research Institute (MAPRI) National Center for Research, Khartoum, Sudan. At MAPRI herbarium, a voucher sample (No. MAP/2017/4) was deposited. The plant material was collected according to national and international guidelines.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Altayb, H.N., Yassin, N.F., Hosawi, S. et al. In-vitro and in-silico antibacterial activity of Azadirachta indica (Neem), methanolic extract, and identification of Beta.d-Mannofuranoside as a promising antibacterial agent. BMC Plant Biol 22, 262 (2022). https://doi.org/10.1186/s12870-022-03650-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-022-03650-5