Abstract
Background
Tef (Eragrostis tef) is a tropical cereal domesticated and grown in the Ethiopian highlands, where it has been a staple food of Ethiopians for many centuries. Food insecurity and nutrient deficiencies are major problems in the country, so breeding for enhanced nutritional traits, such as Zn content, could help to alleviate problems with malnutrition.
Results
To understand the breeding potential of nutritional traits in tef a core set of 24 varieties were sequenced and their mineral content, levels of phytate and protein, as well as a number of nutritionally valuable phenolic compounds measured in grain. Significant variation in all these traits was found between varieties. Genome wide sequencing of the 24 tef varieties revealed 3,193,582 unique SNPs and 897,272 unique INDELs relative to the tef reference var. Dabbi. Sequence analysis of two key transporter families involved in the uptake and transport of Zn by the plant led to the identification of 32 Zinc Iron Permease (ZIP) transporters and 14 Heavy Metal Associated (HMA) transporters in tef. Further analysis identified numerous variants, of which 14.6% of EtZIP and 12.4% of EtHMA variants were non-synonymous changes. Analysis of a key enzyme in flavanol synthesis, flavonoid 3′-hydroxylase (F3’H), identified a T-G variant in the tef homologue Et_s3159-0.29-1.mrna1 that was associated with the differences observed in kaempferol glycoside and quercetin glycoside levels.
Conclusion
Wide genetic and phenotypic variation was found in 24 Ethiopian tef varieties which would allow for breeding gains in many nutritional traits of importance to human health.
Similar content being viewed by others
Background
Tef (Eragrostis tef) is a tropical cereal that has its origins in the Ethiopian highlands, where it was domesticated and has been grown for thousands of years [1]. Globally tef is a minor cereal crop in terms of both production and planted area, with Ethiopia growing an estimated 90% of the annual global tef crop on about three million hectares, which equates to about a quarter of the Ethiopian grain-cultivated area [2]. Compared to other cereals tef is considered a resilient crop. It can withstand adverse weather conditions, growing well at elevations between 1800 and 2200 m above sea level, in regions where there is adequate rainfall [3]. Tef is a staple of Ethiopian diets, providing 11% of the per capita caloric intake and two-thirds of the average Ethiopians daily protein [4].
Tef is widely considered a healthy alternative to cereals such as wheat, maize and rice as it does not contain gluten, is high in slowly digestible starch, rich in calcium (Ca) and polyphenols [5, 6]. However, levels of zinc (Zn) in the grain (in the range of 28–40 mg/kg) are often less than the recommended level of 40 mg/kg necessary to meet human nutritional requirements [1, 7, 8], with further fortification efforts being hampered by low Zn levels in many Ethiopian soils [1].
While breeding efforts to improve tef have been ongoing in Ethiopia since the mid-1950s germplasm advancements have been slow, with only 42 new tef varieties being released [3, 9, 10]. A better understanding of tef nutritional diversity and its genetic control could help drive nutritional gains for this crop. Recent advancements in our understanding of the genetics of tef include a fairly complete genome assembly, the release of gene models, as well as a number of RNASeq datasets which are all publicly available [11]. These allow for translation of knowledge on grain fortification from other species, supporting more rapid advancements in efforts to improve the nutritional potential of tef [12,13,14,15,16].
Several gene families have been identified that support accumulation and transport of various heavy metals within the plant. The Zinc Iron Permease (ZIP) family of transporters, the Heavy Metal Associated (HMA) and Metal Tolerance Protein (MTP) families, and the Natural Resistance-Associated Macrophage protein (NRAMPs) family of transporters have all been shown to be integral in micronutrient transport in plants [17,18,19,20,21]. The transport of Zn from soil to seed can involve a number of these transporters, with Zn primarily being accumulated within the seed embryo and aleurone layer [22]. In rice Zn uptake from the soil is thought to occur through the transporters OsZIP5 and OsZIP9 [23, 24]. Others have suggested that many routes may be available to Zn uptake, as no single transporter is believed to be solely responsible for the uptake of Zn from the soil [25,26,27,28,29]. Several transporter families have also been implicated in the transport of Zn through the plant to the developing grains [18, 30, 31]. These include the HMA family and the ZIP family of transporters, both shown to be able to transport a broad range of metals [18, 19, 26, 32], which can also include the transport of unwanted metals such as cadmium (Cd) and lead (Pb), both of which are detrimental to most plants and animals [17, 19, 26, 32,33,34].
Other factors can also influence the Zn content in both plant leaves and grain. These include phytates, which readily bind cations such as calcium (Ca), iron (Fe) and Zn, phosphate (P) and phytate being found to correlate with Zn concentration in the grain of both wheat and rice [31, 35]. In wheat, nitrogen (N) content has been found to positively correlate with Zn content in the grain [8]. Recent work has shown that macronutrients such as N, P and sulphur (S), can also influence overall micronutrient content [36]. Thus, a holistic approach to nutrient enhancement is required to ensure adequate Zn supply in the human diet.
In this study we set out to understand the phenotypic variation of several nutritional traits in tef varieties commonly grown and consumed in Ethiopia. We used whole genome sequencing and single nucleotide polymorphisms (SNPs) to determine the genetic relationship between these tef varieties. This genomic data was then used to look for relationships between the tef varieties and the nutritional traits to identify those varieties with optimal nutritional potential for breeding. In addition, SNPs within candidate genes underlying nutritional traits of interest were examined to determine linkages with the trait variation.
Materials and methods
Plant material and flour preparation
Twenty-four tef varieties (Additional file 1) were sown during the main cropping season in July 2018, at a seeding rate of 15 kg per ha, at the experimental station of Adet Agricultural Research Center, Ethiopia. Adet (11 o28′ N, 37 o48′ E; 2216 m a.s.l) is located 42 km southwest of Bahir Dar, the capital city of the Amhara regional state, Ethiopia. Agronomic and cultural practices used were those recommended for tef production, and included application of 40 kg N and 60 kg P2O5 per ha [37]. The soil at Adet is brown Nitosol. The 24 tef varieties used in this study are publicly available in Ethiopia from the Adet Agricultural Research Center. The voucher specimen ID numbers are listed in Additional file 1.
Whole tef grains (20 g) of each variety were milled to flour using a Laboratory Hammer Mill (Model: TPS-JXFM110, China) to a sieve size of 0.5 mm, packed in polyethylene bags and stored at room temperature until analysed.
Analyses of nutritional traits
Analyses of all nutritional traits were undertaken at NIAB, UK. Analyses included assessment of elemental concentrations, phenolic compounds, phytate and N content.
Elemental concentrations were assessed using inductive coupled plasma mass spectrometry (ICP-MS). ICP-MS was used to detect the beneficial micronutrients Ca, Cu, Fe, K, Mg, Mn, Mo, P, S, Se, Ti, and Zn, and the detrimental elements Cd and Pb. Approximately 0.2–0.3 g of flour was digested in 5 mL of nitric acid (Sigma) overnight in a 7 mL bijou. The digested sample was transferred to a 50 mL beaker and heated to 115 °C to remove the residual acid, after which 3 mL of H2O2 (Sigma) was added. After H2O2 reduction the remaining powder was dissolved in 15 mL of ddH20 and samples analysed on the ICP-MS using a Thermo-Fisher Scientific iCAP-Q equipped with CCTED (collision cell technology with energy discrimination). Three independent technical replications were run for each tef flour sample. Possible soil contamination was identified where flour samples had greater than 100 mg/kg of Fe, Al and possibly Si, and/ or Ti above 1 μg/g [38, 39].
N content was determined using Dumas analysis. Flour samples were dried at 104 °C for 3 h. One gram of flour was loaded according to the manufacturer’s instructions (Leco TruMacN Dumas gas analyser) to determine N content. Dumas gas analysis was performed on three, technical replicate 1 g aliquots of flour from each variety.
Phenolic compounds were assessed using HPLC. Flour samples of approx. 2.5 g were extracted into 50 ml of ethanol-acetic acid (10% 1 M acetic acid v/v) under reflux conditions for 2 h. Extracts were stored at − 20 °C until analysed. The extracts were prepared for chromatography by centrifugation for 2 min at 13000 rpm, then filtrated through a 0.2 μm filter. The compounds were separated using the Dionex Ultimate 3000 HPLC system. A 150 mm × 4.6 mm × 5 μm × 100 Å Kinetix C18 column was used, with a gradient 0.1% formic acid /acetonitrile mobile phase running at 0.2 ml/minute (gradient of 0.95: 0.05 for 2 min, then 0.72; 0.28 for 18 min, 0.00; 0.10 for 28 min, and then held until 45 min) were used. The column effluent was monitored with a PDA detector between 200 and 600 nm, with data recorded at 254 nm, 280 nm, 340 nm and 520 nm.
Phytate was measured using the commercial Megazyme Phytic Acid Assay Kit (Brey, Ireland) following the manufacturer’s instructions with minor modifications. Approximately 100 mg of flour was digested in 1.8 mL HCl (0.66 M) in 2.2 mL tubes, placed in a rotator mixer overnight with a constant rpm of 20, at room temperature. Three technical replicates were applied for each flour sample.
Genomic DNA isolation and sequencing
DNA was extracted from flour of the 24 tef varieties using Qiagen’s DNAeasy Kit as per the manufacturer’s instructions, including RNase treatment. The same flour samples were used for DNA extraction as were used of nutrient trait analyses. DNA was shipped to Novogene (Cambridge UK) for Illumina Sequencing. Illumina libraries contained on average 350 bp inserts and were sequenced using paired end technology. The estimated genome coverage of each tef variety was at least 25X, based on an estimated size of the genome of 622 MB [11]. Raw reads for all tef varieties tested have been deposited with the ENA under the ArrayExpress accession E-MTAB-8827.
Whole genome sequence assembly and variant calling
Paired-end sequence reads were provided by the sequencing service that had already undergone quality checks, including the removal of over-represented sequences, adapters, and reads with low-quality base scores (at Q > 20). No further analysis using FastQC [40] was therefore required. The paired-end reads were mapped against the indexed reference sequence of the tef variety Dabbi using BWA mem [11, 41]. Output was piped to bam and sorted using SAMtools. PICARD tool was used to assign reads to a single Read Group (option: AddOrReplaceReadGroup). PCR duplicates were flagged using PICARD (option: MarkDuplicates). A dictionary for the reference genome of contig names and sizes was also created using PICARD (option: CreateSequenceDictionary). Variants were identified using the Broad Institute Genome Analysis Tool Kit (GATK4 [42];). GATK (option: Haplotypecaller) was run for each tef variety, using its default parameters, to call variants between the reference Dabbi genome sequence and the variety sequence in both Variant Call Format (VCF) and Genomic VCF (option: –ERC GVCF) modes. Only variants with a quality score of > 20 were considered. The variant SNPs were then analysed using gff3 and bcftools csq [43] to determine whether the SNP was within a coding region, and whether the SNP was synonymous vs non-synonymous.
Using PLINK2 the VCF file was converted into PLINK binary format (bed, bim and fam). A phylogenetic tree was created using SNPhylo and the biallelic SNPs identified across the 24 tef varieties [44]. A principal co-ordinate analysis (PCA) plot was constructed using the same biallelic SNPs and analysed using the R program SNPrelate [45]. FastStructure (with logistic prior, K = 5) was used to infer population structure within the 24 tef varieties using the same SNPs [46].
Identification of zinc iron permease and heavy metal associated transporter family members in tef
The Eragrostis tef genome was searched using the Biomart tool in Ensembl plants (http://plants.ensembl.org/biomart/martview/) to identify predicted genes that contain both the PFAM domain PF02535 and Interpro ID IPR003689, or Interpro ID IPR027256. The PFAM domain PF02535 and Interpro ID IPR003689 are used for the identification of ZIP transporter proteins and Interpro ID IPR027256 for identification of HMA transporter proteins. Candidate ZIP transporter and HMA transporter proteins in Arabidopsis and rice were also identified using Ensembl’s Biomart. The amino acid sequences of these candidate genes were aligned using MUSCLE as part of MEGA X [47] using the default parameters and a phytogenic tree created using the Maximum Likelihood option, with 50 replications used for the determination of bootstrap values. Reciprocal BLAST, using the amino acid sequence of the rice ZIP and HMA genes as the query, was further used to identify putative homologues of each gene in tef using CoGe (https://genomevolution.org/coge/CoGeBlast.pl).
Statistical analysis
Micronutrient, N and phytate levels were tested for normality and homogeneity of variance using Shapiro-Wilk’s test in R, which indicated a normal distribution. Two-way analysis of variance (ANOVA) was used to infer significance between tef varieties using the aov function and TukeyHSD commands in R. A least significant difference of 5% probability level was used as a post-hoc test to determine significance. Results were plotted using R ggplot2 and ggpubr packages [48, 49]. Correlations between micronutrients, N and phytate were calculated in R using the Spearman rank correlation (cor.test) or Kruskal-Wallis test for categorical variables [45].
All statistical analyses of phenolic compound levels were performed using Genstat v.16 (VSN International 2020). The levels of the phenolic compounds were analysed using a modified two-way ANOVA approach, General Linear Regression. The model applied was replicate by variety. Only comparisons having a F probability < 0.001 were considered as statistically significant. The linear relationship between phenolic compound levels were measured using the Pearson correlation coefficient in Excel (Microsoft). Boxplots were generated using R ggplot2.
Results
Comparison of nutritional traits in flour of twenty-four tef varieties
To understand the nutritional potential of 24 tef varieties (Additional file 1) currently grown and consumed in Ethiopia we measured the levels of a number of nutritional traits, comparing the relationship between these traits, and using genomic data to look at the genetic relationship between these 24 tef varieties relative to these nutritional traits. The nutritional traits included elemental micronutrient (Fig. 1 & Additional file 2), nitrogen as a proxy for protein content (Fig. 2A), phytate (Fig. 2D) and a range of phenolic compounds (Additional file 3).
Relationship between Zn, Fe, nitrogen and phytate levels in 24 tef varieties. Nitrogen levels (A). Correlation between Zn and nitrogen content (B). Correlation between Fe and nitrogen content (C). Phytate levels (D). Correlation between Zn and phytate content (E). Correlation between Fe and phytate content (F)
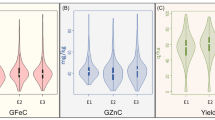
Significant variation between the 24 tef varieties was found for all micronutrients tested (Additional file 2) (p.val < 0.001). Focus on elements essential for human health, Zn and Fe (Fig. 1), showed Zn flour concentrations to range from 14.8 (var Etsub) to 29.2 mg/kg (var. Heber-1) and Fe to range from 22.6 (var Abay) to 684.25 mg/kg (var Yilmana) (Fig. 1A & B). Positive correlations were found between Zn and Ca, Mg, P, S, and to a lesser extent K, while negative correlations were found with Cu, Fe, Mo and Se. A positive correlation was also seen between Zn and Cd (Suppl. Fig. 1). No correlations were found between Pb or Ti and the other elements measured.
Cd is a toxic micronutrient, detrimental to human health. High levels of Cd were seen in many of the tef samples tested, the highest levels of Cd being found in the variety Wellenkomi, having 40.7 μg/kg. Only one of the tef varieties, var. Magna, had Cd levels below the current EU limits for Cd in food products, being below 1 μg/kg (1 ppm) [50].
The N levels within the tef flours were measured as a proxy for protein content [51]. Significant differences were found between the tef varieties, with N values ranging from 1.3 g/100 g in var. Ambo Toke to nearly 2 g/100 g in var. Dagan tef (p val < 0.001) (Fig. 2A). Using the standard conversion of 5.95 this would give protein content in the range of 7.79 to 11.71% [52]. A positive correlation was found between Zn content and N levels in the flours tested (R = 0.59; p val. 0.01; Fig. 2B) but not for Fe and N (R = 0.0096; p val 0.97; Fig. 2C).
Significant differences were found in phytate levels between the 24 tef varieties (p val < 0.01) (Fig. 2D). The phytate levels ranged from 0.83 g/100 g in var. Magna to 2.56 g/100 g in var. Abay. No significant correlations were observed between phytate and Zn (R 0.042 p val. 0.84; Fig. 2E), but a significant negative correlation was found between Fe and phytate (R − 0.54 p val. 0.007; Fig. 2F). There was no significant correlation between overall P levels, measured via ICP-MS, and phytate (R = 0.29; p val. 0.18).
Flour of the tef varieties were also screened for 19 phenolic compounds (Additional file 3). Significant differences (F p val. < 0.001) were found between the 24 tef varieties for 11 phenolic compounds, significant differences between tef varieties not being found for gallic acid. Seven phenolic compounds were not detected. These include cyanidin, cyanidin glycoside, delphinidin, delphinidin glycoside, flavone, pelagonidin and p-coumaric acid.
Kaempferol, quercetin, catechin and myricetin all belong to a group of phenolic compounds commonly known as flavonoids (Fig. 3) [53]. The levels of kaempferol and quercetin were low in all the tef varieties, the majority of these compounds being present as kaempferol glycoside and quercetin glycoside (Fig. 4). The levels of kaempferol glycoside and quercetin glycoside ranged from 4.92 (var. Tseday) to 205.79 μg/g (var. Magna), and 0.46 (var.Felagot) to 105.41 μg/g (var. Quncho), respectively. The levels of catechin ranged from 27.41 (var. Yilmana) to 183.19 μg/g (var. Tseday) (Fig. 4). Catechin is derived from dihydroxyquercetin (Fig. 3) and a precursor of proanthocyanidins, which are thought to give rice its red colouration [53]. The levels of myricetin ranged from 0.82 (var. Were-kiyu) to 17.24 μg/g (var. Dima) (Fig. 4).
Levels of 12 phenols identified in the flours of 24 tef varieties. Each box shows the range of concentrations in μg/g of 12 phenolic compounds measured in the 24 tef varieties. The box defines the upper and lower quartiles. The lines extending vertically, “whiskers”, indicate variability outside the upper and lower quartiles, with points outside those lines, represented by dots, being considered outliers
The levels of t-cinnamic acid ranged from 1.51 μg/g in var. Baset to 24.65 μg/g in var. Dukem. Cinnamic acid is a precursor of ferulic acid, which in turn gives rise to vanillic acid. A strong positive correlation was found between cinnamic and ferulic acids, and a lesser positive correlation with vanillic acid. However, the levels of vanillic acid were low in all 24 tef varieties (Additional file 3, Additional file 4).
Protocatechuic acid and gallic acid are synthesised from a side branch of the shikimate pathway that leads to the synthesis of folates and aromatic amino acids, including phenylalanine (Fig. 3 [53];). The levels of protocatechuic acid ranged from 6.78 μg/g (var. Areka-1) to 44.78 μg/g (var. Felagot) (Additional file 3). The levels of gallic acid were low in all 24 tef varieties, ranging from 0.12 μg/g (var. Dima) to 0.97 μg/g (var. Ambo take (Fig. 4).
Pearson’s correlation analyses were undertaken on the levels of each phenolic compound, phytate and Fe (Additional file 4). A strong, positive correlation was seen between cinnamic acid and ferulic acid (r = 0.78), fitting with the biochemical pathway (Fig. 3), where cinnamic acid is a precursor of ferulic acid. Similarly, a positive association was seen between ferulic acid and vanillic acid (r = 0.51), vanillic acid sitting downstream of ferulic acid. However, positive correlations were also seen between ferulic acid and myricetin (r = 0.61), which may relate to their common precursor cinnamic acid, and between cinnamic acid and quercetin (r = 0.49) and quercetin glycoside (r = 0.50).
The precursors of kaempferol and quercetin, dihydrokaempferol and dihydroquercetin, exist in an equilibrium controlled by a flavonoid 3′-hydroxylase (F3’H) enzyme which converts dihydrokaempferol to dihydroquercetin. This resulted in negative correlations between kaempferol and quercetin (r = − 0.28) and kaempferol glycoside and quercetin glycoside (r = − 0.43); the majority of kaempferol and quercetin being present in the glycosylated state. This was reflected in positive correlations between kaempferol and quercetin glycoside (r = 0.62) and quercetin and kaempferol glycoside (r = 0.45).
Catechin is derived from dihydroquercetin and therefore competes for synthesis with quercetin. Consequently, a negative correlation was observed between the levels of catechin and quercetin (r = − 0.32). However, a positive correlation was seen between catechin and quercetin glycoside (r = 0.52). A positive correlation was also observed between catechin and kaempferol (r = 0.57) and a negative correlation with kaempferol glycoside (r = − 0.51).
In addition to positive correlations with kaempferol (r = 0.62) and catechin (r = 0.52), positive correlations were observed between quercetin glycoside and other phenolic compounds, including ferulic acid (r = 0.65), myricetin (r = 0.56), cinnamic acid (r = 0.50). These correlations may indicate a positive feedback mechanism operating through the biosynthetic pathways leading to quercetin glycoside synthesis.
A negative correlation was found between gallic acid and salicylic acid (r = − 0.43), and between catechin and protocatechuic acid (r = − 0.44), while a positive correlation was observed between protocatechuic acid and Fe content (r = 0.40). No significant correlations were found between phytate levels and any of the phenolic compounds measured in this study (Additional file 4).
Genetic relationship between the twenty-four tef varieties
All 24 tef varieties were sequenced to greater than 25X coverage. Sequences were compared to the reference var. Dabbi across the whole genome. The varieties differed by as few as 1.567 million SNPs in var. Areka-1 relative to Dabbi, to as high as 2.372 million SNPs in var. Yilmana (Additional file 5). There was also considerable variation in INDELs, with Yilmana showing the fewest INDELs at 343,257 and Wellenkomi the highest at 520,234. Overall, 3,193,582 unique SNPs and 897,272 unique INDELs were found, containing a minimum allele frequency of 0.1 within the 24 varieties tested. Considering that the whole tef genome is estimated to be 622 Mb in size, the 2.372 million variants identified in var. Yilmana relative to the reference var. Dabbi, equates to roughly four variants in every kb of the genome.
To understand how the 24 tef varieties relate to each other a phylogenetic tree analysis (Fig. 5A), a principal component analysis (PCA; Fig. 5B), and a structure analysis (Fig. 5C) were carried out using the genome wide SNP data. The phylogenetic tree and PCA separated the 24 tef varieties into 3 groups, however the structure analysis returned two groupings. In general, the tef varieties fell into similar groupings when comparing the phylogenetic tree and PCA, the exception being the varieties Negus and Kora, which fell on the same branch of the tree but in distinct PCA groups.
Genetic variation underlying differences in Zn levels
Zn concentrations were overlaid on the PCA to see if a relationship between flour Zn levels and the tef variety groupings could be identified (Fig. 5B). As no specific association between genetic grouping of tef varieties and Zn concentration were apparent, we chose to look at the genetic variation in specific gene families involved in Zn transport. Two gene families were selected. The ZIP (Zinc Iron Permease) family are involved in uptake of Zn from the soil and the HMA (Heavy metal associated) family of transporters are involved in movement of Zn from roots to seeds.
Zinc Iron permease transporter family in tef
To identify putative ZIP family members the gene models of the Eragrostis tef var. Dabbi sequence in Ensembl plants was searched using the PFAM domain PF02535 and Interpro ID IPR003689. This revealed 32 predicted genes in tef which contained these protein domains. As tef is a tetraploid species these 32 potential ZIP family members is comparable to the 15 in Arabidopsis and 17 in rice [32, 54]. However, some of the predicted genes could be pseudogenes. Only six of the 32 coding regions identified have a good ATG start codon, and two of these six putative ZIP transporters lacked a stop codon. However, as this incomplete sequence data could be due to gaps in the reference var. Dabbi tef sequence we used all 32 putative ZIP sequences in subsequent analyses.
Phylogenetic analysis of the translated, amino acid sequences of the 32 tef ZIP transporters was performed with members of the ZIP families from rice and Arabidopsis Fig. 6). Tef ZIP proteins showed closer linkages with rice ZIP proteins compared to Arabidopsis, both tef and rice being monocots. In most cases there were two tef genes for every rice ZIP gene, which is expected as tef is a tetraploid species, but not all 32 tef ZIP genes showed clear associations with genes found in rice or Arabidopsis, suggesting some divergence. Rice ZIP genes associated with more than two tef genes included OsZIP8, which was associated with four tef genes. Only one of the 32 tef ZIP genes localized with OsZIP3, while three tef genes colocalized with OsIRT2, as well as OsZIP10.
Heavy metal associated transporter family in tef
To identify putative HMA family members the gene models of the Eragrostis tef var. Dabbi sequence in Ensembl plants was searched using the Interpro domain IPR027256. Fourteen HMA proteins were identified in tef compared to eight in Arabidopsis and nine in rice. Phylogenetic analysis of the translated, amino acid sequences of the 14 tef HMA transporters was performed with members of the HMA families from rice and Arabidopsis (Fig. 7). As 14 tef HMA genes was less than expected for this tetraploid species, based on conservation of each of the eight core genes found in both rice and Arabidopsis, the tef Dabbi reference sequence may therefore be incomplete with regards to this family of Zn transporters. In addition, five of the 14 predicted HMA genes in tef did not contain an ATG start codon, suggesting that some sequence maybe missing, or that the gene models are incorrect. Only four rice HMA proteins, OsHMA2, OsHMA5, OsHMA6 and OsHMA9, had two clear homologues within the predicted tef HMA genes. However, no clear homologue for the rice OsHMA3 gene, a major gene involved in Cd tolerance and sequestration [55], was detected.
Identification of variants in ZIP and HMA proteins
Numerous SNPs were found in both the ZIP and HMA tef families of transporters (Additional files 6, Additional file 7). In the 32 EtZIP genes a total of 355 variants were identified in the coding regions relative to the reference var. Dabbi, this included frame shift mutations in twelve of the 32 genes (3.4% of the total mutations) (Additional file 6). Most of the variants were located in an intron (41.4%), followed by synonymous variants in the coding region (22.5%), and then by non-synonymous mutations that result in a change in the amino acid sequence (14.6%). Many of the variants were conserved between the varieties, including a frameshift in both homologues of OsZIP9, with most of the varieties containing the mutated/truncated form of each gene.
In EtHMA transporters 298 variants were identified relative to var. Dabbi (Additional file 7). Most of the variants were again found in introns or represented synonymous mutations in the coding sequence (48 and 16.4%, respectively). Only two of the 14 tef HMA genes had frame shift variants. This included the predicted gene loci Et_s3091–2.42-1.mrna1 and Et_s3193–0.29-1.mrna1. The third most common variants were non-synonymous mutations in the protein coding region (12.4%).
Genetic variation underlying differences in phenolic compounds
Flavonoid 3′-hydroxylase (F3’H) is a key enzyme in the conversion of dihydrokaempferol to dihydroquercetin [53, 56]. To determine whether F3’H in tef was responsible for the differences seen between tef varieties in kaempferol glycoside and quercetin glycoside levels the amino acid sequence of the rice F3’H gene (Os10g0320100) was used to identify possible orthologs in tef. Using an e-value cut off of 1e-10 five putative orthologs of OsF3’H were identified in the tef reference sequence of var. Dabbi: Et_s9738-1.8-1.mrna1, Et_s9399-0.5-1.mrna1, Et_s3159-0.29-1.mrna1, Et_s6352-0.10-1.mrna1 and Et_s15942-0.0-1.mrna1 (Suppl. Fig. 2). Alignment of the amino acid sequences of these five tef genes to their rice putative orthologs showed 58.74 to 78.57% identity. The two tef genes Et_s3159-0.29-1.mrna1 and Et_s15942-0.0-1.mrna1 clustered most closely with Os10g0320100 and were subject to further analysis. SNPs were identified between Et_s3159-0.29-1.mrna1 and Et_s15942-0.0-1.mrna1. Correlations between these SNPs and the levels of kaempferol glycoside and quercetin glycoside in the 24 tef varieties revealed a SNP in Et_s3159–0.29-1.mrna1 that was strongly correlated with kaempferol glycoside and quercetin glycoside levels (p val < 0.001). This SNP resulted in a T → G substitution in the second intron of Et_s3159–0.29-1.mrna1, and therefore does not directly change the coding sequence of the gene (Suppl. Fig. 2). Tef varieties containing the wild-type (WT) “T” allele in the homozygous state had lower levels of kaempferol glycoside than tef varieties containing the mutant “G” SNP, and visa verse, while varieties with a heterozygous SNP had intermediary levels of the two glycosides (Fig. 8). No further associations were found between the other phenolic compounds measured and SNPs in any of the F3’H-type genes in tef.
Haplotype analysis of a F3’H gene in tef. Kaempferol glycoside and quercetin glycoside levels in tef flours given the genotype of the T → G mutation in the second intron of Et_s3159-0.29-1.mrna1. A different letter represents a significant difference (p val < 0.05) between genotypes for either kaempferol glycoside or quercetin glycoside
Discussion
Tef is a staple crop for many Ethiopians, however, due to naturally low Zn levels in soils on which it is grown the grain does not meet the Zn needs of those who consume it [57]. Zn is essential for a healthy immune system and stimulates the activity of many different enzymes. The low level of Zn in tef has therefore led to many Ethiopian children suffering from Zn deficiency [58]. Fe is also an essential element, required for normal blood cell function [59], but has a low bioavailability, the small intestine not readily absorbing large amounts of Fe. However, a number of compounds have been shown to influence Fe absorption in the human gut, including phytate, kaempferol and quercetin [60,61,62].
To understand the breeding potential of tef for Zn and Fe content, as well as several other nutritional factors, grain from 24 tef varieties were assayed for a range of nutritional traits. Analysis of these 24 varieties showed significant variation in all the micronutrients tested, with a twofold difference in the amount of Zn being observed. However, even var. Heber-1, with 29.2 mg/kg of Zn, had levels below the recommended 40 mg/kg of Zn in flour [8]. This may be rectified by Zn fertilization of the soil, but future work would be required to understand how each variety responds to Zn soil supplements, given the complexity of Zn uptake and transportation to the grain.
No correlation was found between Zn and Fe levels in the grains, and is most likely due to different regulatory mechanisms and transporters involved in the movement of each ion through the plant [18, 30]. The inability to identify a homologue of OsHMA3, a gene associated with Cd tolerance and sequestration in rice, may also suggest an underlying cause for some of the high levels of Cd seen in the 24 tef flours. However, it should also be noted that other putative tef genes, including a homolog of OsZIP1 (Et_s3548–1.55-1.mrna1) which is involved in efflux of excess heavy metals, was one of the twelve tef genes with a frame shift variant present in all the tef varieties. This would suggest that multiple genes which sequester Cd in the roots, and keep it away from the grain, may not exist in tef or are non-functional in the modern tef varieties assayed in this study [63].
Sequencing of the 24 tef varieties enabled the identification of several variants which could be used to develop markers for future breeding. Overall, there was a large amount of genetic variation between the varieties compared to the reference var. Dabbi. Although most of these variants were in non-coding regions or synonymous mutations, as seen in the analysis of the ZIP and HMA transporter families. The variation observed is not unexpected as similar levels of variation have been observed in a core set of rice varieties which were recently sequenced [64].
In rice, double mutants of OsZIP5 and OsZIP9 have shown severe Zn deficiency symptoms, suggesting these two genes are the major route for Zn into the rice plant [23, 24]. However, mutants of either gene in isolation does not show major effects on Zn uptake. Mutations in two homologues of OsZIP9, including frame shifts, as well as a frame shift variant in a homologue of OsZIP5 in some varieties, would suggest that the route of Zn into tef, while compromised by these mutations, is not wholly dependent on the homologues of OsZIP5 and OsZIP9. Other transporters maybe the major route for Zn into tef plants. These other routes might also contribute to the high levels of Cd seen in many of the tef varieties. It has been shown that genes involved in Mn and Fe uptake can also transport Cd in vivo whereas high affinity Zn transporters do not appear to have this capability [17, 25, 29, 65, 66].
Comparison of other nutritional traits, including nitrogen (as a proxy for protein), phytate and a number of phenolic compounds, also showed significant variation between the 24 tef varieties. The variation in the phenolic profiles is of importance as some phenolics have been found to alter Fe bioavailability and may therefore present a more effective strategy than Fe fortification for enhancing nutritional outcomes. It is often reported that phytate inhibits Fe bioavailability, so the negative correlation between Fe and phytate levels in tef could prove beneficial. While future research is required to determine whether elevated levels of phenolics that stimulate and inhibit Fe uptake in the human gut can help alleviate the anaemia seen in Ethiopian children [67], the significance of the SNP in the tef F3’H gene Et_s3159-0.29-1.mrna1, which explains a large proportion of the variation in kaempferol glycoside and quercetin glycoside levels, suggest this might be a breeding target to improve Fe bioavailability in tef, as kaempferol glycoside is known to promote Fe absorption while quercetin glycoside inhibits Fe absorption in cell assays [60]. In addition, phenolic compounds have also been found to influence other agronomic traits, including tolerance to both biotic and abiotic stress [16, 60, 61, 68].
With the considerable variation seen for the nutritional traits assessed in this study breeders are in a good position to breed for enhanced nutritional value in tef. The genomic sequence information collected can be used to identify and develop markers linked to target genes and traits. We have yet to test the heritability of these traits and their stability over growing locations and seasons, but marker-assisted selection, particularly within target genes, can now provide a feasible approach to breed for these nutritional traits in tef.
Conclusions
For many subsistence Ethiopian farming families tef is a major crop and source of calories. Yet, as shown in this study, levels of Zn are usually below recommended levels, while levels of Cd exceed EU limits. However, considerable phenotypic and genetic variation for a range of nutritional traits and the genes regulating their levels in planta, exists. This provides considerable potential to determine the relationship between these nutrition phenotypes and identify allelic variants that would allow breeding of new tef varieties with optimal nutritional potential.
Availability of data and materials
Sequencing Data is available at the ENA under ArrayExpress accession E-MTAB-8827.
Abbreviations
- N:
-
Nitrogen
- Zn:
-
Zinc
- P:
-
Phosphate
- Fe:
-
Iron
- Cd:
-
Cadmium
- Pb:
-
Lead
- Ca:
-
Calcium
- S:
-
Sulphur
- K:
-
Potassium
- Mg:
-
Magnesium
- Mn:
-
Manganese
- Mo:
-
Molybdenum
- Se:
-
Selenium
- Ti:
-
Titanium
- Al:
-
Aluminium
- Si:
-
Silicon
- HPLC:
-
High-performance liquid chromatography
- PDA:
-
Photo Diode Array
- HCl:
-
Hydrochloric acid
- Vcf:
-
Variant Call Format
- EU:
-
European Union
- MTP:
-
Metal Tolerance Protein
- ZIP:
-
Zinc Iron Permease
- HMA:
-
Heavy Metal Associated
- NRAMP:
-
Natural Resistance-Associated Macrophage Proteins
- SNP:
-
Single Nucleotide Polymorphism
- INDEL:
-
Insertion or Deletion
- ICP-MS:
-
Inductively Coupled Plasma Mass Spectrometry
- PCA:
-
Principal Component Analysis
- ENA:
-
European Nucleotide Archive
References
Baye K. Teff: nutrient composition and health benefits - IFPRI publications - IFPRI knowledge collections. Int Food Policy Res Inst. 2014:20 [cited 2020 Oct 27]. Available from: https://ebrary.ifpri.org/digital/collection/p15738coll2/id/128334/.
Teff LH, Rising A. Global crop: current status of Teff production and value chain. Open Agric J. 2018;12:185–93 Bentham Science Publishers Ltd.
Fufa B, Behute B, Simons R, Berhe T. Strengthening the Tef value chain in Ethiopia; 2011.
Foodgrain consumption and calorie intake patterns in Ethiopia - IFPRI Publications - IFPRI Knowledge Collections. [cited 2020 Oct 22]. Available from: https://ebrary.ifpri.org/digital/collection/p15738coll2/id/124853
Zhu F. Chemical composition and food uses of teff (Eragrostis tef). Food Chem. 2018;239:402–15 Elsevier Ltd.
Cheng A, Mayes S, Dalle G, Demissew S, Massawe F. Diversifying crops for food and nutrition security - a case of teff. Biol Rev Camb Philos Soc. 2017;92:188–98.
Abebe Y, Bogale A, Hambidge KM, Stoecker BJ, Bailey K, Rosalind GS. Phytate, zinc, iron and calcium content of selected raw and prepared foods consumed in rural Sidama, southern Ethiopia, and implications for bioavailability. J Food Compos Anal. 2007;20:161–8.
Cakmak I, Pfeiffer WH, McClafferty B. Biofortification of durum wheat with zinc and iron. Cereal Chem. 2010;87:10–20.
Kebede W, Tolossa K, Hussein N, Fikre T, Genet Y, Bekele A, et al. Tef (Eragrostis tef) variety ‘Tesfa.’. Ethiop J Agric Sci. 2018;28:107–12.
Mekibib F, Amare M, Bekele A, Jobie T, Merine Y, Tabor G, et al. Plant variety release. Protection and seed quality control directorate. 2016.
VanBuren R, Man Wai C, Wang X, Pardo J, Yocca AE, Wang H, et al. Exceptional subgenome stability and functional divergence in the allotetraploid Ethiopian cereal teff. Nat Commun. 2020;11:1–11 Nature Research.
Nakandalage N, Nicolas M, Norton RM, Hirotsu N, Milham PJ, Seneweera S. Improving rice zinc biofortification success rates through genetic and crop management approaches in a changing environment. Front Plant Sci. 2016;7. https://doi.org/10.3389/fpls.2016.00764.
Klein MA, Grusak MA. Identification of nutrient and physical seed trait QTL in the model legume Lotus japonicus. Genome. 2009;52:677–91.
Arora S, Cheema J, Poland J, Uauy C, Chhuneja P. Genome-wide association mapping of grain micronutrients concentration in aegilops tauschii. Front Plant Sci Frontiers Media S.A. 2019;10. https://doi.org/10.3389/fpls.2019.00054.
Oury FX, Leenhardt F, Rémésy C, Chanliaud E, Duperrier B, Balfourier F, et al. Genetic variability and stability of grain magnesium, zinc and iron concentrations in bread wheat. Eur J Agron. 2006;25:177–85.
Wright TIC, Gardner KA, Glahn RP, Milner MJ. Genetic control of iron bioavailability is independent from iron concentration in a diverse winter wheat mapping population. BMC Plant Biol. 2021;21:1–13 BioMed Central Ltd.
Milner MJ, Mitani-Ueno N, Yamaji N, Yokosho K, Craft E, Fei Z, et al. Root and shoot transcriptome analysis of two ecotypes of Noccaea caerulescens uncovers the role of NcNramp1 in Cd hyperaccumulation. Plant J. 2014;78:398–410.
Milner M, Kochian L. Investigating heavy-metal hyperaccumulation using Thlaspi caerulescens as a model system | annals of botany | Oxford academic. Ann Bot. 2008;102:3–13.
Hussain D, Haydon MJ, Wang Y, Wong E, Sherson SM, Young J, et al. P-type ATPase heavy metal transporters with roles in essential zinc homeostasis in arabidopsis. Plant Cell. 2004;16:1327–39 American Society of Plant Biologists.
Desbrosses-Fonrouge AG, Voigt K, Schröder A, Arrivault S, Thomine S, Krämer U. Arabidopsis thaliana MTP1 is a Zn transporter in the vacuolar membrane which mediates Zn detoxification and drives leaf Zn accumulation. FEBS Lett. 2005;579:4165–74 No longer published by Elsevier.
Dräger DB, Desbrosses-Fonrouge AG, Krach C, Chardonnens AN, Meyer RC, Saumitou-Laprade P, et al. Two genes encoding Arabidopsis halleri MTP1 metal transport proteins co-segregate with zinc tolerance and account for high MTP1 transcript levels. Plant J. 2004;39:425–39 Wiley.
De Brier N, Gomand SV, Donner E, Paterson D, Smolders E, Delcour JA, et al. Element distribution and iron speciation in mature wheat grains (Triticum aestivum L.) using synchrotron X-ray fluorescence microscopy mapping and X-ray absorption near-edge structure (XANES) imaging. Plant Cell Environ. 2016;39:1835–47.
Yang M, Li Y, Liu Z, Tian J, Liang L, Qiu Y, et al. A high activity zinc transporter OsZIP9 mediates zinc uptake in rice. Plant J. 2020;103:1695–709 Wiley.
Tan L, Qu M, Zhu Y, Peng C, Wang J, Gao D, et al. Zinc transporter5 and zinc transporter9 function synergistically in zinc/cadmium uptake1. Plant Physiol. 2020;183:1235–49 American Society of Plant Biologists.
Milner MJ, Craft E, Yamaji N, Koyama E, Ma JF, Kochian LV. Characterization of the high affinity Zn transporter from Noccaea caerulescens, NcZNT1, and dissection of its promoter for its role in Zn uptake and hyperaccumulation. New Phytol. 2012;195:113–23.
Grotz N, Fox T, Connolly E, Park W, Lou GM, Eide D. Identification of a family of zinc transporter genes from Arabidopsis that respond to zinc deficiency. Proc Natl Acad Sci U S A. 1998;95:7220–4 National Academy of Sciences.
Tiong J, McDonald G, Genc Y, Shirley N, Langridge P, Huang CY. Increased expression of six ZIP family genes by zinc (Zn) deficiency is associated with enhanced uptake and root-to-shoot translocation of Zn in barley ( Hordeum vulgare ). New Phytol. 2015;207:1097–109 Blackwell Publishing Ltd.
Palmgren MG, Clemens S, Williams LE, Krämer U, Borg S, Schjørring JK, et al. Zinc biofortification of cereals: problems and solutions. Trends Plant Sci. 2008;13:464–73.
Curie C, Alonso JM, Le Jean M, Ecker JR, Briat JF. Involvement of NRAMP1 from Arabidopsis thaliana in iron transport. Biochem J. 2000;347:749–55 Portland Press Ltd.
Sperotto RA, Ricachenevsky FK, Williams LE, Vasconcelos MW, Menguer PK. From soil to seed: Micronutrient movement into and within the plant. Front Plant Sci Frontiers. 2014;5. https://doi.org/10.3389/fpls.2014.00438.
Magallanes-López AM, Hernandez-Espinosa N, Velu G, Posadas-Romano G, Ordoñez-Villegas VMG, Crossa J, et al. Variability in iron, zinc and phytic acid content in a worldwide collection of commercial durum wheat cultivars and the effect of reduced irrigation on these traits. Food Chem. 2017;237:499–505 Elsevier Ltd.
Milner MJ, Seamon J, Craft E, Kochian LV. Transport properties of members of the ZIP family in plants and their role in Zn and Mn homeostasis. J Exp Bot. 2012;64:369–81.
Papoyan A, Piñeros M, Kochian LV. Plant Cd 2+ and Zn 2+ status effects on root and shoot heavy metal accumulation in Thlaspi caerulescens. New Phytol. 2007;175:51–8 Wiley.
Pence NS, Larsen PB, Ebbs SD, Letham DLD, Lasat MM, Garvin DF, et al. The molecular physiology of heavy metal transport in the Zn/Cd hyperaccumulator Thlaspi caerulescens. Proc Natl Acad Sci U S A. 2000;97:4956–60 National Academy of Sciences.
Stangoulis JCRR, Huynh B-LL, Welch RM, Choi E-YY, Graham RD. Quantitative trait loci for phytate in rice grain and their relationship with grain micronutrient content. Euphytica Springer. 154:289–94.
Kumar S, Kumar S, Mohapatra T. Interaction between macro- and micro-nutrients in plants. Front Plant Sci. 2021;0:753 Frontiers.
Abewa A, Adgo E, Yitaferu B, Alemayehu G, Assefa K, Solomon JKQ, et al. Teff grain physical and chemical quality responses to soil physicochemical properties and the environment. Agron. 2019;9:283 Multidisciplinary Digital Publishing Institute.
Cherney JH, Robinson DL. A comparison of plant digestion methods for identifying soil contamination of plant tissue by TI Analysis1. Agron J. 1983;75:145–7 Wiley.
Jones JB. Plant Tissue Analysis in Micronutrients. In: SSSA Book Series. editor. Mortvedt JJ, Micronutr Agric. 2nd ed. John Wiley & Sons, Ltd; 1991. p. 477–521.
Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010. Available from: http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
Li H, Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics. 2009;25:1754–60.
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303 Cold Spring Harbor Laboratory Press.
Danecek P, McCarthy SA. BCFtools/csq: haplotype-aware variant consequences. Bioinformatics. 2017;33:2037–9.
Lee TH, Guo H, Wang X, Kim C, Paterson AH. SNPhylo: a pipeline to construct a phylogenetic tree from huge SNP data. BMC Genomics. 2014;15:162 BioMed Central.
Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, Weir BS. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics. 2012;28:3326–8 Oxford Academic.
Raj A, Stephens M, Pritchard JK. FastSTRUCTURE: Variational inference of population structure in large SNP data sets. Genetics. 2014;197:573–89.
Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol. 2018;35:1547–9.
Kassambara A. ggplot2. Based Publ ready plots [R Packag ggpubr version 02 5]; 2020.
Computing RF for S. R Core Team (2018). R: a language and environment for statistical computing. Available from: https://www.r-project.org/.
EUR-Lex - 02006R1881-20150521 - EN - EUR-Lex. [cited 2020 Sep 12]. Available from: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX:02006R1881-20150521
Heathcote JG. The protein quality of oats. Br J Nutr. 1950;4:145–54 Cambridge University Press.
Fujihara S, Sasaki H, Aoyagi Y, Sugahara T. Nitrogen-to-protein conversion factors for some cereal products in Japan. J Food Sci. 2008;73:C204–209.
Mbanjo EGN, Kretzschmar T, Jones H, Ereful N, Blanchard C, Boyd LA, et al. The genetic basis and nutritional benefits of pigmented rice grain. Front Genet. 2020;11:229 Frontiers Media S.A.
Chen WR, Feng Y, Chao YE. Genomic analysis and expression pattern of OsZIP1, OsZIP3, and OsZIP4 in two rice (Oryza sativa L.) genotypes with different zinc efficiency. Russ J Plant Physiol. 2008;55:400–9.
Sasaki A, Yamaji N, Ma JF. Overexpression of OsHMA3 enhances Cd tolerance and expression of Zn transporter genes in rice. J Exp Bot. 2014;65:6013–21 Oxford University Press.
Lam PY, Lui ACW, Yamamura M, Wang L, Takeda Y, Suzuki S, et al. Recruitment of specific flavonoid B-ring hydroxylases for two independent biosynthesis pathways of flavone-derived metabolites in grasses. New Phytol. 2019;223:204–19 Blackwell Publishing Ltd.
Tessema M, De Groote H, Brouwer ID, Feskens EJM, Belachew T, Zerfu D, et al. Soil zinc is associated with serum zinc but not with linear growth of children in Ethiopia. Nutrients. 2019;11:221.
Ayana G, Moges T, Samuel A, Asefa T, Eshetu S, Kebede A. Dietary zinc intake and its determinants among Ethiopian children 6–35 months of age. BMC Nutr. 2018;4:30. https://doi.org/10.1186/s40795-018-0237-8.
Sciences NA of S (US) and NRC (US) D of M. The role of iron in hemoglobin synthesis: National Academies Press (US); 1958.
Hart JJ, Tako E, Glahn RP. Characterization of polyphenol effects on inhibition and promotion of Iron uptake by Caco-2 cells. J Agric Food Chem. 2017;65:3285–94 American Chemical Society.
Hart JJ, Tako E, Kochian LV, Glahn RP. Identification of black bean ( Phaseolus vulgaris L.) polyphenols that inhibit and promote Iron uptake by Caco-2 cells. J Agric Food Chem. 2015;63:5950–6.
Jin F, Frohman C, Thannhauser TW, Welch RM, Glahn RP. Effects of ascorbic acid, phytic acid and tannic acid on iron bioavailability from reconstituted ferritin measured by an in vitro digestion-Caco-2 cell model. Br J Nutr. 2009;101:972–81.
Liu XS, Feng SJ, Zhang BQ, Wang MQ, Cao HW, Rono JK, et al. OsZIP1 functions as a metal efflux transporter limiting excess zinc, copper and cadmium accumulation in rice. BMC Plant Biol. 2019;19:1–16 BioMed Central Ltd.
Tanaka N, Shenton M, Kawahara Y, Kumagai M, Sakai H, Kanamori H, et al. Whole-genome sequencing of the NARO world rice core collection (WRC) as the basis for diversity and association studies. Plant Cell Physiol. 2020;61:922–32 Oxford University Press.
Vert G, Grotz N, Dédaldéchamp F, Gaymard F, Guerinot ML, Briat JF, et al. IRT1, an arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell. 2002;14:1223–33.
Lin YF, Hassan Z, Talukdar S, Schat H, Aarts MGM. Expression of the Znt1 zinc transporter from the metal hyperaccumulator noccaea caerulescens confers enhanced zinc and cadmium tolerance and accumulation to arabidopsis thaliana. PLoS One. 2016;11. https://doi.org/10.1371/journal.pone.0149750.
Hailu A. Ethiopian National Micronutrient Survey Report: September 2016: Ethiop Public Heal Institute, Minist Heal; 2016.
Singh P, Arif Y, Bajguz A, Hayat S. The role of quercetin in plants. Plant Physiol Biochem. 2021;166:10–9 Elsevier Masson.
Acknowledgements
The authors would like to thank the Adet Agricultural Research Center for providing the tef samples. The University of Nottingham school of Chemistry for running the ICP-MS samples for elemental content. Helen Appleyard and Marianna Rizzo at NIAB analytical services for analysis for N content analysis of the grains.
Seeds collected
Permission to collect and analyse the milled seeds/flour of the tef varieties documented in this work was obtained from the Ethiopian government prior to research being conducted and samples being sent.
Funding
This work was supported by the Biotechnology and Biological Sciences Council (UK) BBSRC (grant BB/P027458/1) and pump priming grant (GCRF-SA-2020-NIAB). As well as the Cambridge Global Challenges fund. The funding bodies listed here played no role in the design of the study and collection, analysis, and interpretation of data, or in the writing of the manuscript.
Author information
Authors and Affiliations
Contributions
MM, NF, NE, and HAC designed the experiments. MM, NF, NE, HJ performed the experiments. NE undertook the bioinformatic analyses. MM and LB undertook the statistical analyses of the data. The manuscript was written by MM and LB and subsequently revised by all authors. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All the methods were carried out in accordance with relevant guidelines and regulations.
Consent for publication
All authors consent to publication.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1.
Twenty-four teff varieites use in study.
Additional file 2.
Average elemental concentrations of three replicates of teff flour acid digested and measured by ICP-MS, phytate, and N levels measured by Dumas.
Additional file 3.
Average amounts of 12 phenolic compounds found in 24 teff varieties (μg/g flour) and 7 phenolics not found in significant quantities.
Additional file 4.
Correlations between phenolic compunds, phytate and Fe.
Additional file 5.
SNP and INDEL variants relative to var. Dabbi for each of the 24 teff varieties sequenced.
Additional file 6.
Variants identified in the coding regions of teff ZIP type transporters.
Additional file 7.
Variants identified in the coding regions of teff HMA type transporters.
Additional file 8.
Genotype of SNP in the second intron of Et_s3159-0.29-1.mrna1 in the varieties tested. Where WT is a T and mut is a G.
Additional file 9: Fig. 1
. Significant correlations between the elemental concentrations in 24 teff flours. Size of the circle represents the significance level, with the larger the circle the lower the p value. All circles represent a p value less than 0.05. Scale on the left is the range of correlations (r). Fig. 2. Phylogenetic tree showing the relationship between the five teff homologues of the rice F3’H gene Os10t0320100–01.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Ereful, N.C., Jones, H., Fradgley, N. et al. Nutritional and genetic variation in a core set of Ethiopian Tef (Eragrostis tef) varieties. BMC Plant Biol 22, 220 (2022). https://doi.org/10.1186/s12870-022-03595-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-022-03595-9