Abstract
Background
GRAS transcription factor (TF) family is unique and numerous in higher plants with diverse functions that involving in plant growth and development processes, such as gibberellin (GA) signal transduction, root development, root nodule formation, and mycorrhiza formation. Walnut tree is exposed to various environmental stimulus that causing concern about its resistance mechanism. In order to understand the molecular mechanism of walnut to adversity response, a GRAS TF (JrGRAS2) was cloned and characterized from Juglans regia in this study.
Results
A 1500 bp promoter fragment of JrGRAS2 was identified from the genome of J. regia, in which the cis-elements were screened. This JrGRAS2 promoter displayed expression activity that was enhanced significantly by high temperature (HT) stress. Yeast one-hybrid assay, transient expression and chromatin immunoprecipitation (Chip)-PCR analysis revealed that JrDof3 could specifically bind to the DOFCOREZM motif and share similar expression patterns with JrGRAS2 under HT stress. The transcription of JrGRAS2 was induced by HT stress and up-regulated to 6.73-~11.96-fold in the leaf and 2.53-~4.50-fold in the root to control, respectively. JrGRAS2 was overexpressed in Arabidopsis, three lines with much high expression level of JrGRAS2 (S3, S7, and S8) were selected for HT stress tolerance analysis. Compared to the wild type (WT) Arabidopsis, S3, S7, and S8 exhibited enhanced seed germination rate, fresh weight accumulation, and activities of catalase (CAT), peroxidase (POD), superoxide dismutase (SOD) and glutathione-S-transferase (GST) under HT stress. In contrast, the Evans blue staining, electrolyte leakage (EL) rates, hydrogen dioxide (H2O2) and malondialdehyde (MDA) content of transgenic seedlings were all lower than those of WT exposed to HT stress. Furthermore, the expression of heat shock proteins (HSPs) in S3, S7, and S8 was significant higher than those in WT plants. The similar results were obtained in JrGRAS2 transient overexpression walnut lines under normal and HT stress conditions.
Conclusions
Our results suggested that JrDof3 TF contributes to improve the HT stress response of JrGRAS2, which could effectively control the expression of HSPs to enhance HT stress tolerance. JrGRAS2 is an useful candidate gene for heat response in plant molecular breeding.
Similar content being viewed by others
Background
High temperature (HT) stress is one of the most important limiting factors to plant growth and productivity [1]; warming surface temperatures and increasing frequency and duration of widespread droughts threaten the health of natural forests and agricultural crops [2]. The rising temperature may cause a change in the growing periods and the distribution of plants. HT may inactivate major enzymes, disturb protein synthesis, damage proteins and membranes, have major effects on the process of cell divisions, all of these can favor the oxidative damage and seriously limit the plant growth [1, 3]. Other than this, long-term HT stress during the seed filling can result in poor quality and low yield [4,5,6]. For instance, the number of spikes and florets per plant in rice and the seed-set in sorghum were negatively affected by HT stress [7, 8]. Under high night temperature, a decrease in individual grain weight resulted in significant reduction in rice grain production per unit area [9]. Zhao et al. (2017) point out that without effective adaptation, CO2 fertilization, and genetic improvement, each degree-Celsius increase in global mean temperature would, on average, reduce global yields of maize by 7.4%, wheat by 6.0%, rice by 3.2%, and soybean by 3.1% [10]. Therefore, the damage caused by HT to plants should not be underestimated.
The effect of HT varies in different plant species and cultivars, and even at different developmental stages within a species. To enable the production of plants with improved thermotolerance, decoding the mechanisms that which plants cope with HT is very necessary [11]. In recent years, physiological, biochemical, genetic, and molecular studies have revealed a number of vital cellular components and processes involved in thermoresponsive growth and the acquisition of thermotolerance in plants [11]. During these processes, a series of genes are employed that including heat shock proteins (HSPs) and reactive oxygen species (ROS)-scavenging enzymes, which were classified into two groups as follows: (1) The genes involve in heat shock signaling mechanisms that mainly including HSFA1-dependent transcriptional regulation networks, HSFA1-independent transcription regulation networks, Calcium (Ca2+) signaling, ROS signaling, NO signaling, Hydrogen sulfide (H2S) signaling, and unfolded protein response (UPR) [11,12,13,14,15,16,17,18,19,20]. (2) The genes associate with high ambient temperature signaling mechanisms, which contain the coordinated regulation of circadian clock [21], phytohormone signaling [22], and light signaling [23]. The core of these various signaling pathways is partially integrated to the basic helix-loop-helix (bHLH) transcription factor (TF) phytochrome interacting factor 4 (PIF4) [11, 21, 22]. PIF4 is connected with abundant genes such as: ultraviolet (UV) resistance locus 8 (UVR8) [24], constitutively photomorphogenic 1 (COP1) [25], elongated hypocotyl 5 (HY5) [26]. In PIF4-dependent ambient HT responses, PIF4 can activate the expression of auxin biosynthesis-related genes such as cytochrome P450, YUCCA 8 (YUC8), and tryptophan aminotransferase of Arabidopsis 1 (TAA1) via binding to their promoters [27, 28]; PIF4 can synergistically promote the transcription of genes required for hypocotyl elongation such as auxin response factor 6 (ARF6) [29]; In addition, PIF4 can integrate brassinosteroid (BR) and gibberellin (GA) signaling by interacting directly with their central components [29, 30].
In GA signaling, the GRAS TF family is one of the important members, which is unique to higher plants and discovered in recent years. The name of GRAS is derived from the three initially identified members, GA insensitive (GAI), repressor of GA1 (RGA) and scarecrow (SCR) [31]. In addition to play the role in GA signal transduction, studies have demonstrated that GRAS TFs play diverse roles in light signaling, root and meristem development, biotic and abiotic stress responses [32]. According to functional differences and structural characteristics, GRAS TFs were divided into several sub-families such as DELLA, SCR, SHR, SCL3, PAT, and LISCL [33]. Among which, the SCL proteins were considered as members of HT stress response signal pathway. For instance, the levels of cabbage GRAS TF BoSCL13 was increased with heat shock and confirmed as a unique candidate gene for discriminating heat shock tolerance in cabbage breeding [34]. The transcription of AtSCL13 (At4g17230) was increased by heat shock at an early time point after heat treatment [34]. However, the reports on GRAS to HT stress are few, future studies on the specific roles of GRAS TFs in heat tolerance and/or heat response is necessary.
Juglans regia is a nut tree cultivated worldwide and famous for its nutritious fruits [35]. As in all other plant species, walnut tree is sessile and cannot escape the unfavorable environmental conditions [36]. The growth, development, and production of J. regia are all affected by environmental stimulus, such as: high temperature, cold, salinity, and drought stress [37, 38]. However, studies on the stress response mechanism of walnut trees are currently lacking; achieving a better understanding of the mechanisms involved in abiotic stress response of J. regia is timely and essential [39]. In previous studies, we identified a few candidate genes from walnut tree relating to stress response, including some members of GRAS TF family, among which a SCL protein (Named as JrGRAS2) was detected to be induced by HT, and could improve the heat tolerance of yeast [40]. In this study, we further explore the function mechanism of JrGRAS2 response to HT stress, and found JrGRAS2 is a positive factor for plant HT tolerance associating with Dof TF and HSP protein.
Results
Identification and HT stress response of JrGRAS2 promoter
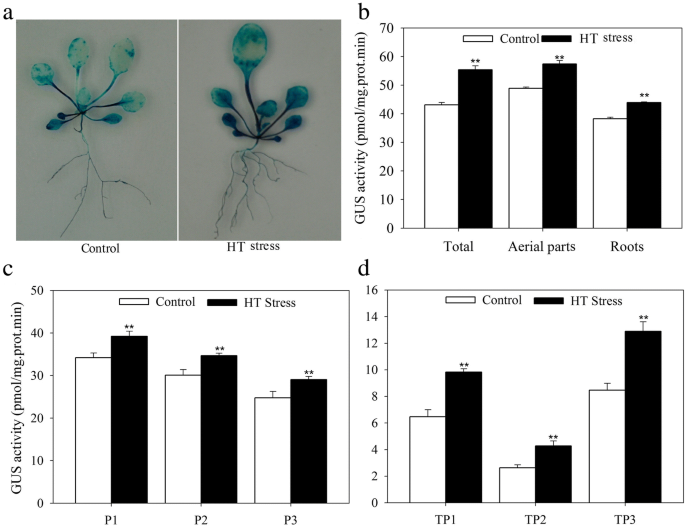
A 1500 bp promoter segment of JrGRAS2 was identified from the J. regia genome that was located in the 874811-870000 (NW_017443591.1) region of the walnut genome [41]. This promoter contains abundant cis-elements which are grouped into some classes, such as class of ‘ABA, Dehydration & salinity (osmotic) stress responsive’ includes the motifs of ABRE, MYB, DRE; class of ‘Miscellaneous’ covers the motifs of DOFCOREZM, RAV1AAT, SEF1MOTIF, POLASIG3, and so on (Additional file 1: Figure S1, Additional file 2: Table S1). The JrGRAS2 promoter fragment was inserted into pCAMBIA1301 vector and then transformed into Arabidopsis and walnut plants, which were further stained to reveal that the promoter caused GUS expression in the leaves and roots. Meanwhile, comparing to control, the GUS activities of JrGRAS2 promoter transgenic plants were significantly induced by HT stress (Fig. 1).
The expression activity of JrGRAS2 promoter under normal and HT stress. The significant differences between the HT stress and normal conditions were marked as two asterisk (**) (p<0.01). a, GUS staining of the JrGRAS2 promoter transformed Arabidopsis plants under normal and HT treatments. b, the GUS activities according to A. c, The GUS activities of three transgenic lines that transformed by JrGRAS2 promoter (P1, P2, P3) under normal and HT stress. d, The GUS activities of JrGRAS2 promoter transient expressed walnut leaves under normal and HT stress. TP1, TP2, TP3, three transient expression of JrGRAS2 promoter walnut lines
JrDof3 acts as the up-stream regulator of JrGRAS2 in HT stress response
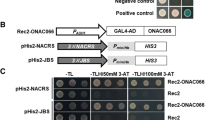
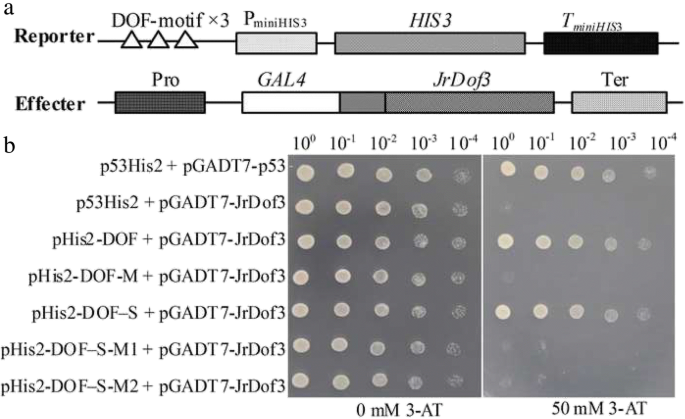
To screen the up-stream regulator of JrGRAS2, the cis-elements distributed in the promoter were analyzed and found that DOFCOREZM motif is the most abundant one (Additional file 2: Table S1). Considering the diverse function of Dof TFs in plant stress response, yeast one-hybrid assays were employed to study the interactions between Dof TFs and DOFCOREZM in the promoter. The results showed that JrDof3 could bind to DOFCOREZM motif, which was verified by the interactions between JrDof3 and the mutated DOFCOREZM motif (pHis2-DOF-M), or promoter segment including the DOFCOREZM motif (pHis2-DOF-S), or promoter segment containing the mutated DOFCOREZM motif (pHis2-DOF-M1), or promoter segment excluding the DOFCOREZM motif (pHis2-DOF-M2) on the solid synthetic drop-out medium (SD)/-Trp-Leu-His plus with 50 mM 3-amino-1, 2, 4-triazole (3-AT) (Fig. 2).
Identification of the up-steam regulator of JrGRAS2 using yeast one-hybrid assays. a, Diagram of the reporter and effecter vectors. Three tandem copies of the DOFCOREZM motif were inserted into the pHis2 vector and used as the reporter construct. The CDS of JrDof3 was cloned into pGADT7-Rec2 and used as the effecter construct. b, The effecter and reporter constructs were co-transformed into the yeast strain Y187. p53His2 + pGADT7-p53, positive control, pGADT7-Rec2 vector that encodes murine p53 fused with GAL4 AD; p53His2 + pGADT7-JrDof3, negative control, pHis2 reporter vector that contains the cis-acting DNA consensus sequence recognized by p53. pHis2-DOF + pGADT7-JrDof3, pHis2-DOF-M + pGADT7-JrDof3, pHis2-DOF-S + pGADT7-JrDof3, pHis2-DOF-M1 + pGADT7-JrDof3, pHis2-DOF-M2 + pGADT7-JrDof3 means the interaction DOFCOREZM motif, mutated DOFCOREZM motif, promoter segment including the DOFCOREZM motif, promoter segment containing the mutated DOFCOREZM motif, promoter segment excluding the DOFCOREZM motif with JrDof3, accordingly. The transformants spotted on TDO plates with 0 mM 3-AT were used as positive controls for transformants growth. Positive transformants were further confirmed by spotting serial dilutions (1/1, 1/10, 1/100, 1/1000, 1/10000) onto TDO plates with 50 mM 3-AT
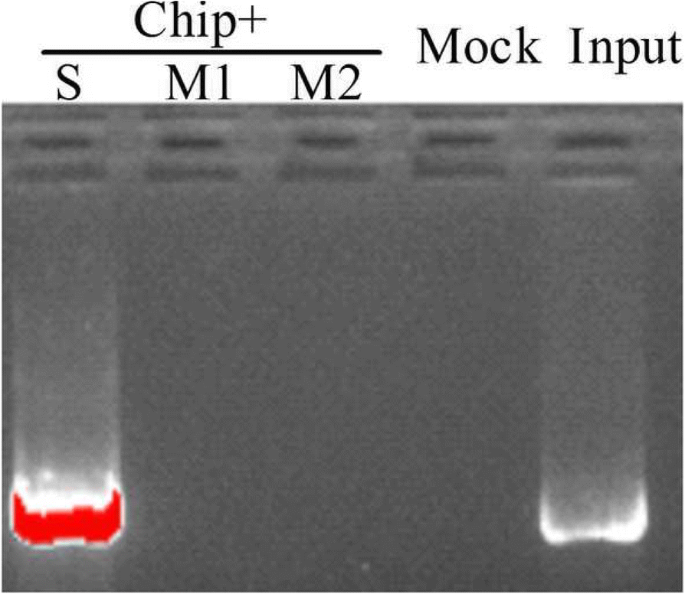
The special binding of JrDof3 to DOFCOREZM motif was confirmed by co-transformation of the reporter --DOFCOREZM motif (pCAM1301-DOF), or mutated DOFCOREZM motif (pCAM1301-DOF-M), or promoter segment including the DOFCOREZM motif (pCAM1301-DOF-S), or promoter segment containing the mutated DOFCOREZM motif (pCAM1301-DOF-M1), or promoter segment excluding the DOFCOREZM motif (pCAM1301-DOF-M2) with the effecter (pROKII-JrDof3) (Fig. 3a), which showed that the GUS activities of the leaves transformed by DOFCOREZM motif or promoter segment including the DOFCOREZM motif were similar to the positive controls and significant higher than those of the negative control and mutated reporters (Fig. 3b). Moreover, the promoter segment including the DOFCOREZM motif (S), or promoter segment containing the mutated DOFCOREZM motif (M1), or promoter segment excluding the DOFCOREZM motif (M2) was chosen for Chip-PCR to analyze the direct binding of JrDof3 to the JrGRAS2 promoter, and Chip+S displayed similar gel electrophoresis strip with input (positive control), while M1 and M2 showed similar results with mock (negative control) (Fig. 4), suggesting that JrDof3 could directly bind to JrGRAS2 promoter and function as an up-stream regulator of JrGRAS2.
Transient co-expression analysis of the interactions obtaining from the yeast one-hybrid assays. a, Diagram of the reporters and effecters. Triple tandem copies of the DOFCOREZM motif were fused with the 35S CaMV-46 minimal promoter and cloned into pCAMBIA1301 for driving the GUS gene which used as the reporter construct. The CDS of JrDof3 was cloned into pROKII under the control of the 35S promoter and used as the effecter construct. b, The GUS activities of the transformed tobacco seedlings were determined. pCAM1301 + pROKII-JrDof3, positive control, pCAMA1301 vector transformed with JrDof3; pROKII + pROKII-JrDof3, negative control, pROKII vector transformed with JrDof3. pCAM1301-DOF + pROKII-JrDof3, pCAM1301-DOF-M + pROKII-JrDof3, pCAM1301-DOF-S + pROKII-JrDof3, pCAM1301-DOF-M1 + pROKII-JrDof3, pCAM1301-DOF-M2 + pROKII-JrDof3, represented the transformation of reporter DOFCOREZM motif, mutated DOFCOREZM motif, promoter segment including the DOFCOREZM motif, promoter segment containing the mutated DOFCOREZM motif, promoter segment excluding the DOFCOREZM motif transformed with the effecter, accordingly. Two asterisk means significant differences (p<0.01) between the negative control and others
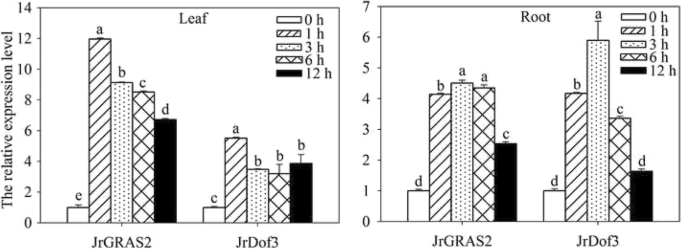
Furthermore, quantitative real-time PCR (qRT-PCR) analysis of the expression of JrDof3 and JrGRAS2 response to HT stress displayed that both of JrDof3 and JrGRAS2 were induced by HT with leaf and root tissue specificity (Fig. 5). In the leaves, the transcription levels of JrDof3 and JrGRAS2 were similar and declined from 1 to 6 h, the expression of JrGRAS2 was 6.73-~11.96-fold and the expression of JrDof3 was 3.20-~5.50-fold of control exposed to 1~ 12 h stress of HT, accordingly. In the roots, JrDof3 and JrGRAS2 showed same expression patterns that they were increased from 1 to 3 h then decreased from 3 to 12 h. The maximum transcription values of JrDof3 and JrGRAS2 were 5.90- and 4.50-fold of control, respectively (Fig. 5). The similar induction of JrDof3 and JrGRAS2 by HT indicated that JrDof3 act as an up-stream regulator of JrGRAS2 in HT stress response.
The expression of JrGRAS2 and JrDof3 in J. regia leaf and root tissue under HT stress. The relative expression level = transcription level under stress treatment/transcription level under control condition. Error bars were obtained from three replicates of qRT-PCR. The lowercase a-e mean the significant differences among the treatment time points (p<0.05)
Overexpression of JrGRAS2 up-regulated plant HT stress tolerance
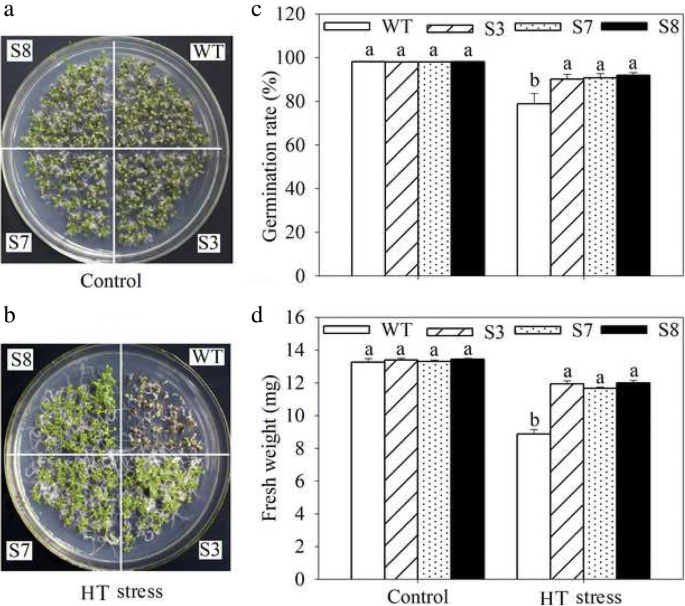
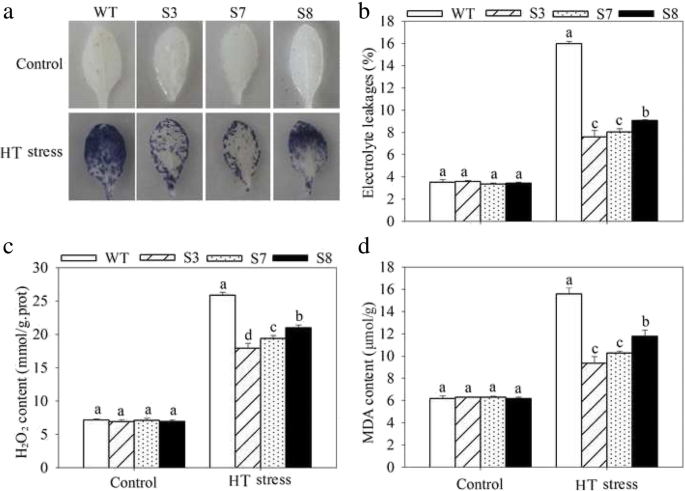
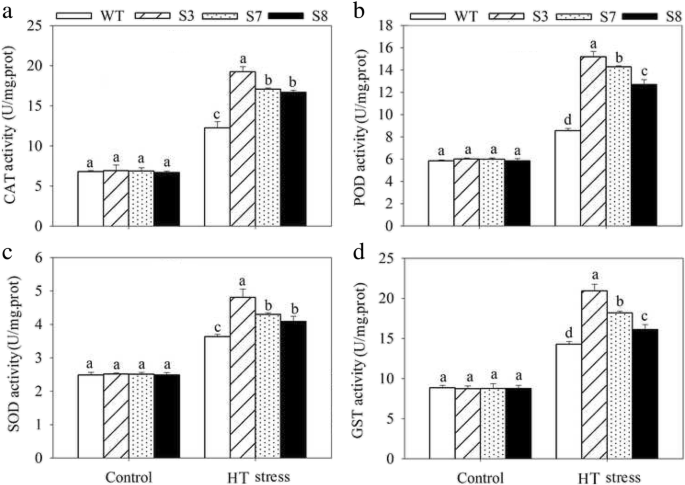
To complete characterize of the HT stress tolerance of JrGRAS2, it was overexpressed in Arabidopsis, and three lines with much higher expression level of JrGRAS2 (S3, S7, and S8) (Additional file 3: Figure S2) were selected for analysis. Germination assays showed that the germination rates of S3, S7, and S8 were 1.14-~1.16-fold of WT exposed to HT treatment. The fresh weight of the germinated seedlings of the transgenic lines was average 1.34-fold of that of WT under HT stress (Fig. 6). The 5-week-old plants of WT, S3, S7, and S8 grow under normal conditions were exposed to 37°C for one-week, then the ROS accumulation was analyzed. Evans blue staining displayed deeper coloring on the leaves of WT, S3, S7, and S8 under HT conditions than under control conditions. What’s more, the staining of WT was deeper than three transgenic lines under HT stress (Fig. 7a). The electrolyte leakage (EL) rates, H2O2 and 3, 3'-Diaminobenzidine (DAB) content were all showed similar trend as the Evans blue staining (Fig. 7). The EL rate of WT was 2.11-, 1.99-, 1.76-fold of that of S3, S7, S8 under HT stress, accordingly; the corresponding H2O2 content of WT was 1.44-, 1.33-, 1.23-fold of that of S3, S7, S8; the MDA content of S3, S7, and S8 was only about 67.10% of that of WT. The activities of the antioxidase including catalase (CAT), peroxidase (POD), superoxide dismutase (SOD), and glutathione-s-transferase (GST) were also tested and showed a reverse tendency to MDA and H2O2 contents (Fig. 8). Under control conditions, WT and three transgenic lines were showed similar CAT, POD, SOD and GST activities, which were all lower compared to themselves under HT stress. However, the activities of CAT, POD, SOD and GST of S3, S7, and S8 were average 1.44-, 1.64-, 1.21- and 1.29-fold higher than those of WT, accordingly (Fig. 8). All these results demonstrated the positive role of JrGRAS2 in plant HT stress tolerance. Moreover, transient expression method revealed that overexpression of JrGRAS2 in walnut also decreased the MDA content and EL rate while increased the SOD and POD activities under HT stress compared to NT and CK lines (Fig. 9), further confirmed the positive role of JrGRAS2 in walnut HT stress resistance.
The germination and growth of JrGRAS2 transgenic plants exposed to high temperature (HT) stress comparing with wild type (WT) Arabidopsis. S3, S7 and S8 were three JrGRAS2 transgenic lines. a-b, germination under normal and HT conditions that sown on the 1/2MS agar medium for 12 d. c, germination percentage of WT, S3, S7 and S8 according to a and b. d, average fresh weight of the germinated seedlings from a and b. The significant differences between WT and transgenic lines were marked with lowercase (p<0.05)
The ROS accumulation in transgenic and WT plants under HT stress. Five-week-old seedlings were treated with HT stress for one-week and used for analysis. a, Evans blue staining; b, EL rate; c, H2O2 content; d, MDA content. The significant differences (p<0.05) between transgenic lines and WT were indicated by lowercase
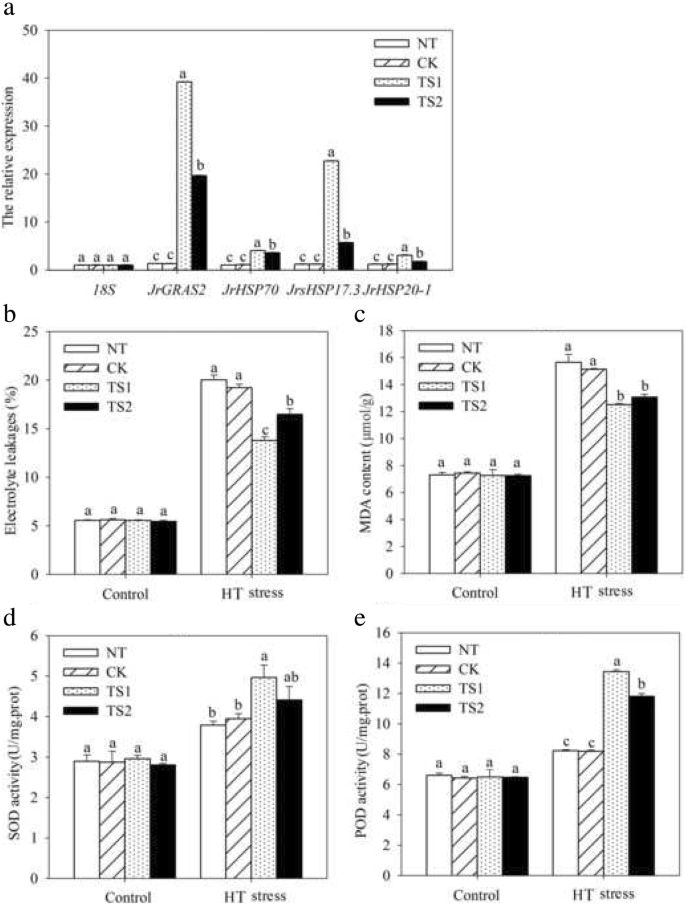
HT stress tolerance analysis by transient overexpression of JrGRAS2 in walnut. NT, the leaves not transformed; CK, the leaves transformed by empty vector; TS1 and TS2 two lines transformed by JrGRAS2. a, the expression level of JrGRAS2 and HSP genes in NT, CK, TS1 and TS2. b-e, the EL rate, MDA content, SOD and POD activities of NT, CK, TS1 and TS2 under normal and HT stress conditions. The significant differences (p<0.05) between transgenic lines and NT were indicated by lowercase
JrGRAS2 improves plant HT tolerance involving in HSP expression
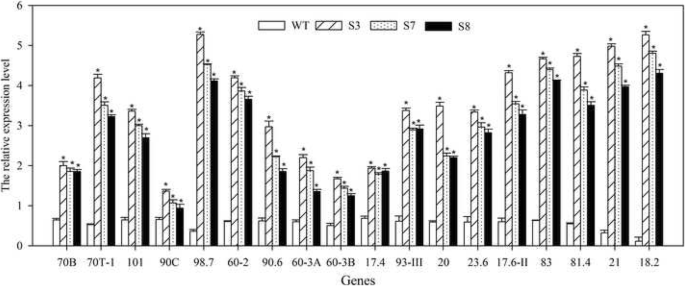
HSP family members are important HT stress response proteins. To understand whether the HT stress regulation of JrGRAS2 relating to the expression of HSP genes, total 18 HSP genes from different subfamilies were chosen, and their transcription were determined in WT, S3, S7, and S8 using qRT-PCR method. The results showed that all the chosen HSPs (AtHSP70B, AtHSP70T-1, AtHSP101, AtHsp90C, AtHSP98.7, AtHSP60-2, AtHsp90.6, AtHSP60-3A, AtHSP60-3B, AtHSP17.4, AtHSP93-III, AtHSP20, AtHSP23.6, AtHSP17.6II, AtHSP83, AtHsp81.4, AtHSP21, and AtHSP18.2) were significantly up-regulated in JrGRAS2 overexpression Arabidopsis plants, especial AtHSP98.7, AtHSP18.2, and AtHSP21, they were the top three induced genes (Fig. 10). Meanwhile, in transient overexpression lines TS1 and TS2, the expression of selected HSPs was enhanced (Fig. 9a), suggesting that HSP proteins participate in HT regulation of JrGRAS2.
The expression of HSPs in JrGRAS2 transgenic plants. The relative expression level is relative to the reference gene. Error bars were obtained from three replicates of qRT-PCR. The significant differences between WT and transgenic lines were marked as an asterisk (p<0.05). 70B, 70T-1, 101, 90C, 98.7, 60-2, 90.6, 60-3A, 60-3B, 17.4, 93-III, 20, 23.6, 17.6II, 83, 81.4, 21, and 18.2 presented the gene of AtHSP70B, AtHSP70T-1, AtHSP101, AtHsp90C, AtHSP98.7, AtHSP60-2, AtHsp90.6, AtHSP60-3A, AtHSP60-3B, AtHSP17.4, AtHSP93-III, AtHSP20, AtHSP23.6, AtHSP17.6II, AtHSP83, AtHsp81.4, AtHSP21, and AtHSP18.2, respectively
Discussion
Walnut is an important economic tree and affected by HT stress as other plant species, whose adaptation and response mechanisms on HT are currently insufficiently studied. Considering the abundant members of GRAS in walnut genome, the potential role of SCL subfamily GRAS TFs in HT stress response [34], and the HT stress response function and molecular mechanism of GRAS remains to be elucidated, in this study, a SCL GRAS was cloned from J. regia (named as JrGRAS2) and the up-stream regulation mechanism involving in HT stress response was characterized. The isolated up-stream promoter was heterologous transformed into Arabidopsis and homologous transient expressed in walnut, which displayed various GUS activities in different tissue parts and were enhanced by HT treatment (Fig. 1). Since the expression activity of the promoter is usually related to its function, for instance, the expression of Tamarix hispida V-ATPase c subunit (ThVHAc1) promoter was up-regulated by CdCl2 and ThVHAc1 was further confirmed as Cd tolerance gene [42]; the promoter expression activity of Cd-resistance gene JrVHAG1 (walnut V-ATPase G subunit) was induced by CdCl2 [43]; transformation of the banana aquaporin family gene MaTIP1;2 promoter into Arabidopsis to assess its function indicated that it responds to both drought and salt stress treatments [44]; it can be learn that JrGRAS2 is a potential HT stress tolerance gene.
The JrGRAS2 promoter is consist of diverse cis-elements those were classified into different subfamilies. In detail, twenty-six, nineteen, thirteen, eight, seven and five different elements were belong to the classes of ‘ABA, Dehydration and salinity (osmotic) stress responsive’, ‘Tissue/organelles specific expression’, ‘Light responsive’, ‘Phytohormone responsive’, ‘Pathogen, elicitor and wound responsive’ and ‘Heat and cold stress related’, respectively (Additional file 2: Table S1), according to the reports from Vivekanand Tiwari et al [45], suggesting the potential abundant regulation function of this promoter. Among all the elements, DOFCOREZM motif is the most one (Additional file 2: Table S1). Meanwhile, Dof TFs were reported as the effective factors in plant stress response, such as salt, drought, cold and heat [46, 47], therefore, DOFCOREZM motif was picked up for yeast one-hybrid assay to screen for the potential up-stream regulators of JrGRAS2, and the results showed that JrDof3 could specially bind to the DOFCOREZM motif (Fig. 2) that functioned as an up-stream regulator of JrGRAS2, which were further confirmed by co-transient expression and Chip-PCR analysis (Figs. 3, 4).
The target gene responds to stress are usually regulated by the upstream regulators binding or unbinding to the motifs in the promoter [43, 45]. Since the promoter expression activity of JrGRAS2 was enhanced by HT (Fig. 1) and JrGRAS2 is speculated as a potential HT tolerance gene, the transcription of JrDof3 and JrGRAS2 exposed to HT treatment was analyzed by qRT-PCR and both showed positive response to HT stress (Fig. 5). For induced transcription is a prediction for potential function in plant stress response, such as: Tamarix ThVHAc1 was up-regulated by CdCl2 then confirmed as a Cd tolerance gene [42]; walnut JrGSTTau1 was induced by cold and further characterized as a chilling tolerance factor [48]; tomato SlGRAS40 was upregulated by D-mannitol or NaCl, which was verified as drought and salt resistance TF [49], we can conclude that the HT stress response of JrGRAS2 is controlled by JrDof3 which may act as an up-stream regulator of JrGRAS2 to either control or act together with JrGRAS2 to participate in plant HT stress response.
To complete confirm the HT stress response function of JrGRAS2, it was overexpressed in Arabidopsis and three transgenic lines S3, S7 and S8 were selected for analysis. Interestingly, the expression of JrGRAS2 is effective to improve plant HT stress tolerance which was demonstrated by germination ability, growth efficiency, ROS accumulation and antioxidant activity (Fig. 6, 7 and 8). And the parallel results were obtained in transient overexpression walnut lines exposed to HT treatment (Fig. 9). The similar results here were observed in the plants overexpression of JrGSTTau1 that the transgenic plants accumulated less ROS, more biomass and higher activities of SOD, POD than WT plant under cold stress [50]; overexpression of the wheat F-Box protein gene TaFBA1 enhanced heat stress tolerance in transgenic tobacco owing to the growth inhibition was reduced and photosynthesis was increased as compared with those in WT plants [51]; Malus sieversii MsHsp16.9 is proved to be a protein chaperone that attenuate plant responses to severe stress via positively regulates antioxidant enzyme activity [52]. Therefore, we believe that JrGRAS2 is a vital HT stress responsive gene for walnut tree in temperature adaption regulation.
Plant TFs usually participate in stress response by regulating downstream related genes [50], for instance, unconventional splicing of wheat TabZIP60 could contribute to heat tolerance in transgenic plants by modulating the expression of ER stress-related genes [50]; Vitis amurensis GRAS TF VaPAT1 confers abiotic stress tolerance via up-regulate stress-related genes such as AtSIZ1, AtCBF1, AtATR1/MYB34, AtMYC2, AtCOR15A, AtRD29A and AtRD29B [53]; Tamarix eukaryotic translation initiation factor 1A (eIF1A) connected to the expression of stress-related genes, TOBLTP, GST, MnSOD, NtMPK9, poxN1 and CDPK15, in salt and drought stress response [54]. Since the HSP proteins were important HT stress-related members [6, 52, 55], the Arabidopsis HSP genes were identified from the TAIR database and analyzed in JrGRAS2 overexpression Arabidopsis plants, whose transcription were enhanced in S3, S7 and S8 compared to those in WT plants (Fig. 10); meanwhile, the HSPs from walnut tree were also up-regulated by JrGRAS2 in transient expression lines TS1 and TS2 (Fig. 9a); these findings were similar to the up-regulation of HSP genes in heat-tolerant csd1, csd2 and ccs plants while reduced in heat-sensitive transgenic plants expressing miR398-resistant forms of CSD1, CSD2 or CCS [56]; overexpression of HsfA1a had positive effects on the tolerance to diverse stressors by promoting inducible of Hsp expression [57]. Therefore, based on these findings, we defined that JrGRAS2 endows plant HT stress tolerance was partially by regulating the expression of HSP genes.
Conclusion
The JrGRAS2 promoter includes abundant stress related cis-elements and could be induced by HT stress, implying the positive role of JrGRAS2 in HT stress response. Yeast one-hybrid, transient expression, Chip-PCR and qRT-PCR assays confirmed that JrDof3 was a potential up-stream regulator of JrGRAS2 for HT stress resistance. Heterologous and homologous overexpression of JrGRAS2 in Arabidopsis and walnut revealed that JrGRAS2 is an effective TF for plant HT stress tolerance, which was involved in growth, ROS scavenging and antioxidant metabolism. These results indicated that JrGRAS2 is an important candidate gene for plant HT stress tolerance in molecular breeding, and it will offer new insights to reveal the adverse stimulus adaptation mechanism of walnut trees.
Methods
Plant materials and treatments
Two-year-old grafted ‘Xiangling’ (a genotype of J. regia widely planted in China, in this study, the seedlings were obtained from Walnut Experimental Station, Northwest A & F University) seedlings were grown in a greenhouse with the relative humidity 70±5%, temperature 22±2°C, illumination cycle 14/10 h) [48], and treated with 37°C for 0 (control), 1, 3, 6, and 12 h. The leaves and roots were harvested independently and frozen in liquid nitrogen, then stored at -80°C for total DNA and RNA isolation, which were used as the template of promoter cloning and qRT-PCR analysis, accordingly. The treatment at each time point was applied three times and each treatment contained 9 seedlings.
Identification and expression activity of the JrGRAS2 promoter
The J. regia leaves was used to extract the genomic DNA by CTAB (cetyltrimethylammonium bromide) method with after ground in liquid nitrogen and cleaned in a 0.14 M NaCl solution. The DNA quality was confirmed by electrophoresing in 1.0 % agarose gel and staining with ethidium bromide (EB). The JrGRAS2 promoter was identified from the walnut genome [41], and amplified by PCR reaction from the J. regia DNA. The PCR amplification parameters were set as follows: 30 s at 94°C followed by 35 cycles at 94°C for 30 s, 58°C for 30 s, 72°C for 90 s, and at 72°C for 7 min extending. The 20 μL PCR reaction mixture was generated according to manufacturer instructions (Takara Ex Taq®, Takara, Dalian, China). The cis-elements in the JrGRAS2 promoter were categorized into different groups after the analysis using the online programs PLACE [58] and PLANTCARE [59]. The expression activity of JrGRAS2 promoter (JrGRAS2-P) was functionally validated by a transgenic approach: The 35S promoter was replaced by the JrGRAS2 promoter to drive the expression of β-glucuronidase (GUS) gene in a pCAMBIA1301 vector to generate a recombinant construct pCAM-JrGRAS2-P, which was used for Arabidopsis plant transformation using Agrobacterium-mediated floral dip method [60]. Five-week-old transgenic seedlings were used to study the expression activity and level through GUS activity determination and staining [42, 61] under normal and HT stress (37°C). Meanwhile, the pCAM-JrGRAS2-P was transient transformed into walnut leaves using Agrobacterium-mediated method. The EHA105(pCAM-JrGRAS2-P) cells were grown to OD600=0.8~1.0, then diluted to OD600=0.05~0.1 with 1/2MS (Murashige and Skoog) liquid medium plus with 100 μM acetosyringone (AS). The three-month-old J. regia leaves were immersed in this solution and incubated for 8~10 h at 25°C with 40~50 rpm shaking, rinsed third with 1/2MS and incubated in fresh 1/2MS plus with 100 μM AS for 52~55 h. Fresh 1/2MS plus with 100 μM AS was added immediately to keep the OD600<1.0. Then the GUS activities of the transformed leaves treated by 25 and 37°C were determined. Every treatment was replicated three times and each replicate contained at least 15 seedlings.
Characterization of the potential upstream regulator of JrGRAS2
The core sequence of DOFCOREZM motif is "AAAG", and total 26 DOFCOREZM elements were found in the JrGRAS2 promoter (Additional file 2: Table S1, Additional file 1: Figure S1). Yeast one-hybrid assays were employed to identify the up-stream TFs capable of recognizing the DOFCOREZM motif. Three tandem copies of "AAAG" were cloned into pHis2 vector (pHis2-DOF) (Fig. 2a) [43]. The Dof TFs were identified from the J. regia transcriptome and then cloned into the pGADT7-Rec2 vector to generate a cDNA library for use in yeast one-hybrid assays [43]. The interactions between the DOFCOREZM motif and positive clones were further confirmed by examining the binding of mutated motif or promoter segments to the potential Dof TFs. I.e, the DOFCOREZM "AAAG" was mutated to "CCCA" and inserted into pHis2 (pHis2-DOF-M); JrGRAS2 promoter fragments including the DOFCOREZM motif (pHis2-DOF-S), containing the mutated DOFCOREZM motif (pHis2-DOF-M1), and excluding the DOFCOREZM motif (pHis2-DOF-M2) were all independently cloned into pHis2 [43]; Then the interactions between pHis2-DOF-M, or pHis2-DOF-S, or pHis2-DOF-M1, or pHis2-DOF-M2 and candidate positive clones were verified on the TDO plates with 50 mM 3-AT. The construct of p53His2 was set as a control in the yeast one-hybrid assays [42, 61].
Furthermore, above interactions were examined in tobacco seedlings by transient co-expression method [42, 43]. The DOFCOREZM motif (DOF), DOF-M, DOF-S, DOF-M1, DOF-M2 were each fused with a CaMV35S-46 minimal promoter to construct recombinants, which were further independently cloned into the pCAMBIA1301 vector to drive the GUS gene expression (reporters) (Fig. 3a) [43]. The ORF of the screened TF-- JrDof3 was cloned into pROKII vector under the control of a 35S promoter (pROKII-JrDof3) to generate the effecter (Fig. 3a) [43], which was transiently co-transformed with each of the reporters in tobacco leaves using Agrobacterium-mediated transformation method, then GUS activities of the co-transformed tobacco leaves were measured to evaluate the interactions [61, 62]. Every co-transformation was replicated three times and each replicate contained at least 20 leaves. Meanwhile, the direct binding of JrDof3 to the JrGRAS2 promoter was analyzed by Chip-PCR using the JrGRAS2 promoter segment containing the DOFCOREZM motif (S), or containing the mutated DOFCOREZM motif (M1), or excluding the DOFCOREZM motif (M2). The Chip assay was performed according to instructions of ChIP Assay Kit (Beyotime Biotechnology, Shanghai, China) (http://www.beyotime.com) and the published method [63]. All the used primers are listed in Additional file 4: Table S2.
RNA isolation and HT stress response analysis of JrGRAS2 and JrDof3
Total RNA of each sample was isolated using the CTAB method and reverse-transcribed into cDNA [64], which was used as the template of qRT-PCR after diluting to 1/10 of the original concentration with sterile water. The 18S rRNA was used as a reference gene [65]. The qRT-PCR was performed in a CFX96 TouchTM Real-Time PCR Detection System (Bio-Rad Laboratories, Redmond, WA) [48]. The 20 μL reaction mixture contained 10 μL of SYBR Green Real-time PCR Master Mix (CWBIO), 0.5 μM of each forward and reverse primer (Additional file 4: Table S2), 2 μL cDNA template (equivalent to 100 ng of total RNA). The amplification was applied using the following cycling parameters: one cycle of 94°C for 30 s, followed with 44 cycles at 94°C for 12 s, 60°C for 30 s, 72°C for 40 s, and at 81°C for 1 s plate reading. Three independent experiments were applied to ensure the reproducibility of qRT-PCR results. The relative expression levels were calculated based on the threshold cycle according to the 2-ΔΔCT method [66].
HT stress tolerance analysis of JrGRAS2 in transgenic Arabidopsis plants
The open reading frame (ORF) of JrGRAS2 was amplified using the primers of JrGRAS2-F/-R (Additional file 4: Table S2), which was further confirmed by sequencing. The JrGRAS2 amplified ORF fragment was digested by XbaI and KpnI and then cloned into pROKII vector under the control of a CaMV35S promoter to construct 35S::JrGRAS2. The EHA105 harboring recombinant 35S::JrGRAS2 was cultivated and used for transformation of Arabidopsis plants. Kanamycin-resistant transformed seedlings were detected by PCR, whose expression level was analyzed by qRT-PCR. Three transgenic lines with highest expression of JrGRAS2 (Line S3, S7, and S8) were chosen for further analysis.
Firstly, the seed germination ability and growth performance exposed to HT were analyzed. The seeds of WT, S3, S7, and S8 were sown on 1/2MS agar medium under 24°C or 37°C for 12 d. Then the germination rate and fresh weight were recorded, respectively. Next, the ROS accumulation was assessed. The 5-week-old plants of WT, S3, S7, and S8 grow under normal conditions were transferred to 37°C for another one-week, then the leaves were stained by Evans blue, and the corresponding EL rate, H2O2 and MDA contents were tested. Lastly, the activities of the antioxidases including CAT, POD, SOD, and GST were determined under the treatments as ROS accumulation assessment.
Transient expression analysis of JrGRAS2 to HT stress
To further confirm the HT stress response ability of JrGRAS2, the recombinant 35S::JrGRAS2 was transient transformed into walnut. The EHA105 (35S::JrGRAS2) and EHA105 (pROKII) (CK) cells were grown to OD600=0.8, then diluted to OD600=0.05 in 1/2MS liquid medium supplementing with 100 μM AS. Then the three-mouth-old J. regia leaves were transformed as the method using for pCAM-JrGRAS2-P transient transformation [67]. The non-transformed (NT) and CK lines were used as control. The expression level of JrGRAS2 was checked to confirm the transformation. The EL rate, MDA content, SOD activity and POD activity were tested. The transcription of walnut HSP genes (JrHSP70, JrsHSP17.3, JrHSP20-1) were analyzed to understand the regulation of JrGRAS2 whether relating to HSPs. All experiments were performed three times, each replicate contained 15 seedlings.
Transcription analysis of the HSPs regulating by JrGRAS2
The HSP proteins were identified from the Arabidopsis genome online database --TAIR (The Arabidopsis Information Resource, http://www.arabidopsis.org/). Total 18 HSP genes from different subfamilies were selected, they are AtHSP70B, AtHSP70T-1, AtHSP101, AtHsp90C, AtHSP98.7, AtHSP60-2, AtHsp90.6, AtHSP60-3A, AtHSP60-3B, AtHSP17.4, AtHSP93-III, AtHSP20, AtHSP23.6, AtHSP17.6II, AtHSP83, AtHsp81.4, AtHSP21, and AtHSP18.2. Their expression levels were confirmed in JrGRAS2 transgenic Arabidopsis plants using qRT-PCR. Actin2 was used as an internal control. All the primers and gene accession number were included in Additional file 5: Table S3.
Statistical analysis
All experiments were repeated three times, all of the data were analyzed using the Statistical Package for Social Science (SPSS) (SPSS, Chicago, Illinois), the sample variability is reported as standard deviation (S.D.). The differences between the transgenic and WT lines were evaluated using Tukey’s multiple comparison test with the significance level set at p<0.05.
Abbreviations
- ABA:
-
Abscisic acid
- CAT:
-
Catalase
- CTAB:
-
Cetyltrimethylammonium ammonium bromide
- EL:
-
Electrolyte leakage
- GST:
-
Glutathione-S-transferase
- HSP:
-
Heat shock protein
- HT:
-
High temperature
- MDA:
-
Malondialdehyde
- MS:
-
Murashige and skoog
- POD:
-
Peroxidase
- qRT-PCR:
-
Quantitative real-time PCR
- ROS:
-
Reactive oxygen species
- S.D.:
-
Standard deviation
- SD:
-
Synthetic drop-out medium
- SOD:
-
Superoxide dismutase
- TF:
-
Transcription factor
- 3-AT:
-
3-amino-1, 2, 4-triazole
References
Fahad S, Bajwa AA, Nazir U, Anjum SA, Farooq A, Zohaib A, Sadia S, Nasim W, Adkins S, Saud S, et al. Crop production under drought and heat stress: plant responses and management options. Front Plant Sci. 2017;8(1147). https://doi.org/10.3389/fpls.2017.01147. eCollection 2017.
Sampaio Filho IJ, Jardine KJ, de Oliveira RCA, Gimenez BO, Cobello LO, Piva LRO, Candido LA, Higuchi N, Chambers JQ. Below versus above ground plant sources of abscisic acid (ABA) at the heart of tropical forest response to warming. Int J Mol Sci. 2018;19(7). https://doi.org/10.3390/ijms19072023.
SMERTENKO A, DRÁBER P, VIKLICKÝ V, OPATRNÝ Z. Heat stress affects the organization of microtubules and cell division in Nicotiana tabacum cells. Plant Cell Environ. 1997;20(12):1534–42.
Friedrich T, Faivre L, Baurle I, Schubert D. Chromatin-based mechanisms of temperature memory in plants. Plant Cell Environ. 2018;19(10):13373.
Wang QL, Chen JH, He NY, Guo FQ. Metabolic reprogramming in chloroplasts under heat stress in plants. Int J Mol Sci. 2018;19(3). https://doi.org/10.3390/ijms19030849.
Hossain MA, Li ZG, Hoque TS, Burritt DJ, Fujita M, Munne-Bosch S. Heat or cold priming-induced cross-tolerance to abiotic stresses in plants: key regulators and possible mechanisms. Protoplasma. 2018;255(1):399–412.
Prasad PVV, Boote KJ, Allen LH. Adverse high temperature effects on pollen viability, seed-set, seed yield and harvest index of grain-sorghum [Sorghum bicolor (L.) Moench] are more severe at elevated carbon dioxide due to higher tissue temperatures. Agricultural and Forest Meteorology. 2006;139(3):237–51.
Fahad S, Hussain S, Saud S, Khan F, Hassan S. Amanullah, et al.: Exogenously applied plant growth regulators affect heat-stressed rice pollens. Journal of Agronomy and Crop Science. 2016;202(2):139–50.
Fahad S, Hussain S, Saud S, Hassan S, Tanveer M, Ihsan MZ, Shah AN, Ullah A, Nasrullah KF, et al. A combined application of biochar and phosphorus alleviates heat-induced adversities on physiological, agronomical and quality attributes of rice. Plant Physiology and Biochemistry. 2016;103:191–8.
Zhao C, Liu B, Piao S, Wang X, Lobell DB, Huang Y, Huang M, Yao Y, Bassu S, Ciais P, et al. Temperature increase reduces global yields of major crops in four independent estimates. Proc Natl Acad Sci U S A. 2017;114(35):9326–31.
Li B, Gao K, Ren H, Tang W. Molecular mechanisms governing plant responses to high temperatures. J Integr Plant Biol. 2018;20(10):12701.
Liu HC, Liao HT, Charng YY. The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant Cell Environ. 2011;34(5):738–51.
Guan Q, Yue X, Zeng H, Zhu J. The protein phosphatase RCF2 and its interacting partner NAC019 are critical for heat stress-responsive gene regulation and thermotolerance in Arabidopsis. Plant Cell. 2014;26(1):438–53.
Inda ME, Vandenbranden M, Fernandez A, de Mendoza D, Ruysschaert JM, Cybulski LE. A lipid-mediated conformational switch modulates the thermosensing activity of DesK. Proc Natl Acad Sci U S A. 2014;111(9):3579–84.
Choudhury FK, Rivero RM, Blumwald E, Mittler R. Reactive oxygen species, abiotic stress and stress combination. Plant J. 2017;90(5):856–67.
Yao Y, He RJ, Xie QL, Zhao XH, Deng XM, He JB, Song L, He J, Marchant A, Chen XY, et al. ETHYLENE RESPONSE FACTOR 74 (ERF74) plays an essential role in controlling a respiratory burst oxidase homolog D (RbohD)-dependent mechanism in response to different stresses in Arabidopsis. New Phytol. 2017;213(4):1667–81.
Niu L, Liao W. Hydrogen peroxide signaling in plant development and abiotic responses: crosstalk with nitric oxide and calcium. Front Plant Sci. 2016;7(230). https://doi.org/10.3389/fpls.2016.00230.
Hussain A, Mun BG, Imran QM, Lee SU, Adamu TA, Shahid M, Kim KM, Yun BW. Nitric oxide mediated transcriptome profiling reveals activation of multiple regulatory pathways in Arabidopsis thaliana. Front Plant Sci. 2016;7(975). https://doi.org/10.3389/fpls.2016.00975.
Hancock JT, Whiteman M. Hydrogen sulfide signaling: interactions with nitric oxide and reactive oxygen species. Ann N Y Acad Sci. 2016;1365(1):5–14.
Zhang SS, Yang H, Ding L, Song ZT, Ma H, Chang F, Liu JX. Tissue-specific transcriptomics reveals an important role of the unfolded protein response in maintaining fertility upon heat stress in Arabidopsis. Plant Cell. 2017;29(5):1007–23.
Nomoto Y, Kubozono S, Yamashino T, Nakamichi N, Mizuno T. Circadian clock- and PIF4-controlled plant growth: a coincidence mechanism directly integrates a hormone signaling network into the photoperiodic control of plant architectures in Arabidopsis thaliana. Plant Cell Physiol. 2012;53(11):1950–64.
Choi H, Oh E. PIF4 Integrates multiple environmental and hormonal signals for plant growth regulation in Arabidopsis. Mol Cells. 2016;39(8):587–93.
Lorenzo CD, Sanchez-Lamas M, Antonietti MS, Cerdan PD. Emerging hubs in plant light and temperature signaling. Photochem Photobiol. 2016;92(1):3–13.
Ma D, Li X, Guo Y, Chu J, Fang S, Yan C, Noel JP, Liu H. Cryptochrome 1 interacts with PIF4 to regulate high temperature-mediated hypocotyl elongation in response to blue light. Proc Natl Acad Sci U S A. 2016;113(1):224–9.
Hayes S, Sharma A, Fraser DP, Trevisan M, Cragg-Barber CK, Tavridou E, Fankhauser C, Jenkins GI, Franklin KA. UV-B Perceived by the UVR8 photoreceptor inhibits plant thermomorphogenesis. Curr Biol. 2017;27(1):120–7.
Gangappa SN, Kumar SV. DET1 and HY5 Control PIF4-Mediated Thermosensory Elongation Growth through Distinct Mechanisms. Cell Rep. 2017;18(2):344–51.
Franklin KA, Lee SH, Patel D, Kumar SV, Spartz AK, Gu C, Ye S, Yu P, Breen G, Cohen JD, et al. Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc Natl Acad Sci U S A. 2011;108(50):20231–5.
Sun J, Qi L, Li Y, Chu J, Li C. PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating Arabidopsis hypocotyl growth. PLoS Genet. 2012;8(3):29.
Oh E, Zhu JY, Bai MY, Arenhart RA, Sun Y, Wang ZY. Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. elife. 2014;27(3):03031.
Wei Z, Yuan T, Tarkowska D, Kim J, Nam HG, Novak O, He K, Gou X, Li J. Brassinosteroid biosynthesis is modulated via a transcription factor cascade of COG1, PIF4, and PIF5. Plant Physiol. 2017;174(2):1260–73.
Pysh LD, Wysocka-Diller JW, Camilleri C, Bouchez D, Benfey PN. The GRAS gene family in Arabidopsis: sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 1999;18(1):111–9.
Bolle C. The role of GRAS proteins in plant signal transduction and development. Planta. 2004;218(5):683–92.
Tian C, Wan P, Sun S, Li J, Chen M. Genome-wide analysis of the GRAS gene family in rice and Arabidopsis. Plant Mol Biol. 2004;54(4):519–32.
Park HJ, Jung WY, Lee SS, Song JH, Kwon SY, Kim H, Kim C, Ahn JC, Cho HS. Use of heat stress responsive gene expression levels for early selection of heat tolerant cabbage (Brassica oleracea L.). Int J Mol Sci. 2013;14(6):11871–94.
Abdallah IB, Tlili N, Martinez-Force E, Rubio AG, Perez-Camino MC, Albouchi A, Boukhchina S. Content of carotenoids, tocopherols, sterols, triterpenic and aliphatic alcohols, and volatile compounds in six walnuts (Juglans regia L.) varieties. Food Chem. 2015;173:972–8.
Hidalgo J, Casas RR, AM M. Environmental unpredictability and inbreeding depression select for mixed dispersal syndromes. BMC Evol Biol. 2016;16(71):016–0638.
Vahdati K, Lotfi N, Kholdebarin B, Hassani D, Amiri R, Mozaffari MR, Leslie C. Screening for drought-tolerant genotypes of persian walnuts (Juglans regia L.) during seed Germination. HortScience. 2009;44(7):1815–9.
Lotfi N, Vahdati K, Kholdebarin B, Ashrafi EN. Germination, mineral composition, and ion uptake in walnut under salinity conditions. HortScience. 2009;44(5):1352–7.
Xu Z, Zhao Y, Ge Y, Peng J, Dong M, Yang G. Characterization of a vacuolar H+-ATPase G subunit gene from Juglans regia (JrVHAG1) involved in mannitol-induced osmotic stress tolerance. Plant Cell Rep. 2017;36(3):407–18.
LiYuan-Su X-L, Shuwen-Chen T-Z, Fangfang-Zhang G-Y. The expression and function analysis of walnut JrGRAS2 gene under heat stress. Bull Bot Res. 2018;38(01):125–31.
Martinez-Garcia PJ, Crepeau MW, Puiu D, Gonzalez-Ibeas D, Whalen J, Stevens KA, Paul R, Butterfield TS, Britton MT, Reagan RL, et al. The walnut (Juglans regia) genome sequence reveals diversity in genes coding for the biosynthesis of non-structural polyphenols. Plant J. 2016;87(5):507–32.
Yang G, Wang C, Wang Y, Guo Y, Zhao Y, Yang C, Gao C. Overexpression of ThVHAc1 and its potential upstream regulator, ThWRKY7, improved plant tolerance of Cadmium stress. Sci Rep. 2016;6(18752). https://doi.org/10.1038/srep18752.
Xu Z, Ge Y, Zhang W, Zhao Y, Yang G. The walnut JrVHAG1 gene is involved in cadmium stress response through ABA-signal pathway and MYB transcription regulation. BMC Plant Biol. 2018;18(1):018–1231.
Song S, Xu Y, Huang D, Miao H, Liu J, Jia C, Hu W, Valarezo AV, Xu B, Jin Z. Identification of a novel promoter from banana aquaporin family gene (MaTIP1;2) which responses to drought and salt-stress in transgenic Arabidopsis thaliana. Plant Physiol Biochem. 2018;128:163–9.
Tiwari V, Patel MK, Chaturvedi AK, Mishra A, Jha B. Functional characterization of the tau class glutathione-S-transferases gene (SbGSTU) promoter of salicornia brachiata under salinity and osmotic stress. PLoS One. 2016;11(2). https://doi.org/10.1371/journal.pone.0148494.
Zang D, Wang L, Zhang Y, Zhao H, Wang Y. ThDof1.4 and ThZFP1 constitute a transcriptional regulatory cascade involved in salt or osmotic stress in Tamarix hispida. Plant Mol Biol. 2017;94(4-5):495–507.
Ma J, Li MY, Wang F, Tang J, Xiong AS. Genome-wide analysis of Dof family transcription factors and their responses to abiotic stresses in Chinese cabbage. BMC Genomics. 2015;16(33):015–1242.
Yang G, Xu Z, Peng S, Sun Y, Jia C, Zhai M. In planta characterization of a tau class glutathione S-transferase gene from Juglans regia (JrGSTTau1) involved in chilling tolerance. Plant Cell Rep. 2016;35(3):681–92.
Liu Y, Huang W, Xian Z, Hu N, Lin D, Ren H, Chen J, Su D, Li Z. Overexpression of SlGRAS40 in Tomato Enhances Tolerance to Abiotic Stresses and Influences Auxin and Gibberellin Signaling. Front Plant Sci. 2017;8(1659). https://doi.org/10.3389/fpls.2017.
Geng X, Zang X, Li H, Liu Z, Zhao A, Liu J, Peng H, Yao Y, Hu Z, Ni Z, et al. Unconventional splicing of wheat TabZIP60 confers heat tolerance in transgenic Arabidopsis. Plant Sci. 2018;274:252–60.
Li Q, Wang W, Zhang G, Liu Y, Wang Y. Wheat F-Box Protein Gene TaFBA1 Is Involved in Plant Tolerance to Heat Stress. Front Plant Sci. 2018;9(521). https://doi.org/10.3389/fpls.2018.00521.
Yang M, Zhang Y, Zhang H, Wang H, Wei T, Che S, Zhang L, Hu B, Long H, Song W, et al. Identification of MsHsp20 gene family in Malus sieversii and functional characterization of MsHsp16.9 in heat tolerance. Front Plant Sci. 2017;8(1761). https://doi.org/10.3389/fpls.2017.01761.
Yuan Y, Fang L, Karungo SK, Zhang L, Gao Y, Li S, Xin H. Overexpression of VaPAT1, a GRAS transcription factor from Vitis amurensis, confers abiotic stress tolerance in Arabidopsis. Plant Cell Rep. 2016;35(3):655–66.
Yang G, Yu L, Wang Y, Wang C, Gao C. The translation initiation factor 1A (TheIF1A) from Tamarix hispida Is regulated by a Dof transcription factor and increased abiotic stress tolerance. Front Plant Sci. 2017;8(513). https://doi.org/10.3389/fpls.2017.00513.
Vishwakarma H, Junaid A, Manjhi J, Singh GP, Gaikwad K, Padaria JC. Heat stress transcripts, differential expression, and profiling of heat stress tolerant gene TaHsp90 in Indian wheat (Triticum aestivum L.) cv C306. PLoS One. 2018;13(6). https://doi.org/10.1371/journal.pone.0198293
Guan Q, Lu X, Zeng H, Zhang Y, Zhu J. Heat stress induction of miR398 triggers a regulatory loop that is critical for thermotolerance in Arabidopsis. Plant J. 2013;74(5):840–51.
Qian J, Chen J, Liu YF, Yang LL, Li WP, Zhang LM. Overexpression of Arabidopsis HsfA1a enhances diverse stress tolerance by promoting stress-induced Hsp expression. Genet Mol Res. 2014;13(1):1233–43.
Higo K, Ugawa Y, Iwamoto M, Higo H. PLACE: a database of plant cis-acting regulatory DNA elements. Nucleic Acids Res. 1998;26(1):358–9.
Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouze P, Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30(1):325–7.
Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation ofArabidopsis thaliana. Plant J. 1998;16(6):735–43.
Zheng L, Liu G, Meng X, Liu Y, Ji X, Li Y, Nie X, Wang Y. A WRKY gene from Tamarix hispida, ThWRKY4, mediates abiotic stress responses by modulating reactive oxygen species and expression of stress-responsive genes. Plant Mol Biol. 2013;82(4-5):303–20.
Jefferson R. The GUS reporter gene system. Nature. 1989;342(6251):837.
Guo H, Wang L, Yang C, Zhang Y, Zhang C, Wang C. Identification of novel cis-elements bound by BplMYB46 involved in abiotic stress responses and secondary wall deposition. J Integr Plant Biol. 2018;60(10):1000–14.
Yang G, Wang Y, Xia D, Gao C, Wang C, Yang C. Overexpression of a GST gene (ThGSTZ1) from Tamarix hispida improves drought and salinity tolerance by enhancing the ability to scavenge reactive oxygen species. Plant Cell, Tissue and Organ Culture (PCTOC). 2014;117(1):99–112.
Xu F, Deng G, Cheng SY, Zhang WW, Huang XH, Li LL, Cheng H, Rong XF, Li JB. Molecular cloning, characterization and expression of the phenylalanine ammonia-lyase gene from Juglans regia. Molecules. 2012;17(7):7810–23.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–8.
Zhai M, Sun Y, Jia C, Peng S, Liu Z, Yang G. Over-expression of JrsHSP17.3 gene from Juglans regia confer the tolerance to abnormal temperature and NaCl stresses. J Plant Biol. 2016;59(5):549–58.
Acknowledgements
Not applicable
Funding
The current study was supported by Central University Basic Research Funds Project of China (2452016057, 2452015171), Inaugurating Program of Northwest A & F University for Doctoral Staff (2452015295), Natural Science Basic Research Project of Shaanxi Province (2018JQ3066), National Natural Science Foundation of China (31700332, 31800510), Special Financial Grant from the China Postdoctoral Science Foundation (2017T100782). The funding agency was not involved in the design of the study, collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.
Author information
Authors and Affiliations
Contributions
GY wrote the paper, analyzed all the data and fund part for the experiments. XG completed the Arabidopsis transformation, promoter identification and Chip-PCR. KM did the yeast one-hybrid assay, transient co-expression analysis and tested the physiological index. DL isolated the DNA, RNA and carried out the qRT-PCR experiments. CJ and MZ analysis and interpretation part of the data. MZ and ZX fund partial for the current study. GY and ZX designed the study, and ZX inspected and re-analyzed all the data. All authors have read and approved the final version.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
All the authors declare that they have no conflict of interest.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Figure S1. The JrGRAS2 promoter sequence and main cis-elements existing in the promoter that predicted by PLACE and PLANTCARE. (JPG 2941 kb)
Additional file 2:
Table S1. All the motifs existed in the promoter that predicted and classified according to the online programs of PLACE and PLANTCARE. (PDF 89 kb)
Additional file 3:
Figure S2. The expression level of JrGRAS2 in transgenic Arabidopsis. 1-12, twelves transgenic lines. (JPG 126 kb)
Additional file 4:
Table S2. The primers used for pHIS2, pGADT7-Rec2, pCAMBIA1301, and pROKII recombinant vector construction, and qRT-PCR analysis of JrGRAS2, JrDof3 as well as 18S rRNA gene. (PDF 138 kb)
Additional file 5:
Table S3. The primers used for qRT-PCR analysis of HSP genes. (PDF 133 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Yang, G., Gao, X., Ma, K. et al. The walnut transcription factor JrGRAS2 contributes to high temperature stress tolerance involving in Dof transcriptional regulation and HSP protein expression. BMC Plant Biol 18, 367 (2018). https://doi.org/10.1186/s12870-018-1568-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-018-1568-y