Abstract
Background
Coagulase-negative Staphylococcus species are an emerging cause of intramammary infection, posing a significant economic and public health threat. The aim of this study was to assess the occurrence of coagulase-negative Staphylococcus species in bovine milk and dairy farms in Northwestern Ethiopia and to provide information about their antibiotic susceptibility and virulence gene profiles.
Methods
The cross-sectional study was conducted from February to August 2022. Coagulase-negative Staphylococcus species were isolated from 290 milk samples. Species isolation and identification were performed by plate culturing and biochemical tests and the antimicrobial susceptibility pattern of each isolate was determined by the Kirby-Bauer disc diffusion test. The single-plex PCR was used to detect the presence of virulent genes. The STATA software version 16 was used for data analysis. The prevalence, proportion of antimicrobial resistance and the number of virulent genes detected from coagulase-negative Staphylococcus species were analyzed using descriptive statistics.
Results
Coagulase-negative Staphylococcus species were isolated in 28.6%, (95% CI: 23.5–34.2) of the samples. Of these, the S. epidermidis, S. sciuri, S. warneri, S. haemolyticus, S. simulans, S. chromogens, S. cohnii, and S. captis species were isolated at the rates of 11, 5.2, 3.4, 3.1, 3.1, 1, 1, and 0.7% respectively. All the isolates showed a high percentage (100%) of resistance to Amoxicillin, Ampicillin, and Cefotetan and 37.5% of resistance to Oxacillin. The majority (54.2%) of coagulase-negative isolates also showed multidrug resistance. Coagulase-negative Staphylococcus species carried the icaD, pvl, mecA, hlb, sec, and hla virulent genes at the rates of 26.5%, 22.1%, 21.7%, 9.6%, 9.6% and 8.4% respectively.
Conclusion
The present study revealed that the majority of the isolates (54.2%) were found multidrug-resistant and carriage of one or more virulent and enterotoxin genes responsible for intramammary and food poisoning infections. Thus, urgent disease control and prevention measures are warranted to reduce the deleterious impact of coagulase-negative species. To the best of our knowledge, this is the first study in Ethiopia to detect coagulase-negative Staphylococcus species with their associated virulent and food poisoning genes from bovine milk.
Similar content being viewed by others
Background
Staphylococcus species are the major pathogen of lactating dairy cows causing a significant reduction in milk yield and posing risk to the public health [27, 45]. It can be classified as coagulase-positive and coagulase-negative Staphylococcus species based on their ability to clot the rabbit plasma, a critical diagnostic step in clinical microbiology [44].
Coagulase-negative Staphylococcus (CoNS) species were previously considered less pathogenic because they were only reported in sub-clinical mastitis cases and thus received less attention. However, a growing body of evidence suggests that CoNS species have been isolated from clinical mastitis infections, and they are regarded as emerging pathogens of bovine intramammary infections [13, 46, 59]. The most common CoNS species reported from intramammary infections are S. chromogens, S. epidermidis, S. haemolyticus, S. sciuri, S. simulans, S. cohnii, S. xylosus, S. warneri, S. captis and S. equorum [8, 35, 47].
Several virulence factors, such as the enterotoxin (SE) genes (sea to seQ), the toxic shock syndrome toxin-1 gene (tsst-1), clumping factor (clfA to clfD), intracellular adhesion gene (icaA and icaD), hemolysin toxin gene (hla, hlb, hld, Y-hlg) the panton valentine leukocidin (pvl) gene and genes encoding drug resistance (mecA and mecC) were originally identified and characterized in Staphylococcus aureus; but now these virulent genes are detected in CoNS species. These virulent factors are responsible for the colonization and pathogenesis of the mammary gland as well as food poisoning [24, 25, 30, 38].
A lot of reports were available on S. aureus as a cause of mastitis and food intoxication; however, information on virulence factors of CoNS species for intramammary infection and food poisoning was scarce globally [37]. Furthermore, the level of occurrence and antibiotic resistance patterns of these bacterial species are not well studied in Ethiopia. In fact, no published reports are available on the virulent genes of CoNS species from bovine milk. Therefore, the study aimed to determine the antimicrobial susceptibility and virulent gene profile of coagulase-negative Staphylococcus species from bovine milk samples.
Materials and methods
Study area
The study was carried out in Gondar City (Fig. 1); which is located 728 km northwest of Addis Ababa, at 12° North latitude, 37° 28` East longitude, with altitudes ranging from 1802 to 2200 m above sea level. According to the Gondar City livestock and fishery resource office, 2022 report during commencement of the study, the city consists of 42,929 cattle, 16,090 sheep, 7,614 goats, 6,461 donkeys, 516 horses, 35 mules, 13,468 bee colonies, and 126,061 poultries. The dairy cows are managed in both an intensive and semi-intensive systems [9].
Study design, and sampling methods
A cross-sectional study was designed to collect milk samples from February to August 2022 from Zebu and cross-Holstein Friesian breed lactating cows. The sample size was determined based on Slovon’s formula [3].
n = N/(1 + N*e2)
Where, n = is the number of lactating cows selected for sample collection, N = the total number of lactating cows found in the study area, and e = the margin of error. In Gondar town, around 1060 lactating cows were found at the farm level and a 0.05 margin of error (95% CI) was taken for the study. Therefore, based on the above formula the milk samples were collected from 290 lactating cows. The dairy farms were chosen purposively based on the herd size; farms having three and more lactating cows were considered for sample collection and the milk samples were collected from all lactating cows in selected dairy farms.
The milk samples were collected following the standard procedures outlined in the National Mastitis Council [43]. The animals selected for milk sample collection were properly examined for the presence of cardinal signs of inflammation through visual inspection and palpation of the udder and visual inspection of milk for the presence of flakes, clots, and discoloration. A composite milk of about 10 ml was collected from all quarters of lactating cows using a universal bottle after discarding the first three milk streams and transported to the University of Gondar, Veterinary Microbiology Laboratory for bacteriological analysis.
Isolation and identification protocol
The milk samples taken from lactating cows were cultured directly on mannitol salt agar (MSA) (Alpha Chemica, India) and incubated aerobically at 37 °C for 24–48 h. The cultures that produced pink and white colonies on mannitol salt agar were considered either Staphylococcus species or Micrococcus species. Thus, the oxidation fermentation test (OF) test was performed according to procedures outlined by Varghese and Joy [58] to differentiate the Staphylococcus species from the Micrococcus species.
Those Staphylococcal colonies that grew on the mannitol salt agar; were positive for catalase tests, fermentative on the OF media, had characteristic cluster cocci shape on Gram stain were further differentiated into coagulase-positive and negative Staphylococcus species based on their ability to clot the rabbit plasma. Thus, the tube coagulase test was performed by mixing 4–5 pure staphylococcal colonies grown on tryptone soya agar (HiMedia, Laboratories, Ltd., India) with 0.5 mL of the rabbit plasma (National Veterinary Institute, Debre Zeit, Ethiopia) according to Fernandes Queiroga Moraes et al. [22].
A Pure colony of the CoNS species was sub-cultured on blood agar (HiMedia Laboratories Pvt. Ltd., India) containing 7% sheep blood to observe the hemolysis pattern and colony morphology. The identification of CoNS was performed based on the colony characteristics on mannitol salt agar, hemolysis characteristics on blood agar, the ability to utilize urea and maltose and sucrose sugar fermentation tests. The oxidase test was also performed by rubbing four to five pure colonies grown on nutrient agar with a cotton swab soaked with the oxidase reagent (Table 1).
Phenotypically isolated and purified CoNS species isolates were preserved in tryptone soya (HiMedia Laboratories Pvt. Ltd, India) broth mixed with 25% glycerol at -20◦C until molecular analysis started.
Antimicrobial susceptibility pattern of coagulase-negative Staphylococcus species
The antimicrobial susceptibility pattern of CoNS species isolates was examined using the Kirby Bauer disc diffusion methods, following the guidelines of the clinical laboratory standard institute [12]. The standardized bacterial inoculum was prepared by dissolving the CoNS species colony in 0.85% normal saline until the turbidity of the suspension was equivalent to 0.5 Mac Farland standards. A sterile cotton swab was soaked to the bacterial suspension and streaked onto the Mueller–Hinton Agar (MHA) plate (HiMedia Laboratories, Pvt. Ltd., India). The plates were left dry for 5 min before disc placement.
The antimicrobial discs (all from Oxoid, UK), with the following disc concentration; Gentamicin (GEN,10 µg), Sulphamethoxazole-trimethoprim (SXT, 2 µg), Erythromycin (E, 10 µg), Ampicillin (AMP, 10 µg), Amoxicillin (AMX, 10 µg), Penicillin G (P10, 1U), Cefotetan (CTT, 30 µg), Oxacillin (OXC, 1 µg), Tetracycline (TE, 30 µg) were used for antimicrobial susceptibility tests. The diameter of the zone of inhibition produced by each antimicrobial disc was compared with the clinical laboratory standard institute [12] standards to determine whether the CoNS species isolates were susceptible or resistant to particular antimicrobial discs. Coagulase-negative Staphylococcus species isolates were considered multidrug resistant (MDRS) when the isolates showed resistance to three or more antimicrobial classes [16]. Phenotypic methicillin resistance of CoNS species isolates was checked using Oxacillin discs.
Virulent gene detection
Bacterial DNA extraction
A pure colony of CoNS species isolates were cultured for 24 h in brain heart infusion broth (BHI) (HiMedia Laboratories Pvt. Ltd, India). One ml of the broth was transferred to 1.5 ml of the Eppendorf tube and spun at 10,000 rpm for 10 min. After discarding the supernatant, the bacterial pellets were washed with 1 ml of phosphate buffer saline (PBS) by spinning at 10,000 rpm for 10 min and discarding the supernatant completely. The bacterial DNA extraction was performed using the EZ-10 spin column genomic DNA minipreps kit (Bio Basic Inc, Canada) according to the manufacturer’s instructions. The quality of extracted DNA was checked with NanoDrop 2000 spectrophotometer.
Polymerase chain reaction assay
Single-plex polymerase chain reaction assay was performed for the detection of 14 virulent genes including; the six enterotoxins, SE genes, (sea, seb, sec, sed, see, seh), the toxic shock syndrome toxin-1 (tsst-1), intercellular adhesion gene D (icaD), hemolysin toxin (hla and hlb, Y-hlg), the panton valentine leukocidin (pvl) gene, as well as genes encoding drug-resistant (mecA and mecC) using gene-specific oligonucleotide primers (Table 2).
The PCR reaction mixture of 20 µl was prepared by adding a similar amount of 2 µl of 10x reaction buffer (Solis BioDyne, Estonia), 0.2 µl of dNTPs, 0.2 µl of FIREPol DNA polymerase (SolisBioDyne, Estonia) for all PCR reaction; but the amount of MgCl2, primers, bacterial genomic DNA and nuclease-free water added to each PCR reaction was vary (Table 3).
Prima 96 thermocycler (HiMedia, Laboratories Pvt. Ltd., India) was used for the amplification of virulent genes in CoNS species isolates with an initial denaturation at 95 oC for 5 min, denaturation at 95 oC for 30 s, extension at 72 oC for 1 min and final extension at 72 oC for 8 min. All virulent gene amplification was performed for 35 PCR cycles except pvl gene amplification which was performed for 37 PCR cycles.
The electrophoresis of each PCR amplified product was performed using 1.5% agarose gel (HiMedia Laboratories Pvt. Ltd., India) and stained with ethidium bromide. A molecular ladder of 100 bp was used to compare the size of the amplicon product. The gel product was examined by the gel documentation system (UVITEC, Cambridge, UK) under ultraviolet (UV) illumination.
Data management and analysis
The data was recorded in a Microsoft Excel spreadsheet. The STATA version 16 software was used to analyze the data. Descriptive statistics were used to analyze the isolation rate, proportion of antimicrobial susceptibility and virulent gene profile of the CoNS species isolates.
Results
Species isolation and identification
The milk samples were collected from 290 lactating cows. The overall isolation rate of CoNS species in the study area was 28.62%, 83/290 (95% CI: 23.5–34.2). The isolation rate of each CoNS species from the milk sample described in (Table 4).
Antibiogram profiles of the coagulase-negative Staphylococcus species
The phenotypic methicillin resistance capacity of each CoNS species was tested using Oxacillin antibiotic discs; thus, 37.3% of CoNS species showed phenotypic methicillin resistance to Oxacillin whereas 21.7% of CoNS isolates showed genotypic methicillin resistance.
The CoNS species isolates showed 51.8, 27.7, 49.4, 37.3, 43.4, and 39.8% resistance to Penicillin G, Gentamycin, Erythromycin, Oxacillin, sulphamethoxazole-trimethoprim, and Tetracycline discs respectively. The CoNS species isolates also showed 100% resistance to Ampicillin, Amoxicillin, and Cefotetan antimicrobials discs (Table 5).
The present study also revealed, 54% of CoNS species isolates showed multidrug resistance. All isolates of the S. simulans, S. chromogens, S. cohnii, S. captis species showed multidrug resistance towards three or more antimicrobial classes (Fig. 2).
Virulent Genes of coagulase-negative Staphylococcus species
Among the 14 virulent genes tested in CoNS species isolates, six virulent genes; namely the icaD, pvl, mecA, hlb, sec and hla genes were detected at the rates of 26.5, 22.1, 21.7, 9.6, 9.6 and 8.4% respectively from 83 CoNS species isolates (Fig. 3).
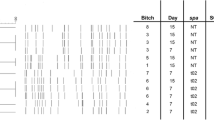
Lane M: Molecular marker; PC: positive control; NC: negative control. A and C, lanes 1 to 4 are positive tests for sec and hla genes respectively. B and F, lanes 1–5 indicate positive tests for hlb and mecA genes respectively. E, Lanes 1–6 are positive test results for pvl gene. D, lanes 2, 4,5,8,9 are positive and lanes 1,3,6,7 are negative for the icaD gene
All CoNS species isolates except the S. captis were found to harbor one or more virulent genes. Genes coding for food poisoning (sec) were detected from S. haemolyticus, S. sciuri and S. warneri species isolates. The icaD genes were the most frequently detected followed by mecA and the pvl genes. The present study also revealed, except for S. captis the intracellular adhesion (icaD) genes were detected in all CoNS species isolates. The S. epidermidis isolates were carried only the icaD and mecA genes; whereas the S. simulans and S. cohnii species were found harboring five virulent genes (Table 6).
Discussion
Coagulase-negative staphylococcal infection is an emerging disease of lactating dairy cows and causes a significant reduction in milk yield and threatens public health [17, 30]. Virulent gene detection from coagulase-negative Staphylococcus species is the first report in Ethiopia.
The present study revealed that CoNS species were isolated at a rate of 28.6%, (83/290), 95% CI: 23.5–34.2). The result was lower than the report of Enquebaher et al. [19] in the Tigray region, Zeryehun and Abera [61] in selected districts of Eastern Harrarghe, Balemi et al. [6] in two pastoral districts Southern Ethiopia who reported 51.6%, 34.2% and 39% prevalence respectively, and higher than the reports of Gizaw et al. [28], Abunna et al. [1], Fesseha et al. [23] who reported 9.6%, 15% and 12.5% prevalence of CoNS species in Oromia, in and around Addis Ababa and in Modjo Town and Suburbs, Central Oromia respectively. A higher isolation rate of CoNS species from milk might be associated with the CoNS being prominent biofilm producers [29, 36] which enhances the bacteria to resist antibiotics and evade immune system mounting. In addition to this, CoNS are mainly recovered from subclinical mastitis, which helps the pathogen remain undetected and not treated with antimicrobials.
The current study revealed all CoNS species isolates showed 100% (95% CI: 95.6–99.6) resistance to Amoxicillin, Ampicillin, and Cefotetan antimicrobials. The result was relatively higher as compared to the report of Phophi et al. [46] who reported 90% and 89% of CoNS species resistance to ampicillin and penicillin antibiotics. In the current study, 54.2%,45/83( 95% CI: 42.9–65.2) of the CoNS species showed MDRS. The finding was relatively higher compared with the report of Phophi et al. [46], Mahato et al. [37] who reported 51% and 45.16% MDRS and lower than the report of Gizaw et al. [28] Central Oromia who reported 87.5% prevalence in CoNS species.
A large number (37.5%, 95% CI; 26.97–48.6) of coagulase-negative Staphylococcus species isolates were shown phenotypic methicillin resistance as compared to its genotypic (21.7%, CI: 13.4–32.1) methicillin resistance. The result was different from the report of Mahato et al. [37] in India reported 91.5% of CoNS isolates harboring the mecA genes; however, only 85.5% of the isolates were resistant to Oxacillin. The difference in prevalence of coagulase-negative Staphylococcus species developing multidrug resistance in the current study and other scholars’ reports might be associated with variations in disease epidemiology and the spread of drug-resistant CoNS species. Prolonged and inappropriate use of antimicrobials, creates selective pressure by depleting innocuous bacterial species and favors the flourishing of the pathogenic Staphylococcus species which in turn spread from one species to the other species through horizontal gene transfer [10, 57]. The difference in drug resistance capacity of the CoNS species isolates may also be attributed to variations in veterinary service delivery and farm hygiene which have a great contribution to the spread of drug-resistant CoNS species.
The CoNS species causes a wide range of human and animal diseases, and its pathogenicity is primarily due to a combination of toxin-mediated virulence, invasive capacity, and antibiotic resistance. The enterotoxin genes are responsible for the most potent staphylococcal food poisoning [11, 49].
The enterotoxin C (sec) gene was the only classical enterotoxin gene detected in CoNS species isolates in the present study. The finding was in agreement with the report of Homsombat et al. [33], Banaszkiewicz et al. [7], Nasaj et al. [40] who reported higher enterotoxin C (sec) gene prevalence in CoNS species-induced bovine mastitis. However, the result was different from the reports of Chajecka-Wierzchowska et al. [11], Helak et al. [32] who reported no classical enterotoxin genes detected from CoNS isolates.
The hla and hlb genes were detected in CoNS species at the rates of 8.4% and 9.6% respectively. The finding was much lower than the report of Pinheiro et al. [48] in Brazil, Nasaj et al. [41] in India reported 91.7% and 47.3%, 94.6% and 92.9% carriage of hla and hlb genes respectively. The discrepancy in the prevalence of the hemolysin genes might be associated with the difference in disease epidemiology [4], sample number, and sample source as well as the laboratory methods used for CoNS species virulent gene detection.
The mecA gene was the second most frequently detected (21.7%, 18/83) virulent gene in CoNS species isolates. The result infers a significant number of CoNS species harboring the methicillin resistance (mecA) gene, which poses a great health problem in animals and humans. The result was relatively in agreement with Seker et al. [51] in Turkey described 25% carriage of the mecA gene in CoNS species isolated from bovine milk. However, the finding was lower than the report of Xu et al. [60] in UK, Shrestha et al. [54] in Nepal, Ibadin et al. [34] in Nigeria, reported 29.5%, 70.7%, 30.5% carriage of mecA gene in CoNS isolates. A relatively lower report was made by Taponen et al. [56] in Finland from bovine mastitis milk, who reported 5% mecA gene carriage in CoNS species isolates.
The pvl gene was detected in 15/83, 18.07% of CoNS species isolates. The result was relatively higher than reports in India by Mahato et al. [37] who reported 6.5% pvl gene prevalence in CoNS species isolated from bovine milk and lower than Seker et al. [51] in Turkey reported 30.8% of CoNS species isolates were found carriage pvl gene.
Biofilm formation which is coded by the intracellular adhesion (icaD) gene is the major cause of drug resistance and persistent intramammary infections in bovine [52, 53]. Coagulase-negative Staphylococcus species were found to harbor 26.5% (22/83) of the icaD gene. The result was in agreement with the report of Gajewska and Chajecka-Wierzchowska [26] in central Poland showed carriage of 21.4% icaD genes. However, the result was lower than the report of Felipe et al. [21] on Argentinean dairy farms reporting 73.2% carriage of icaD in CoNS.
The difference in virulent gene profile in CoNS species between the present study and other reports might be associated with variations in the epidemiology of the study sites [4]. The knowledge gap on the rational use of drugs between veterinary practitioners might also play a great role in the spread of drug-resistant CoNS species [46]. Variations in CoNS species isolates virulent gene acquisition via horizontal gene transfer might also be another reason for the difference in the number of virulent genes detected from CoNS species isolates.
Conclusions
Coagulase-negative Staphylococcus species were isolated from both apparently healthy and clinical mastitis-infected cow’s milk; which poses high risk to milk consumers and persons in contact with them. A large number of coagulase-negative Staphylococcus species isolates were found multidrug-resistant (54.2%) and carriage of methicillin resistance genes. Various virulence and food poisoning factors were detected from CoNS species isolates; but, the pathogenic impact of these virulence factors for intramammary infections is not well studied. Usage of antimicrobial like Gentamycin and Oxacillin and proper boiling of milk before consumption is advisable to reduce the impact of coagulase-negative Staphylococcus species. Performing virulent gene protein expression, case-control studies and whole-genome sequencing is the limitation of this study and should be studied in the future.
Data availability
The raw data generated during the study was attached along with this manuscript as a supporting file and further information on the data generated can be obtained upon the request of the corresponding author.
Abbreviations
- CLSI:
-
Clinical Laboratory Standard Institute
- CoNS:
-
Coagulase-negative Staphylococcus
- CSA:
-
Central statistical agency
- IMI:
-
Intramammary infection
- MDRS:
-
Multidrug resistance Staphylococcus
- MHA:
-
Muller Hinton Agar
- MSA:
-
Mannitol Salt Agar
- NMC:
-
National Mastitis Council
References
Abunna F, Adugna B, Tufa TB, Ayana D, Gutema FD, Waktole H, et al. Detection and Antimicrobial Resistance of Staphylococcus species from Chicken, Chicken Litter, and humans in Addis Ababa, Ethiopia. Veterinary Med Int. 2022;2022:1–8.
Acosta AC, Oliveira PRF, Albuquerque L, Silva IF, Medeiros ES, Costa MM, et al. Frequency of Staphylococcus aureus virulence genes in milk of cows and goats with mastitis. Pesquisa Veterinária Brasileira. 2018;38(11):2029–36.
Adhikari GP. Calculating the sample size in quantitative studies. Scholars’ J. 2021;4:14–29.
Akova M. Epidemiology of antimicrobial resistance in bloodstream infections. Virulence. 2016;7(3):252–66.
Andrade NC, Laranjo M, Costa MM, Queiroga MC. Virulence factors in Staphylococcus Associated with Small Ruminant mastitis: Biofilm Production and Antimicrobial Resistance genes. J Antibiot. 2021;10(6):633.
Balemi A, Gumi B, Amenu K, Girma S, Gebru M, Tekle M et al. Prevalence of Mastitis and Antibiotic Resistance of Bacterial isolates from CMT Positive Milk Samples Obtained from dairy cows, camels, and goats in two Pastoral districts in Southern Ethiopia. Anim (Basel). 2021;11(6).
Banaszkiewicz S, Walecka-Zacharska E, Schubert J, Tabis A, Krol J, Stefaniak T, et al. Staphylococcal enterotoxin genes in coagulase-negative Staphylococci-Stability, expression, and genomic context. Int J Mol Sci. 2022;23(5):2560.
Bhavana RN, Chaitanya RK. Identification of coagulase negative staphylococcal species from bovine mastitis in India. Iran J Veterinary Res. 2022;23(4):358.
Bihon A, Syoum A, Assefa A. Assessment of risk factors and isolation of Staphylococcus aureus and Escherichia coli from bovine subclinical mastitic milk in and around Gondar, Northwest Ethiopia. Trop Anim Health Prod. 2019;51(4):939–48.
Boamah VE, Agyare C, Odoi H, Adu F, Gbedema SY, Dalsgaard A. Prevalence and antibiotic resistance of coagulase-negative Staphylococci isolated from poultry farms in three regions of Ghana. Infect Drug Resist. 2017;10:175–83.
Chajecka-Wierzchowska W, Gajewska J, Wisniewski P, Zadernowska A. Enterotoxigenic potential of coagulase-negative staphylococci from ready-to-eat food. Pathogens. 2020;9(9).
CLSI. Performance standards for Antimicrobial susceptibility testing. USA; 2021.
Condas LAZ, De Buck J, Nobrega DB, Carson DA, Roy JP, Keefe GP, et al. Distribution of non-aureus staphylococci species in udder quarters with low and high somatic cell count, and clinical mastitis. J Dairy Sci. 2017;100(7):5613–27.
Costa FN, Belo NO, Costa EA, Andrade GI, Pereira LS, Carvalho IA, et al. Frequency of enterotoxins, toxic shock syndrome toxin-1, and biofilm formation genes in Staphylococcus aureus isolates from cows with mastitis in the Northeast of Brazil. Trop Anim Health Prod. 2018;50(5):1089–97.
Cunha ML, Sinzato YK, Silveira LV. Comparison of methods for the identification of coagulase-negative staphylococci. Mem Inst Oswaldo Cruz. 2004;99(8):855–60.
Dakal TC, Kumar A, Majumdar RS, Yadav V. Mechanistic basis of antimicrobial actions of silver nanoparticles. Front Microbiol. 2016;7:1831.
De Buck J, Ha V, Naushad S, Nobrega DB, Luby C, Middleton JR, et al. Non-aureus staphylococci and Bovine Udder Health: current understanding and knowledge gaps. Front Vet Sci. 2021;8:658031.
Dos Santos DC, Lange CC, Avellar-Costa P, Dos Santos KR, Brito MA, Giambiagi-deMarval M. Staphylococcus chromogenes, a coagulase-negative Staphylococcus species that can clot plasma. J Clin Microbiol. 2016;54(5):1372–5.
Enquebaher T, Siv S, Knut R, Taran S, Judith AN. Staphylococcus aureus and other Staphylococcus species in milk and milk products from Tigray region, Northern Ethiopia. Afr J Food Sci. 2015;9(12):567–76.
Ewida RM, Al-Hosary AAT. Prevalence of enterotoxins and other virulence genes of Staphylococcus aureus caused subclinical mastitis in dairy cows. Vet World. 2020;13(6):1193–8.
Felipe V, Morgante CA, Somale PS, Varroni F, Zingaretti ML, Bachetti RA, et al. Evaluation of the biofilm forming ability and its associated genes in Staphylococcus species isolates from bovine mastitis in Argentinean dairy farms. Microb Pathog. 2017;104:278–86.
Fernandes Queiroga Moraes G, Cordeiro LV, de Andrade Júnior FP. Main laboratory methods used for the isolation and identification of Staphylococcus species. Rev Colomb Cienc Quím Farm. 2021;50(1):5–28.
Fesseha H, Mathewos M, Aliye S, Wolde A. Study on prevalence of bovine mastitis and Associated Risk Factors in Dairy Farms of Modjo Town and Suburbs, Central Oromia, Ethiopia. Vet Med (Auckl). 2021;12:271–83.
Franca A, Gaio V, Lopes N, Melo LDR. Virulence factors in Coagulase-Negative Staphylococci. Pathogens. 2021;10(2):170.
Fursova KK, Shchannikova MP, Loskutova IV, Shepelyakovskaya AO, Laman AG, Boutanaev AM, et al. Exotoxin diversity of Staphylococcus aureus isolated from milk of cows with subclinical mastitis in Central Russia. J Dairy Sci. 2018;101(5):4325–31.
Gajewska J, Chajecka-Wierzchowska W. Biofilm formation ability and Presence of Adhesion genes among coagulase-negative and coagulase-positive Staphylococci isolates from raw cow’s milk. Pathogens. 2020;9(8):654.
Gebremedhin EZ, Ararso AB, Borana BM, Kelbesa KA, Tadese ND, Marami LM, et al. Isolation and identification of Staphylococcus aureus from milk and milk products, Associated Factors for Contamination, and their Antibiogram in Holeta, Central Ethiopia. Vet Med Int. 2022;2022:6544705.
Gizaw F, Kekeba T, Teshome F, Kebede M, Abreham T, Hayishe H, et al. Distribution and antimicrobial resistance profile of coagulase-negative staphylococci from cattle, equipment, and personnel on dairy farm and abattoir settings. Heliyon. 2020;6(3):e03606.
Goetz C, Tremblay YDN, Lamarche D, Blondeau A, Gaudreau AM, Labrie J, et al. Coagulase-negative staphylococci species affect biofilm formation of other coagulase-negative and coagulase-positive staphylococci. J Dairy Sci. 2017;100(8):6454–64.
Grazul M, Balcerczak E, Sienkiewicz M. Analysis of the Presence of the virulence and regulation genes from Staphylococcus aureus (S. Aureus) in Coagulase Negative Staphylococci and the influence of the staphylococcal cross-talk on their functions. Int J Environ Res Public Health. 2023;20(6).
Grosjean S, Rank EL. Identifying species of coagulase-negative Staphylococci using a Microliter plate panel. Lab Med. 1985;16(11):688–92.
Helak I, Daczkowska-Kozon EG, Dlubala AA. Short communication: enterotoxigenic potential of coagulase-negative staphylococci isolated from bovine milk in Poland. J Dairy Sci. 2020;103(4):3076–81.
Homsombat T, Boonyayatra S, Awaiwanont N, Pichpol D. Effect of temperature on the expression of classical enterotoxin genes among Staphylococci Associated with Bovine Mastitis. Pathogens. 2021;10(8):975.
Ibadin EE, Enabulele IO, Muinah F. Prevalence of mecA gene among staphylococci from clinical samples of a tertiary hospital in Benin City, Nigeria. Afr Health Sci. 2017;17(4):1000–10.
Jenkins SN, Okello E, Rossitto PV, Lehenbauer TW, Champagne J, Penedo MCT, et al. Molecular epidemiology of coagulase-negative Staphylococcus species isolated at different lactation stages from dairy cattle in the United States. PeerJ. 2019;7:e6749.
Lee YJ, Lee YJ. Characterization of Biofilm Producing Coagulase-negative Staphylococci isolated from Bulk Tank Milk. Vet Sci. 2022;9(8):430.
Mahato S, Mistry HU, Chakraborty S, Sharma P, Saravanan R, Bhandari V. Identification of variable traits among the Methicillin resistant and sensitive Coagulase negative staphylococci in milk samples from Mastitic Cows in India. Front Microbiol. 2017;8:1446.
Martins KB, Faccioli PY, Bonesso MF, Fernandes S, Oliveira AA, Dantas A, et al. Characteristics of resistance and virulence factors in different species of coagulase-negative staphylococci isolated from milk of healthy sheep and animals with subclinical mastitis. J Dairy Sci. 2017;100(3):2184–95.
Moraveji Z, Tabatabaei M, Shirzad Aski H, Khoshbakht R. Characterization of hemolysins of Staphylococcus strains isolated from human and bovine, southern Iran. Iran J Veterinary Res. 2014;15(4):326–30.
Nasaj M, Saeidi Z, Tahmasebi H, Dehbashi S, Arabestani MR. Prevalence and distribution of resistance and enterotoxins/enterotoxin-like genes in different clinical isolates of coagulase-negative Staphylococcus. Eur J Med Res. 2020;25(1):48.
Nasaj M, Saeidi Z, Asghari B, Roshanaei G, Arabestani MR. Identification of hemolysin encoding genes and their association with antimicrobial resistance pattern among clinical isolates of coagulase-negative Staphylococci. BMC Res Notes. 2020;13(1):68.
Neelam, Jain VK, Singh M, Joshi VG, Chhabra R, Singh K, et al. Virulence and antimicrobial resistance gene profiles of Staphylococcus aureus associated with clinical mastitis in cattle. PLoS ONE. 2022;17(5):e0264762.
NMC. Microbiological procedures for the diagnosis of bovine udder infection and determination of milk quality. 4th ed. National Mastitis Council, USA; 2004.
Ogodo AC, Agwaranze DI, Daji M, Aso RE. Microbial techniques and methods: Basic techniques and microscopy. Analytical Techniques in Biosciences: Elsevier; 2022. pp. 201 – 20.
Omwenga I, Aboge GO, Mitema ES, Obiero G, Ngaywa C, Ngwili N et al. Staphylococcus aureus enterotoxin genes detected in milk from various livestock species in northern pastoral region of Kenya. 2019;103:126–32.
Phophi L, Petzer IM, Qekwana DN. Antimicrobial resistance patterns and biofilm formation of coagulase-negative Staphylococcus species isolated from subclinical mastitis cow milk samples submitted to the Onderstepoort Milk Laboratory. BMC Vet Res. 2019;15(1):420.
Piessens V, Van Coillie E, Verbist B, Supre K, Braem G, Van Nuffel A, et al. Distribution of coagulase-negative Staphylococcus species from milk and environment of dairy cows differs between herds. J Dairy Sci. 2011;94(6):2933–44.
Pinheiro L, Brito CI, de Oliveira A, Martins PY, Pereira VC, da Cunha Mde L. Staphylococcus epidermidis and Staphylococcus haemolyticus: Molecular Detection of Cytotoxin and Enterotoxin genes. Toxins (Basel). 2015;7(9):3688–99.
Podkowik M, Park JY, Seo KS, Bystron J, Bania J. Enterotoxigenic potential of coagulase-negative staphylococci. Int J Food Microbiol. 2013;163(1):34–40.
Quinn PJ, Markey BK, Leonard FC, Hartigan P, Fanning S, FitzPatrick ES. Veterinary microbiology and microbial disease. 2nd ed: John Wiley & Sons, UK; 2011. 1985 p.
Seker E, Ozenc E, Turedi OK, Yilmaz M. Prevalence of mecA and pvl genes in coagulase negative staphylococci isolated from bovine mastitis in smallholder dairy farms in Turkey. J Anim Biotechnol. 2022:1–6.
Seng R, Kitti T, Thummeepak R, Kongthai P, Leungtongkam U, Wannalerdsakun S, et al. Biofilm formation of methicillin-resistant coagulase negative staphylococci (MR-CoNS) isolated from community and hospital environments. PLoS ONE. 2017;12(8):e0184172.
Shrestha LB, Bhattarai NR, Khanal B. Antibiotic resistance and biofilm formation among coagulase-negative staphylococci isolated from clinical samples at a tertiary care hospital of eastern Nepal. Antimicrob Resist Infect Control. 2017;6:89.
Shrestha LB, Bhattarai NR, Rai K, Khanal B. Antibiotic resistance and mecA Gene characterization of coagulase-negative Staphylococci isolated from clinical samples in Nepal. Infect Drug Resist. 2020;13:3163–9.
Stepanovic S, Dakic I, Hauschild T, Vukovic D, Morrison D, Jezek P, et al. Supplementary biochemical tests useful for the differentiation of oxidase positive staphylococci. Syst Appl Microbiol. 2007;30(4):316–8.
Taponen S, Nykasenoja S, Pohjanvirta T, Pitkala A, Pyorala S. Species distribution and in vitro antimicrobial susceptibility of coagulase-negative staphylococci isolated from bovine mastitic milk. Acta Vet Scand. 2016;58:12.
Tello A, Austin B, Telfer TC. Selective pressure of antibiotic pollution on bacteria of importance to public health. Environ Health Perspect. 2012;120(8):1100–6.
Varghese N, Joy PP. Microbiology Laboratory Manual. India: Kerala Agricultural University, Pineapple Research Station.; 2014. 1–78 p.
Wald R, Hess C, Urbantke V, Wittek T, Baumgartner M. Characterization of Staphylococcus species isolated from bovine quarter milk samples. Anim (Basel). 2019;9(5):200.
Xu Z, Mkrtchyan HV, Cutler RR. Antibiotic resistance and mecA characterization of coagulase-negative staphylococci isolated from three hotels in London, UK. Front Microbiol. 2015;6:947.
Zeryehun T, Abera G. Prevalence and bacterial isolates of Mastitis in dairy farms in selected districts of Eastern Harrarghe Zone, Eastern Ethiopia. J Vet Med. 2017;2017:6498618.
Acknowledgements
All authors offer heartfelt gratitude and respect to Gondar University Microbiology and Addis Ababa University, Institute of Biotechnology laboratory unit for providing materials and technical support during the laboratory work.
Funding
The authors didn’t receive funds for this work.
Author information
Authors and Affiliations
Contributions
YAG- participated in writing the proposal and preparation of the manuscript, TST- contributed to study concept and manuscript edition, SLA- contributed to manuscript edition and write up, AMB- contributed to manuscript edition and laboratory work, MAB- contributed in laboratory work and all authors are read and approved for publication.
Corresponding author
Ethics declarations
Ethical approval and consent to Participate
The University of Gondar’s ethical review committee evaluated ethical issues related to research work and approved its ethical soundness and acceptability under reference no CVMAS.sc-03/22 written on 05 January 2022. Thus, the University of Gondar sent a letter of cooperation to the study area. Furthermore, an official permission letter was obtained from the livestock and fishery production office to conduct the study. The purpose and procedures of the study were properly explained to the farm owner and study participants and milk samples were collected from dairy farms, whose owners showed a willingness to participate and informed consent was obtained from all of them. All methods were performed in accordance with the relevant guidelines and regulations.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Getahun, Y.A., Abey, S.L., Beyene, A.M. et al. Coagulase-negative staphylococci from bovine milk: Antibiogram profiles and virulent gene detection. BMC Microbiol 24, 263 (2024). https://doi.org/10.1186/s12866-024-03415-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12866-024-03415-0