Abstract
Background
Haemophilus influenzae (Hi) is an emerging cause of early onset neonatal sepsis, but mechanisms of transmission are not well understood. We aimed to determine the prevalence of vaginal carriage of Hi in reproductive age women and to examine behavioral and demographic characteristics associated with its carriage.
Methods
We performed a secondary analysis of stored vaginal lavage specimens from a prospective cohort study of nonpregnant reproductive-age women. After extraction of bacterial genomic DNA, samples were tested for the presence of the gene encoding Haemophilus protein d (hpd) by quantitative real-time polymerase chain reaction (PCR) using validated primers and probe. PCR for the V3-V4 region of the 16 S rRNA gene (positive control) assessed sample quality. Samples with cycle threshold (CT) value < 35 were defined as positive. Sanger sequencing confirmed the presence of hpd. Behavioral and demographic characteristics associated with vaginal carriage of Hi were examined.
Results
415 samples were available. 315 (75.9%) had sufficient bacterial DNA and were included. 14 (4.4%) were positive for hpd. There were no demographic or behavioral differences between the women with Hi vaginal carriage and those without. There was no difference in history of bacterial vaginosis, vaginal microbiome community state type, or presence of Group B Streptococcus in women with and without vaginal carriage of Hi.
Conclusion
Hi was present in vaginal lavage specimens of 4.4% of this cohort. Hi presence was unrelated to clinical or demographic characteristics, though the relatively small number of positive samples may have limited power to detect such differences.
Similar content being viewed by others
Background
Worldwide, sepsis and other infections cause approximately 15% of neonatal deaths [1]. Neonatal sepsis is divided into early-onset sepsis (EOS), defined as onset of symptoms before 7 days of life, and late-onset sepsis (LOS), which occurs between 7 days and 3 months of life. Many neonatal sepsis deaths occur on the first day of life, and preterm infants are at particularly high risk for sepsis and its sequelae.
EOS is frequently caused by vertical transmission of bacteria through infected amniotic fluid or from the mother’s vaginal canal during labor and delivery. LOS is generally thought to be the result of either vertical transmission or horizontal transmission from caregivers or the environment. In the United States, the most common pathogens for both EOS and LOS are Group B Streptococcus (GBS) and Escherichia coli, though with universal GBS screening and intrapartum antibiotic prophylaxis (IAP) the number of EOS cases caused by GBS has decreased [2, 3]. Concern remains that other pathogens resistant to antibiotics used in IAP may emerge as more frequent causes of neonatal sepsis.
Haemophilus influenzae (Hi) type b, a respiratory pathogen, was once a common cause of invasive bacterial disease in childhood, but with widespread vaccination, it has become a rare cause of invasive disease in the United States [4]. Both typeable and nontypeable strains of Haemophilus influenzae remain responsible for adult and neonatal pneumonia and can also cause severe female reproductive tract infection, when the organism’s presence in the vagina leads to upper genital tract infection through a break in anatomical barriers such as after surgery or delivery. In recent years there has been an increase in reported cases of neonatal sepsis due to Haemophilus influenzae [5,6,7]. The majority of these cases are EOS, suggesting vertical transmission as a potential source of infection. This hypothesis is additionally supported by another study reporting a significantly higher rate of invasive Hi infection in pregnant women [8].
The maternal vaginal microbiota represents a potential source of Hi in cases of neonatal infection, by vertical transmission during parturition. However, little is known about Hi in the vagina. It may be a transient vaginal colonizer, or it may be introduced from a respiratory or oropharyngeal source. Prevalence estimates range from 1.8/ 1000 in Scandinavian women in pregnancy9 to 7.3% in women with preterm premature rupture of membranes in Chile [10]. The rate of vaginal carriage of E. coli, which is known to be transmitted perinatally, is estimated at 13–32% [11,12,13] while GBS is estimated to be present in the vaginal microbiota of 18–40% of women [14]. Notably, the rate of Hi vaginal carriage in the U.S. in both pregnant and non-pregnant individuals has not been studied.
In order to further understand which women and neonates are at risk for sepsis caused by Hi, we evaluated the rate of vaginal carriage of Hi in a cohort of nonpregnant women.
Methods
Samples and parent study
We analyzed samples from a previously reported prospective study of 432 nonpregnant reproductive-age women. The Bacterial Vaginosis–Improved Diagnosis by ELISA and Sequencing (BV-IDEAS) study enrolled nonpregnant women aged 18–55 years seeking primary gynecologic care in New York City from July 2010-June 2012. After obtaining informed consent, 5 mL sterile saline vaginal lavage specimens were collected, refrigerated for transport (< 6 h), and stored at − 80 °C as previously described [15]. GBS status, determined by polymerase chain reaction (PCR) of vaginal lavage specimens, and vaginal microbiota community state subtypes have been reported for this cohort [15, 16]. Additionally, participants provided demographic information by self-report, including age, race, ethnicity, education level, income level, and behavioral characteristics including history of bacterial vaginosis, recent treatments with antibiotics or antifungals, and sexual practices. The original study was approved by the Institutional Review Boards at Columbia University Medical Center and Weill Cornell Medical College, and participants providing consent for future studies were included in the current analysis.
DNA extraction and PCR
DNA extraction was performed on 500 µL of lavage specimen using the MagMax CORE Nucleic Acid Purification Kit on a Kingfisher Flex Purification System (ThermoFisher Scientific, Waltham, MA) after pretreatment with proteinase K, mutanolysin, and 1% lysozyme.
Samples were tested for the presence of the gene encoding Haemophilus protein d (hpd) by quantitative real-time polymerase chain reaction (PCR) using a validated primer/ probe set [17]. Each reaction mixture contained 1 µl of primer/probe mix with final concentrations of 500 nM primer and 250 nM probe, 10 µl of TaqMan Universal Master Mix II without UNG, 4 µl of RNAse-free PCR grade water, and 5 µl of extracted DNA. PCR conditions were as follows: 50.0°C for 2 min, 95.0°C for 10 min, followed by 95.0°C for 15s and 60.0°C for 1 min for 50 cycles. PCR was performed in 96-well plates on an Applied Biosystems StepOne Real-Time PCR System. Fluorescence threshold was set at 0.2 and CT values less than 35 were considered positive. A positive control of Hi genomic DNA extracted using the MoBio Powersoil DNA extraction kit (Qiagen) was included on each run. Negative controls with master mix, primer, probe and no template were also included.
To assess sample quality, PCR for the V4-V5 region of the 16 S rRNA gene was performed using specific primers and probe as described [18]. The same conditions were used for PCR as described above. PCR for the 16 S rRNA gene and the hpd gene was performed on the same run for each sample. Samples were determined to have sufficient bacterial content if the CT was < 35 for the 16 S rRNA PCR.
Sequencing
PCR products from samples that had sufficient bacterial content and were positive for hpd were run on a 2% agarose gel. Bands corresponding to hpd were cut from the gel and DNA was extracted using a QIAquick gel extraction kit (Qiagen, Valencia, CA). Sanger sequencing of the PCR products of positive samples was performed (Genewiz, South Plainfield, NJ) to confirm the presence of hpd.
Statistical analysis
Demographic and behavioral characteristics of women with and without vaginal carriage of Hi were compared. Missing data are due to participant non-response to the survey questions in the parent study. Vaginal microbiome community state types and presence of GBS was also compared between groups. Chi-square, Fisher’s exact test, and t-test were used with p < 0.05 set as the level of statistical significance. All statistical analysis was performed using SPSS Version 24 (IBM Corp, Armonk, NY).
Results
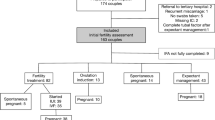
452 subjects were included in the original cohort. Of those, 415 had provided consent for future analysis and had samples were available for this study. 315 (75.9%) had sufficient bacterial DNA present, as determined by real time PCR with CT < 35 for the 16 S rRNA gene and were included in the analysis. 14 samples (4.4%) were positive for hpd by real-time PCR with CT < 35, which was confirmed by Sanger sequencing of the PCR products. Study flow diagram is shown in Fig. 1.
There were no demographic differences between the women with Hi vaginal carriage and those without (Table 1). There was no difference in presence of GBS, history of bacterial vaginosis, or vaginal microbiome community state type in women with and without vaginal carriage of Hi (Table 2). Additionally there were no statistically significant differences in sexual behaviors such as receptive oral sex noted (Table 2). Characteristics of participants’ partners are reported in Table 3. No difference was seen in partner characteristics in the two groups.
Discussion
Haemophilus influenzae DNA was present in vaginal lavage specimens of 4.4% of nonpregnant women. The presence of Hi was unrelated to clinical or demographic characteristics in this cohort, though the relatively small number of positive samples may have limited power to detect such differences.
The rate of Hi vaginal carriage in this study is similar to the rates seen in the literature in pregnant and non-pregnant women. In pregnancy, the rate of vaginal carriage of Hi in Denmark in 1989 was 1.8/ 1000 women presenting in labor, [9] while in a study from Chile in 1992–1998, Hi was isolated from the vaginal culture specimens of 7.3% of 110 women with PPROM [10]. In both studies the presence of Hi was determined by culture and confirmed by PCR of the 16 S rRNA gene. A study of 510 pregnant women in Italy found the overall prevalence of carriage of the Haemophilus genus to be 9% using culture-based methods confirmed by sequencing the full-length 16 S rRNA gene. However, only H. parainfluenzae, H. pittmaniae and H. haemolyticus were present with no Hi detected [19].
Outside of pregnancy, the rate of vaginal carriage of Hi appears to be similar. A study of 216 nonpregnant women in Australia using a multiplex PCR assay for 14 microbial species and the Hi gyrR gene found that Hi was present in 5.1% of the vaginal swab samples [20, 21]. An individual participant meta-analysis of the vaginal microbiome data of 1,163 women noted the presence of Hi at low levels, though the prevalence of Hi from the 16 S rRNA microbiome sequencing data was not reported [22]. In our previously reported sequencing analysis, Hi was not present at detectable levels in the current samples [15]. We speculate that levels of hpd DNA were likely too low to be detected by the sequencing pipeline that was used in that report. Targeted PCR based amplification as performed in this study allowed identification of hpd at lower levels.
This study has several strengths. It is a large cohort with detailed information about individual clinical and behavioral characteristics, and validated primers and probe from the CDC were used for evaluation of the presence of Hi. The hpd protein that was targeted is specific to Hi and more accurately identifies Hi than the 16 S rRNA or the culture-based techniques used in most prior studies. Sanger sequencing was performed on samples that were positive for hpd to confirm its presence, decreasing the risk that contamination or background signal led to false positives. The use of PCR enhances the ability to detect low numbers of Hi as compared to vaginal microbiome studies utilizing 16 S rRNA sequencing. While this ability to detect low levels of Hi is a strength of our study, these levels may in fact be too low to cause clinically significant infection. Therefore, we are unable to draw any conclusions about the infectivity of this bacterial species based on this data, and additional research is needed to further elucidate the ability of Hi to cause reproductive tract and neonatal infection.
There are also several limitations to the study. The number of samples positive for Hi is low, limiting our ability to make meaningful conclusions regarding demographic and clinical risk factors for the vaginal carriage of Hi. The samples may have degraded during storage, causing some to have a low bacterial content. We included only samples with sufficient bacterial content (16 S rRNA CT < 35) but there may have been Hi present in samples that were excluded. It is unlikely that samples with or without Hi present would degrade at different rates, thus sample degradation is unlikely to have biased our conclusions. There may also be some selection bias present as we only analyzed the samples of women who consented to future research. Though unlikely, there may have been a different prevalence of Hi in women who declined future study participation. The samples used in this analysis were collected by vaginal lavage, which may not correlate exactly with vaginal swabs as were used in other studies. However, since the prevalence of Hi in our population is similar to that seen in other studies, this method of collection of vaginal samples does not appear to limit the applicability of our study.Finally, it is possible that some samples in this study that were positive for hpd may in fact contain H. haemolyticus. The ability to differentiate the two species by culture-based or molecular techniques is limited and rare cases of invasive infection due to H. haemolyticus have been reported. Though H. haemolyticus is typically negative for the hpd gene, sequencing has shown that it may be present in some strains, making H. haemolyticus very difficult to distinguish from Hi [23,24,25].
Conclusion
Haemophilus influenzae was present in vaginal lavage specimens of 4.4% of nonpregnant women. The presence of Hi was unrelated to clinical or demographic characteristics in this cohort, though the relatively small number of positive samples may have limited power to detect such differences. Further studies should assess for the presence of Hi as a colonizing organism in pregnant women and well neonates. Persistence of vaginal carriage throughout pregnancy, transmissibility of Hi to the neonate and the percentage of exposed neonates who become clinically ill all remain unknown and should be assessed.
Data Availability
The dataset used during the current study is available from the corresponding author on reasonable request.
References
Oza S, Lawn JE, Hogan DR, Mathers C, Cousens SN. Neonatal cause-of-death estimates for the early and late neonatal periods for 194 countries: 2000–2013. Bull World Health Organ Jan. 2015;1(1):19–28. https://doi.org/10.2471/BLT.14.139790.
Active Bacterial Core Surveillance Report, Emerging Infections Program Network, Group B Streptococcus., 2015. Centers for Disease Control and Prevention. 2015.
Bauserman MS, Laughon MM, Hornik CP, et al. Group B Streptococcus and Escherichia coli infections in the intensive care nursery in the era of intrapartum antibiotic prophylaxis. Pediatr Infect Dis J Mar. 2013;32(3):208–12. https://doi.org/10.1097/INF.0b013e318275058a.
Briere EC, Rubin L, Moro PL, Cohn A, Clark T, Messonnier N. Prevention and control of haemophilus influenzae type b disease: recommendations of the advisory committee on immunization practices (ACIP). MMWR Recomm Rep Feb. 2014;28(Rr–01):1–14.
Collins S, Litt DJ, Flynn S, Ramsay ME, Slack MP, Ladhani SN. Neonatal invasive Haemophilus influenzae disease in England and Wales: epidemiology, clinical characteristics, and outcome. Clin Infect Dis Jun. 2015;15(12):1786–92. https://doi.org/10.1093/cid/civ194.
Soeters HM, Blain A, Pondo T, et al. Current epidemiology and Trends in Invasive Haemophilus influenzae Disease-United States, 2009–2015. Clin Infect Dis Aug. 2018;31(6):881–9. https://doi.org/10.1093/cid/ciy187.
Oliver SE, Rubis AB, Soeters HM, et al. Epidemiology of invasive nontypeable Haemophilus influenzae disease-United States, 2008–2019. Clin Infect Dis Feb. 2023;1. https://doi.org/10.1093/cid/ciad054.
Collins S, Ramsay M, Slack MP, et al. Risk of invasive Haemophilus influenzae infection during pregnancy and association with adverse fetal outcomes. JAMA Mar. 2014;19(11):1125–32. https://doi.org/10.1001/jama.2014.1878.
Schonheyder H, Ebbesen F, Grunnet N, Ejlertsen T. Non-capsulated Haemophilus influenzae in the genital flora of pregnant and post-puerperal women. Scand J Infect Dis. 1991;23(2):183–7. https://doi.org/10.3109/00365549109023398.
Martínez MA, Ovalle A, Ulloa MT, Vidal RM. Role of Haemophilus influenzae in intra-amniotic infection in patients with preterm rupture of membranes. Eur J Clin Microbiol Infect Dis Dec. 1999;18(12):890–2. https://doi.org/10.1007/s100960050425.
Amstey MS, Lewin E, Colaice J. Vaginal colonization with invasive Escherichia coli during pregnancy. Am J Obstet Gynecol. 1980;137(5):534–535. https://doi.org/10.1016/0002-9378(80)90691-2.
Sáez-López E, Guiral E, Fernández-Orth D, et al. Vaginal versus obstetric infection Escherichia coli isolates among pregnant women: Antimicrobial Resistance and Genetic Virulence Profile. PLoS ONE. 2016;11(1):e0146531. https://doi.org/10.1371/journal.pone.0146531.
Moradi Y, Eshrati B, Motevalian SA, Majidpour A, Baradaran HR. A systematic review and meta-analysis on the prevalence of Escherichia coli and extended-spectrum β-lactamase-producing Escherichia coli in pregnant women. Arch Gynecol Obstet Feb. 2021;303(2):363–79. https://doi.org/10.1007/s00404-020-05903-w.
Regan JA, Klebanoff MA, Nugent RP. The epidemiology of group B streptococcal colonization in pregnancy. Vaginal infections and Prematurity Study Group. Obstet Gynecol Apr. 1991;77(4):604–10.
Rosen GH, Randis TM, Desai PV, et al. Group B Streptococcus and the vaginal microbiota. J Infect Dis Sep. 2017;15(6):744–51. https://doi.org/10.1093/infdis/jix395.
Khatami A, Randis TM, Tavares L, Gegick M, Suzman E, Ratner AJ. Vaginal co-colonization with multiple Group B Streptococcus serotypes. Vaccine Jan. 2019;14(3):409–11. https://doi.org/10.1016/j.vaccine.2018.12.001.
Laboratory Methods for the Diagnosis of Meningitis Caused by Neisseria meningitidis, Streptococcus pneumoniae, and Haemophilus influenzae. Centers for Disease Control and Prevention. Updated April 15., 2016. Accessed July 1, 2019.
Srinivasan S, Hoffman NG, Morgan MT, et al. Bacterial communities in women with bacterial vaginosis: high resolution phylogenetic analyses reveal relationships of microbiota to clinical criteria. PLoS ONE. 2012;7(6):e37818. https://doi.org/10.1371/journal.pone.0037818.
Cardines R, Daprai L, Giufre M, Torresani E, Garlaschi ML, Cerquetti M. Genital carriage of the genus Haemophilus in pregnancy: species distribution and antibiotic susceptibility. J Med Microbiol Jul. 2015;64(7):724–30. https://doi.org/10.1099/jmm.0.000083.
McKechnie ML, Hillman RJ, Jones R, et al. The prevalence of urogenital micro-organisms detected by a multiplex PCR-reverse line blot assay in women attending three sexual health clinics in Sydney, Australia. J Med Microbiol Jul. 2011;60(Pt 7):1010–6. https://doi.org/10.1099/jmm.0.029108-0.
McKechnie ML, Hillman R, Couldwell D, et al. Simultaneous identification of 14 genital microorganisms in urine by use of a multiplex PCR-based reverse line blot assay. J Clin Microbiol Jun. 2009;47(6):1871–7. https://doi.org/10.1128/jcm.00120-09.
van de Wijgert J, Verwijs MC, Gill AC, Borgdorff H, van der Veer C, Mayaud P. Pathobionts in the vaginal microbiota: individual Participant Data Meta-Analysis of three sequencing studies. Front Cell Infect Microbiol. 2020;10:129. https://doi.org/10.3389/fcimb.2020.00129.
Anderson R, Wang X, Briere EC, et al. Haemophilus haemolyticus isolates causing clinical disease. J Clin Microbiol Jul. 2012;50(7):2462–5. https://doi.org/10.1128/JCM.06575-11.
Marti S, Puig C, de la Campa AG, et al. Identification of Haemophilus haemolyticus in clinical samples and characterization of their mechanisms of antimicrobial resistance. J Antimicrob Chemother Jan. 2016;71(1):80–4. https://doi.org/10.1093/jac/dkv307.
Pickering J, Richmond PC, Kirkham LA. Molecular tools for differentiation of non-typeable Haemophilus influenzae from Haemophilus haemolyticus. Front Microbiol. 2014;5:664. https://doi.org/10.3389/fmicb.2014.00664.
Acknowledgements
None.
Funding
This work was supported by funds from New York University Grossman School of Medicine.
Author information
Authors and Affiliations
Contributions
All authors contributed to study conception and design. Data collection and analysis was performed by Meghana Limaye. The first draft of the manuscript was written by Meghana Limaye and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The original study was approved by the Institutional Review Boards at Columbia and Cornell, and the current study was approved by the Institutional Review Board at NYU (i16-00780), approved 8/24/16. All methods were performed in accordance with the relevant guidelines and regulations.
Consent for publication
Not applicable.
Competing interests
AJR has served as a consultant for Pfizer and on a compassionate use advisory board for CompAC (NYU/Janssen), both outside the scope of this manuscript. All other authors report no conflicts of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Limaye, M.A., Brubaker, S., Randis, T.M. et al. Vaginal carriage of Haemophilus influenzae in a non-pregnant reproductive-age population. BMC Microbiol 23, 141 (2023). https://doi.org/10.1186/s12866-023-02885-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12866-023-02885-y