Abstract
Background
Catabolite control protein A (CcpA) regulates the transcription of lactate dehydrogenase and pyruvate formate-lyase in Streptococcus bovis, but knowledge of its role in response to different pH is still limited. In this study, a ccpA-knockout strain of S. bovis S1 was constructed and then used to examine the effects of ccpA gene deletion on the growth and fermentation characteristics of S. bovis S1 at pH 5.5 or 6.5.
Results
There was a significant interaction between strain and pH for the maximum specific growth rate (μmax) and growth lag period (λ), which caused a lowest μmax and a longest λ in ccpA-knockout strain at pH 5.5. Deletion of ccpA decreased the concentration and molar percentage of lactic acid, while increased those of formic acid. Strains at pH 5.5 had decreased concentrations of lactic acid and formic acid compared to pH 6.5. The significant interaction between strain and pH caused the highest production of total organic acids and acetic acid in ccpA-knockout strain at pH 6.5. The activities of α-amylase and lactate dehydrogenase decreased in ccpA-knockout strain compared to the wild-type strain, and increased at pH 5.5 compared to pH 6.5. There was a significant interaction between strain and pH for the activity of acetate kinase, which was the highest in the ccpA-knockout strain at pH 6.5. The expression of pyruvate formate-lyase and acetate kinase was higher in the ccpA-knockout strain compared to wild-type strain. The lower pH improved the relative expression of pyruvate formate-lyase, while had no effect on the relative expression of acetate kinase. The strain × pH interaction was significant for the relative expression of lactate dehydrogenase and α-amylase, both of which were highest in the wild-type strain at pH 5.5 and lowest in the ccpA-knockout strain at pH 6.5.
Conclusions
Overall, low pH inhibited the growth of S. bovis S1, but did not affect the fermentation pattern. CcpA regulated S. bovis S1 growth and organic acid fermentation pattern. Moreover, there seemed to be an interaction effect between pH and ccpA deletion on regulating the growth and organic acids production of S. bovis S1.
Similar content being viewed by others
Introduction
Generally, high-concentrate diets based on grains are applied in modern intensive ruminant production to increase production. However, ruminants fed on high-concentrate diets display increased yields of volatile fatty acids (VFA) and lactic acid in the rumen and decreased rumen pH, which may cause subacute ruminal acidosis [1, 2]. In this process, the concentration of lactic acid is much lower than VFA because there is a balance between the production of lactic acid and its conversion into VFA. Once the balance is broken, lactic acid begins to accumulate in the rumen. Because lactic acid is a stronger acid than VFA (pKa 3.9 vs 4.7–4.9), its accumulation often brings about a downward spiral in ruminal pH, which may induce rumen lactic acidosis [3]. Previous studies demonstrated that an initial overgrowth of Streptococcus bovis is the major cause of increasing lactic acid and declining ruminal pH, which will result in the inability of most ruminal bacteria, and the acid-tolerant Lactobacilli becoming predominant [4, 5]. Therefore, S. bovis is considered the major etiologic agent of rumen lactic acidosis, and suppressing its overgrowth and overproduction of lactic acid is vital to prevent rumen lactic acidosis when animals are supplied with a high grain diet.
S. bovis is a gram-positive bacterium that utilizes starch and soluble sugars as substrate in the rumen, thereby producing lactic acid, formic acid, acetic acid and ethanol. Several previous studies have shown that the fermentation products of S. bovis are regulated by the pH, carbon source and substrate concentration [6,7,8,9,10]. Our previous work with S. bovis S1 demonstrated that the carbohydrate source (soluble starch vs. glucose) had a major effect on lactic acid production due to the transcriptional regulation of metabolic genes [10]. Furthermore, we evaluated the relative importance of pH and starch concentration in fermentation characteristics of S. bovis S1 and found that the fermentation of the strain was more sensitive to the pH changes [11]. In addition to the environmental factors, the fermentation pattern of S. bovis is also controlled by the catabolite control protein A (CcpA) [12], which is a pleiotropic regulatory protein in low-GC gram-positive bacteria involved in carbon and nitrogen metabolism, biofilm formation, toxic gene expression, and other physiological processes. Several studies have reported that CcpA activates or inhibits transcriptional expression of the target gene via binding to the specific sequence of its target gene, such as catabolite response element (cre) [13,14,15]. The regulation of target genes by CcpA is not only associated with heat-stable protein (HPr) [16,17,18] and small molecule compounds such as fructose-1,6-diphosphate (FDP) and 6-phosphate glucose [19], but also affected by the environmental factors, such as substrate type, oxygen presence and pH [20,21,22]. Based on the above literatures and our previous work, we hypothesized that CcpA transcriptionally regulates the fermentation pattern of S. bovis S1, which might be is affected by pH. To test this hypothesis, we constructed the ccpA gene knockout mutant of S. bovis S1 using homologous recombination technology, and used it to investigate the effect of ccpA gene deletion on the growth and fermentation characteristics of S. bovis S1 at different pH.
Materials and methods
Bacterial strains
Strain S. bovis S1 (CCTCC AB 2016240) used in this study was previously isolated from the rumen fluid of Saanen dairy goats (late lactation) in our laboratory [10]. Its ccpA-disrupted mutant was constructed by homologous recombination as follows. First, DNA fragments corresponding to the upstream (1053 bp fragment; primer pairs ccpA Up F/ccpA Up R, Table 1) and downstream (1101 bp fragment; primer pairs ccpA Down F/ccpA Down R, Table 1) sequences of ccpA were amplified by PCR using S. bovis S1 genomic DNA as a template. The erythromycin resistance gene erm was amplified with the primers erm F and erm R (Table 1). The PCR product was purified using a PCR purification kit (Qiagen, Beijing, China) according to the manufacturer’s instructions. The amplified fragments were respectively cloned into EcoRI, BamHI and SacI restriction sites of pUC19 vector to generate pUC19-ccpA up-erm-ccpA down (pCE). The recombinant vector pCE was electroporated into S. bovis S1 cells using an electroporation system at 2.5 kV, 200 Ω, and 25 μF. Finally, knockout mutants were selected on MRS plates containing 1 μg/mL erythromycin at 37 °C for 3–4 days.
The result of knockout was validated by PCR and DNA sequencing. Briefly, genomic DNA of S. bovis S1 wild-type strain and its mutant strain were extracted with the TIANamp Bacteria DNA Kit (DP302, Tiangen Biotech Co. Ltd., Beijing, China) and used as the templates. Primers used for verification are shown in Table 1, which were designed based on the internal sequence of the ccpA gene (ccpA1-F/ccpA1-R) or across the upstream and downstream sequences of the ccpA gene (ccpA2-F/ccpA2-R). PCR analysis was performed using the 2 × Taq PCR MasterMix II Kit (KT211–02, Tiangen Biotech Co. Ltd., Beijing, China) according to the manufacturer’s instructions. The amplified PCR products were electrophoresed on a 1% (wt/vol) agarose gel. Sequence analysis for wt2 and ko2, which were the products of primer ccpA2 amplified from S. bovis S1 wild-type strain and the ccpA-knockout strain respectively, was performed by Shanghai Sangon Biological Engineering Technology and Services Co., Ltd. for further verification.
Experimental design
The experiment treatments were arranged as a 2 × 2 factorial design: two strains, which were S. bovis S1 wild-type strain and its ccpA-disrupted mutant respectively, were cultured in the media at pH 5.5 or pH 6.5. Specific operations are as follows. First, the seed cultures of S. bovis S1 wild-type strain and its mutant strain were cultured in a modified MRS medium [10] in an anaerobic workstation (DG250, Don Whitley Scientific, England) at 37 °C. Next, the seed cultures of both strains (at exponential phase) were transferred with 1% (v/v) inoculum into 200 mL anaerobic serum bottles containing 100 mL basal medium, respectively. The basal medium was prepared according to Chen et al. [10], and it contained: 0.45 g/L KH2PO4, 0.45 g/L K2HPO4, 0.9 g/L NaCl, 0.9 g/L (NH4)2SO4, 0.12 g/L CaCl2·2H2O, 0.19 g/L MgSO4·7H2O, 1.0 g/L tryptone, 1.0 g/L yeast extract, 0.6 g/L cysteine hydrochloride, and 3.0 g/L soluble starch (Cat#G8300, Solarbio, Beijing, China). The original pH of the medium was 4.2, which was adjusted to 5.5 and 6.5 using 10% NaOH (w/v) before sterilization. The cultivation was performed in an anaerobic thermostat shaker at 37 °C and 160 rpm, and the pH of the medium was monitored using a pH meter (SevenExcellence-S470, Mettler-Toledo, Switzerland) and adjusted to 5.5 or 6.5 by continuous titration with 10% NaOH (w/v) using an injection pump (TYD02–10, Leadfluid Technology Co., Ltd., Baoding, China). Three replicates were set for each treatment.
Sample collection
Cell growth were monitored by measuring OD values at 600 nm every hour using SpectraMax M5 plate reader (Molecular Devices Corporation, USA). The cultures of each sample were harvested in duplicate when they reached the exponential growth phase. A portion of the samples was quickly placed in liquid nitrogen and stored at − 80 °C for enzyme activity assay, while the remaining samples were centrifuged at 13,400×g for 2 min at 4 °C. The obtained cell pellets were quickly frozen in liquid nitrogen for 15 min, then stored at − 80 °C for further RNA isolation; the supernatant was filtered using a 0.22 μm filter membrane and stored at − 80 °C for the determination of organic acids.
Determination of organic acids and enzyme activity
A high performance liquid chromatographer (HPLC, Shimadzu, Japan) equipped with an acclaim OA column (Sepax Carbomix H-NP) and a UV detector was used to determine concentrations of organic acids (lactic acid, formic acid, and acetic acid) in the supernatant. The column temperature was kept at 55 °C, and the mobile phase was 2.5 mM H2SO4 with the flow rate set at 0.5 mL/min. Organic acids were then measured with a UV detector set at 210 nm. Commercial assay kits (Comin Biotechnology Co., Ltd., Suzhou, China) were used to measure the activities of α-amylase (Cat. No. DFMA-1-Y), lactate dehydrogenase (Cat. No. LDH-1-Y), and acetate kinase (Cat. No. ACK-1-Y). Before the measurement, 2 mL of bacterial fluid was centrifuged at 8000×g for 10 min at 4 °C and the cell pellets were resuspended in the extracting solution from the kit. To lyse the bacterial cells, the suspension was then sonicated on an ice bath using a VCX-130 Sonicator (Sonics, USA) for 10 min with 30 s pulse on and 30 s pulse off, at 100 W. Unbroken cells were removed by centrifugation at 8000×g for 10 min at 4 °C, and the supernatant was collected for the determination of enzyme activities according to the corresponding kit instructions.
RNA extraction and RT-qPCR analysis
Before RNA extraction, 200 μL of bacterial fluid was centrifuged at 13,400×g for 2 min at 4 °C and the cell pellets were incubated with 180 μL of lysozyme solution (3 mg/mL) at room temperature for 10 min to break the cell walls. Total RNA was extracted from S. bovis S1 wild-type and mutant strains using the RNAprep Pure Bacteria Kit (DP430, Tiangen Biotech Co. Ltd., Beijing, China) according to the manufacturer’s instructions. The quantity and quality of total RNA were evaluated using Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA) and agarose gel electrophoresis, respectively. Next, total RNA was reverse-transcribed to cDNA in a 20 μL reaction mixture using the Primer-Script™ Reagent Kit (TaKaRa Biotechnology Co. Ltd., Dalian, China) according to the manufacturer’s instructions. The reverse transcriptase quantitative PCR (RT-qPCR) was performed with the ABI Step-One-Plus RT-PCR system (ABI 7500, Applied Biosystems, Foster City, CA) using the TB Green Premix Ex Taq™ II Kit (TaKaRa Biotechnology Co. Ltd., Dalian, China). The reaction was first performed in a 20 μL reaction solution containing 10 μL 2 × TB Green Premix Ex Taq II, 1.6 μL primer, 1 μL cDNA, 0.4 μL 50 × ROX, and 7.0 μL RNase-free water. The RT-qPCR conditions were as follows: 95 °C for 30 s, followed by 40 cycles of the amplification at 95 °C for 5 s and 60 °C for 34 s. Primers of α-amy (gene encoding α-amylase), ldh (gene encoding lactate dehydrogenase), pfl (gene encoding pyruvate formate lyase), and ack (gene encoding acetate kinase) were designed using Primer-Blast and the 16S rRNA was used as an internal reference gene (Table 1). Relative gene expression was normalized to expression of the 16S rRNA gene and calculated using the 2-△△CT method.
Data analysis
Results were expressed as means and SEMs. All data were analyzed as a 2 × 2 factorial design by ANOVA using GLM procedures of SPSS (SPSS 25.0, IBM, USA) software. The statistical model included the effects of strain (wild-type strain or ccpA-knockout strain) and pH (6.5 or 5.5), as well as their interaction. Graphs were drawn using GraphPad Prism 8. The logistic function model [24] was used to non-linearly fit the growth curves of bacteria. The specific model is: “Ln(yt/y0) = D/(1 + exp.((4 × μmax/D)(λ-t) + 2))”, where yt is OD value at time t, y0 is initial OD value, t is hour of the incubation, μmax is the maximum specific growth rate, D is the limit of ln (yt/y0), and λ is the growth lag period (h).
Results
Verification of ccpA gene knockout
The results of PCR verification of ccpA gene knockout are shown in Fig. 1. Figure 1A shows the expected sizes and targets of the PCR products, which was confirmed by the results of gel electrophoresis as shown in Fig. 1B (the full-length picture is provided in the Supplementary file). These results indicated that the ccpA gene was deleted successfully in the ccpA-knockout strain. In addition, the DNA sequencing results of wt2 and ko2 further confirmed that the ccpA deletion strain was constructed.
The result of PCR verification for wild-type strain (wt) and ccpA-knockout strain (ko) of S. bovis S1. A Diagram of PCR verification for genomic structure of wild-type strain (wt) and ccpA-knockout strain (ko). ccpA 1 was designed based on the internal sequence of the ccpA gene, with a size of about 426 bp; ccpA 2 was designed across the upstream and downstream sequences of the ccpA gene, with a size of about 1827 bp for wild-type strain and 1985 bp for ccpA-knockout strain. B The result of the PCR verification. wt1: a 426 bp PCR product of ccpA 1 amplified in wild-type strain; ko1: no PCR product of ccpA 1 amplified in ccpA-knockout strain; wt2: a 1827 bp PCR product of ccpA 2 amplified in wild-type strain; ko2: a 1985 bp PCR product of ccpA 2 amplified in ccpA-knockout strain
Growth characteristics
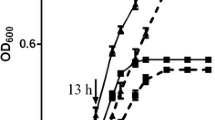
Figure 2 shows the growth curve of S. bovis S1wild-type strain and ccpA deletion strain at different pH. When the pH was 6.5, the wild-type strain reached the stationary phase at 3 h and its OD value was 0.59, while the ccpA-deficient strain reached the stationary phase at 5 h and its OD value was 0.56. Under the pH of 5.5, the wild-type strain reached the stationary phase at 6 h and its OD value was 0.55, while the ccpA-deficient strain reached the stationary phase at 7 h and its OD value was 0.53.
Nonlinear fitting of the growth curves of S. bovis S1wild-type strain and ccpA-deficient strain at different pH were constructed using the logistic function model. Results showed that the R2 of the fitting equations were all above 0.99. The corresponding fitting parameters are shown in Table 2. There was a significant interaction between strain and pH for the growth lag period (λ) (P < 0.01) and maximum specific growth rate (μmax) (P < 0.05). Compared with the wild-type strain at pH 6.5, the μmax of other groups were reduced (P < 0.05), and the ccpA-deficient strain at pH 5.5 had the lowest μmax (P < 0.05). The λ values of ccpA-deficient strain at both pH were increased compared with those of the wild-type strain, and the ccpA-deficient strain at pH 5.5 had the highest λ (P < 0.01).
Organic acids production
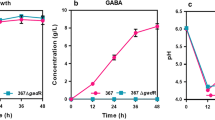
Table 3 shows the organic acid production characteristics of S. bovis S1 wild-type strain and ccpA deletion strain at different pH. Results indicated that both strains produced lactic acid, formic acid, and acetic acid at different pH. Among them, lactic acid made up the largest proportion of organic acids followed by formic acid, with acetic acid being the least abundant. The interaction between strain and pH affected the concentration of total organic acids (P < 0.05) and acetic acid (P < 0.01), and the acetic acid molar percentage to total organic acids (P < 0.05). Compared with the wild-type strain at pH 6.5, the total organic acids concentrations of ccpA-deficient strain at pH 6.5 were increased, but those of both strains at pH 5.5 were decreased. The acetic acid concentration and its molar percentage to total organic acids of ccpA-deficient strain were higher than those of wild-type strain at both pH 6.5 and pH 5.5, with the highest at pH 6.5. The significant strain × pH interaction was not observed for lactic acid and formic acid concentration, and their molar percentages to total organic acids. Deletion of ccpA gene decreased the lactic acid concentration (37.38 vs 40.15 mM) and its molar percentage to total organic acids (73.83% vs 81.66%), while increased the formic acid concentration (8.07 vs 5.46 mM) and molar percentage (15.93% vs 11.10%) compared to the wild-type strain (P < 0.01). Strains at pH 5.5 had decreased concentrations of lactic acid (37.59 vs 39.94 mM; P < 0.01) and formic acid (6.51 vs 7.01 mM; P < 0.01), while an increased lactic acid molar percentage to total organic acids (78.06% vs 77.43%; P < 0.05) compared to those at pH 6.5.
Enzyme activity and gene expression
Table 4 shows the enzyme activities of S. bovis S1 wild-type strain and ccpA-deficient strain at different pH. The interaction between strain and pH did not affect the activities of α-amylase and lactate dehydrogenase. Lower activities of α-amylase (32.74 vs 37.07 U/L) and lactate dehydrogenase (18.75 vs 22.06 U/L) were observed for ccpA-deficient strain compared to the wild-type strain (P < 0.01), and higher activities of these two enzymes (38.67 vs 31.14 U/L and 21.65 vs 19.16 U/L) were observed at pH 5.5 than at pH 6.5 (P < 0.01). There was a significant interaction between strain and pH for the activity of acetate kinase, which was the highest in the ccpA-deficient strain at pH 6.5 (P < 0.05).
Figure 3 shows results of the relative expressions of α-amy, ldh, pfl, and ack of S. bovis S1 wild-type strain and ccpA-deficient strain at different pH. The strain × pH interaction was significant for the relative expression of ldh (P < 0.05) and α-amy (P < 0.01), both of which were the highest in the wild-type strain at pH 5.5 and the lowest in the ccpA-deficient strain at pH 6.5. The interaction between strain and pH did not affect the relative expression of pfl and ack. The relative expressions of pfl and ack in the ccpA-deficient strain were increased compared to the wild-type strain (P < 0.01). The lower pH improved the relative expression of pfl (P < 0.01), while did not affect the relative expression of ack.
Relative mRNA expressions of α-amy, ldh, ack and pfl in wild-type strain (wt) and ccpA-knockout strain (ko) of S.bovis S1 at different pH. Values are the means (n = 3), with standard deviation indicated by vertical bars. α-amy, α-amylase; ldh, lactate dehydrogenase; ack, acetokinase; pfl, pyruvate formate lyase
Discussion
Ruminal pH generally fluctuates within a physiological range of about 5.5–7.0 in a 24-h period, which is driven by the amount of fermentable carbohydrate in each meal, innate buffering capacity of animals, and the absorption rate of organic acids [25, 26]. However, the rate of organic acids production is greater than absorption and the buffering capacity is also limited by inadequate salivary secretion when ruminants consume a large amount of rapidly fermentable (non-fiber) carbohydrates. As a result, ruminal pH will drop below its physiological level, which influences microbial composition and fermentation [3, 27]. It is worth noting that pH is an important environmental factor for bacteria, which not only affects the bacterial growth and fermentation rate, but also the final yield and purity of fermentation products [28]. In the rumen, the growth and organic acid fermentation pathways of most bacterial are affected by ruminal pH [27, 29], even for acid-tolerant species such as S. bovis and Lactobacillus [7].
S. bovis can grow in the pH range of 4.5–6.7 with the highest growth rate at pH 6.4 [6]. In this study, the maximum specific growth rate of S. bovis S1 wild-type strain was greater at pH 6.5 than pH 5.5, indicating that the low pH inhibited the growth of S. bovis S1. This finding is consistent with the characteristic of this strain found by Chen et al. [11], and ccpA deletion does not alter this characteristic. The absence of ccpA, at both pH 6.5 and 5.5, caused an extended lag phase and decreased maximum specific growth rate of S. bovis S1, which was consistent with the results obtained in other lactic acid-producing bacteria, such as Lactobacillus bulgaricus [21] and Lactobacillus casei [30]. However, the growth differences between the wild-type strain and ccpA-disrupted mutant of S. bovis 12 U1 have not been previously observed [12], which is inconsistent with present findings. This discrepancy may be attributed to the different growth and metabolic functions of strains isolated from different environments, even if they belong to the same species [31]. Besides, the significant interaction between strain and pH was observed for λ and μmax in this study, which caused a lowest maximum specific growth rate and a longest lag phase in the ccpA-deficient strain at pH 5.5. The finding suggests a possible synergic effect between low pH and ccpA deletion on suppressing the growth of S. bovis S1, which is probably because that both deletion of ccpA and low pH could inhibit its growth, and the gene expression of ccpA in S. bovis S1 wild-type strain is influenced by the pH [11]. The results of present study imply that deletion ccpA gene would have a better effect on controlling the overgrowth of S. bovis S1 in rumen when ruminal pH is low, which need to be verified in the further study. We did not observe significant differences in maximum specific growth rate between wild-type strain at pH 5.5 and ccpA-disrupted mutant at pH 6.5, indicating that ccpA deletion inhibited the growth of S. bovis S1 only if the strain grown at the same pH condition.
The organic acid fermentation pattern of S. bovis is regulated by extracellular pH and growth rate [32, 33]. An earlier study found that at pH 6.7, lactic acid was the primary fermentation product when S. bovis JB1 was grown with a relatively fast growth rate, while it changed to formic acid, acetic acid and ethanol fermentation when the strain was grown slower in continuous culture [34]. As the pH dropped to 4.7, S. bovis JB1 mainly produced lactic acid even at slow growth rate, which is because the intracellular pH at this condition is similar to the optimal pH of S. bovis LDH [8, 34, 35]. However, the fermentation pattern of S. bovis S1 in the present study did not change when the extracellular pH dropped from 6.5 to 5.5, and the molar percentage of lactic acid is more than 80% of total organic acids at both pH, which is similar to the outcome previously observed in our lab [11]. The use of a batch culture and possibly insufficiently low extracellular pH of the current study may provide some explanation for the unaltered fermentation pattern of S. bovis S1 as extracellular pH dropped. Although the fermentation pattern was not affected by the extracellular pH in the present study, low pH reduced the final concentrations of lactic acid, formic acid and total organic acids of S. bovis S1 wild-type strain. Similarly, this pattern of changes was also found after ccpA knockout. The production of organic acids is associated with the specific activity and amount of enzymes. In Streptococcal, the LDH has a requirement for FDP [36], and the concentration of FDP produced by S. bovis S1 at pH 5.5 is lower than pH 6.5 [11], which may be the reason for the decreased lactic acid production of this study even though the activity of LDH and the ldh gene expression in both strains were significantly higher at pH 5.5 than pH 6.5.
Inactivation of the ccpA gene significantly decreased lactic acid production, and increased the production of formic acid and acetic acid of S. bovis S1 growing at both pH 6.5 and pH 5.5. The findings are consistent with the results of Asanuma et al. [12]. Subsequently, we further analyzed the activity and gene expression of related enzymes involved in the organic acids production pathway. The activity and gene expression of enzyme for lactic acid production (LDH) were reduced, and enzymes for formic acid and acetic acid production (PFL and ACK) were increased in the mutant strain compared to the wild-type strain. This observation may explain the altered production of organic acids in ccpA-disrupted mutants. Similar results were found in other lactic acid bacteria. For example, Asanuma et al. [12] found that the ratio of formic acid to lactic acid in S. bovis 12 U1 increased significantly after ccpA gene deletion, and this change was directly associated with changes in related enzyme activities and gene expression. The transcription of the las operon encoding phosphofructokinase (pfk), pyruvate kinase (pk), and lactate dehydrogenase (ldh) in L. lactis was reduced by 75% in the ccpA deletion mutant strain compared to the wild-type strain, which increased ethanol and acetic acid and decreased lactic acid production [37]. Moreover, the activities of LDH, PK, and PFK in L. bulgaricus were significantly reduced after ccpA gene deletion, which resulted in reduced lactic acid yield [21]. In S. mutans, CcpA has been demonstrated to regulate the transcription of ackA (acetate kinase A) and pta (phosphotransacetylase) in response to pH [22]. In the present study, there was a significant interaction between strain and pH for the concentration of total organic acids and acetic acid production, which led to the highest total organic acids and acetic acid production in ccpA-disrupted mutant at pH 6.5. Moreover, a significant interaction between strain and pH was observed for the activity of acetate kinase and gene expressions of ldh and α-amy. The interaction effect between pH and ccpA gene deletion on these indicators is possibly caused by the fact that the expression of ccpA gene is regulated by extracellular pH in S. bovis S1 [11]. However, further experiment will be needed to verify this speculation.
Conclusion
Low pH inhibits the growth of S. bovis S1, and lowers the production of organic acids but does not alter the production pattern despite the increased activities of α-amylase and LDH. The deletion of ccpA gene inhibits the growth of S. bovis S1 and regulates the organic acid fermentation pattern towards lower lactic acid and higher formic acid and acetic acid, suggesting that CcpA is probably involved in the carbon metabolism of S. bovis S1. Moreover, there seems to be an additive effect between pH and ccpA deletion on regulating the growth and organic acids production of S. bovis S1. The mechanism underlying this interaction remains to be clarified.
Availability of data and materials
The sequence data during the current study are available in the [figshare] repository, [https://doi.org/10.6084/m9.figshare.14779206].
Abbreviations
- α-amy :
-
α-amylase
- ack :
-
Acetic acid kinase
- CcpA:
-
Catabolite control protein A
- cre :
-
Catabolite response element
- FDP:
-
Fructose-1,6-diphosphate
- HPr:
-
Heat-stable protein
- ldh :
-
Lactate dehydrogenase
- OD:
-
Optical density
- pfl :
-
Pyruvate formate lyase
- pta :
-
Phosphotransacetylase
- SEM:
-
Standard error of mean
References
Mao SY, Zhang RY, Wang DS, Zhu WY. Impact of subacute ruminal acidosis (SARA) adaptation on rumen microbiota in dairy cattle using pyrosequencing. Anaerobe. 2013;24:12–9.
Pan X, Xue F, Nan X, Tang Z, Wang K, Beckers Y, et al. Illumina sequencing approach to characterize thiamine metabolism related bacteria and the impacts of thiamine supplementation on ruminal microbiota in dairy cows fed high-grain diets. Front Microbiol. 2017;8:1818.
Nagaraja TG, Titgemeyer EC. Ruminal acidosis in beef cattle: the current microbiological and nutritional outlook. J Dairy Sci. 2007;90(Suppl 1):E17–38.
McAllister TA, Cheng KJ, Rode LM, Forsberg CW. Digestion of barley, maize, and wheat by selected species of ruminal bacteria. Appl Environ Microbiol. 1990;56(10):3146–53.
Wang HR, Pan XH, Wang C, Wang MZ, Yu LH. Effects of different dietary concentrate to forage ratio and thiamine supplementation on the rumen fermentation and ruminal bacterial community in dairy cows. Anim Prod Sci. 2015;55(2):189–93.
Therion JJ, Kistner A, Kornelius JH. Effect of pH on growth rates of rumen amylolytic and lactilytic bacteria. Appl Environ Microbiol. 1982;44(2):428–34.
Russell JB, Dombrowski DB. Effect of pH on the efficiency of growth by pure cultures of rumen bacteria in continuous culture. Appl Environ Microbiol. 1980;39(3):604–10.
Asanuma N, Hino T. Effects of pH and energy supply on activity and amount of pyruvate formate-lyase in Streptococcus bovis. Appl Environ Microbiol. 2000;66(9):3773–7.
Asanuma N, Hino T. Fructose bisphosphate aldolase activity and glycolytic intermediate concentrations in relation to lactate production in Streptococcus bovis. Anaerobe. 2002;8(1):1–8.
Chen L, Luo Y, Wang H, Liu S, Shen Y, Wang M. Effects of glucose and starch on lactate production by newly isolated Streptococcus bovis S1 from Saanen goats. Appl Environ Microbiol. 2016;82(19):5982–9.
Chen L, Liu S, Wang H, Wang M, Yu L. Relative significances of pH and substrate starch level to roles of Streptococcus bovis S1 in rumen acidosis. AMB Express. 2016;6:80.
Asanuma N, Yoshii T, Hino T. Molecular characterization of CcpA and involvement of this protein in transcriptional regulation of lactate dehydrogenase and pyruvate formate-lyase in the ruminal bacterium Streptococcus bovis. Appl Environ Microbiol. 2004;70(9):5244–51.
Nicholson WL, Park YK, Henkin TM, Won M, Weickert MJ, Gaskell JA, et al. Catabolite repression-resistant mutations of the Bacillus subtilis alpha-amylase promoter affect transcription levels and are in an operator-like sequence. J Mol Biol. 1987;198(4):609–18.
Turinsky AJ, Grundy FJ, Kim J-H, Chambliss GH, Henkin TM. Transcriptional activation of the Bacillus subtilis ackA gene requires sequences upstream of the promoter. J Bacteriol. 1998;180(22):5961–7.
Moir-Blais TR, Grundy FJ, Henkin TM. Transcriptional activation of the Bacillus subtilis ackA promoter requires sequences upstream of the CcpA binding site. J Bacteriol. 2001;183(7):2389–93.
Jones BE, Dossonnet V, Kuster E, Hillen W, Deutscher J, Klevit RE. Binding of the catabolite repressor protein CcpA to its DNA target is regulated by phosphorylation of its corepressor HPr. J Biol Chem. 1997;272(42):26530–5.
Lorca GL, Chung YJ, Barabote RD, Weyler W, Schilling CH, Saier MH. Catabolite repression and activation in Bacillus subtilis: dependency on CcpA, HPr, and HprK. J Bacteriol. 2005;187(22):7826–39.
Chauvaux S. CcpA and Hpr(ser-p): mediators of catabolite repression in Bacillus subtilis. Res Microbiol. 1996;147(6–7):518–22.
Schumacher MA, Seidel G, Hillen W, Brennan RG. Structural mechanism for the fine-tuning of CcpA function by the small molecule effectors glucose 6-phosphate and fructose 1,6-bisphosphate. J Mol Bio. 2007;368(4):1042–50.
Chen C, Wang L, Lu Y, Yu H, Than H. Comparative transcriptional analysis of Lactobacillus plantarum and its CcpA-knockout mutant under galactooligosaccharides and glucose conditions. Front Microbiol. 2019;10:1584.
Li C, Sun JW, Zhang GF, Liu LB. Effect of the absence of the ccpa gene on growth, metabolic production, and stress tolerance in lactobacillus delbrueckii ssp bulgaricus. J Dairy Sci. 2016;99(1):104–11.
Kim JN, Burne RA. CcpA and Cody coordinate acetate metabolism in Streptococcus mutans. Appl Environ Microbiol. 2017;83(7):e03274–16.
Asanuma NK, Kanada Y, Arai K, Yoshizawa IT, Nino T. Molecular characterization and significance of phosphoenolpyruvate carboxykinase in a ruminal bacterium, Streptococcus bovis. J Gen Appl Microbiol. 2010;56:121–7.
Perni S, Andrew PW, Shama G. Estimating the maximum growth rate from microbial growth curves: definition is everything. Food Microbiol. 2005;22(6):491–5.
Krause KM, Oetzel GR. Understanding and preventing subacute ruminal acidosis in dairy herds: a review. Anim Feed Sci Tech. 2006;126(3–4):215–36.
Nocek JE, Allman JG, Kautz WP. Evaluation of an indwelling ruminal probe methodology and effect of grain level on diurnal pH variation in dairy cattle. J Dairy Sci. 2002;85(2):422–8.
Hook SE, Steele MA, Northwood KS, Dijkstra J, France J, Wright A-DG, et al. Impact of subacute ruminal acidosis (SARA) adaptation and recovery on the density and diversity of bacteria in the rumen of dairy cows. FEMS Microbiol Ecol. 2011;78(2):275–84.
Zhu Y, Yang ST. Effect of pH on metabolic pathway shift in fermentation of xylose by Clostridium tyrobutyricum. J Biotechnol. 2004;110(2):143–57.
Cook GM, Russell JB. The effect of extracellular pH and lactic acid on pH homeostasis in Lactococcus lactis and Streptococcus bovis. Curr Microbiol. 1994;28(3):165–8.
Esteban CD, Kerstin M, Monedero V, et al. Complementation of △CcpA mutant of Lactobacillus casei with CcpA mutants affected in the DNA- and cofactor-binding domains. Microbiology. 2004;150(3):613–20.
Wang SL, Wu Q, Nie Y, Wu JF, Xu Y. Construction of synthetic microbiota for reproducible flavor compound metabolism in Chinese light-aroma-type liquor produced by solid-state fermentation. Appl Environ Microbiol. 2019;85(10):e03090–18.
Chow JM, Russell JB. Effect of ionophores and pH on growth of Streptococcus bovis in batch and continuous culture. Appl Environ Microbiol. 1990;56(6):1588–93.
Asanuma N, Iwamoto M, Hino T. Structure and transcriptional regulation of the gene encoding pyruvate formate-lyase of a ruminal bacterium, Streptococcus bovis. Microbiology-Uk. 1999;145:151–7.
Russell JR, Hino T. Regulation of lactate production in Streptococcus bovis: a spiraling effect that contributes to rumen acidosis. J Dairy Sci. 1985;68(7):1712–21.
Russell JB, Dombrowski DB. Effect of pH on the efficiency of growth by pure cultures of rumen bacteria in continuous culture. Appl Environ Microbiol. 1980;39(3):604–10.
Wolin MJ. Fructose-1,6-diphosphate requirement of Streptococcal lactic dehydrogenases. Science. 1964;146:775–7.
Luesink E. Transcriptional activation of the glycolytic las operon and catabolite repression of the gal operon in Lactobacillus lactis are mediated by the catabolite contron protein CcpA. Mol Microbiol. 1998;30(4):789–98.
Acknowledgements
Not Applicable.
Funding
This research was funded by the National Natural Science Foundation of China (NSFC No. 31872988, No. 31572429) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PADA).
Author information
Authors and Affiliations
Contributions
YQJ and HRW designed the research; YQJ, YTF and YZ performed research; CW contributed analytic tools; YQJ and CW analyzed data; YQJ wrote the manuscript; CW and HRW provided critical comments, ME revised the language of the manuscripy. All the authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Jin, Y., Wang, C., Fan, Y. et al. Regulation of CcpA on the growth and organic acid production characteristics of ruminal Streptococcus bovis at different pH. BMC Microbiol 21, 344 (2021). https://doi.org/10.1186/s12866-021-02404-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12866-021-02404-x