Abstract
Background
Klebsiella pneumoniae is the most frequent KPC-producing bacteria. The blaKPC gene is frequently embedded in Tn4401 transposon, and less frequently in non-Tn4401 elements (NTEKPC) variants I-III. The first case of KPC in the UC-CHRISTUS Clinical Hospital was detected in Pseudomonas aeruginosa. Soon after this event, KPC was detected in 2 additional Pseudomonas aeruginosa, 3 Escherichia coli, 3 Enterobacter cloacae, 3 Klebsiella pneumoniae, and 1 Citrobacter freundii, isolated from 6 different patients. We aimed to elucidate the possible mechanisms of genetic transfer and dissemination of the blaKPC gene among isolates of this multispecies outbreak. A molecular epidemiology analysis of the above mentioned clinical isolates (n = 13) through Multi-Locus Sequence Typing, plasmid analysis, Pulsed-Field Gel-Electrophoresis, and Whole-genome sequencing (WGS) was performed.
Results
High-risk sequence types were found: K. pneumoniae ST11, P. aeruginosa ST654, and E. cloacae ST114. All enterobacterial isolates were not clonal except for 3 E. coli isolated from the same patient. WGS analysis in 6 enterobacterial isolates showed that 4 of them had blaKPC embedded in a novel variant of NTEKPC designated NTEKPC-IIe. Upstream of blaKPC gene there was a 570 pb truncated blaTEM-1 gene followed by an insertion sequence that was 84% similar to ISEc63, a 4473 bp element of the Tn3 family. Downstream the blaKPC gene there was a truncated ISKpn6 gene, and the inverted repeat right sequence of Tn4401. The ISec63-like element together with the blaKPC gene plus Tn4401 remnants were inserted in the Tra operon involved in conjugative transfer of the plasmid. This NTE was carried in a broad host-range IncN plasmid. P. aeruginosa isolates carried blaKPC gene embedded in a typical Tn4401b transposon in a different plasmid, suggesting that there was no plasmid transfer between Enterobacteriaceae and P. aeruginosa as initially hypothesized.
Conclusions
Most enterobacterial isolates had blaKPC embedded in the same NTEKPC-IIe element, suggesting that this multispecies KPC outbreak was due to horizontal gene transfer rather than clonal spread. This poses a greater challenge to infection control measures often directed against containment of clonal spread.
Similar content being viewed by others
Background
Carbapenem-resistant bacteria are a serious public health threat worldwide. Carbapenems are used as last resort antibiotics in infections caused by multidrug-resistant bacteria and carbapenemases are threatening this valuable therapeutic agent [1]. KPC is the most clinically significant class A carbapenemase because KPC-producing bacteria are susceptible to only a few antibiotics (colistin, aminoglycosides, tigecycline, and ceftazidime/avibactam) and patients infected with them have poor outcomes [2]. KPC carbapenemase was first reported in the United States in 1996 in a Klebsiella pneumoniae isolate [3] and a few years later it became endemic in regions like United States, Italy, Israel and Colombia [1]. To date, there are more than 30 KPC variants described, with KPC-2 and KPC-3 being the most frequently encountered [4]. K. pneumoniae is by far the most frequent species carrying blaKPC. However, it has been described in several other species of Enterobacteriaceae [5]. Albeit less frequently, KPC-producing Pseudomonas aeruginosa has also been described [6, 7].
The worldwide dissemination of KPC-producing K. pneumoniae has been associated with the successful spread of a specific genetic lineage designated clonal group 258 (CG258). This CG contains 43 different sequence types (STs), with ST258, and ST512 being the predominant ones. Indeed, ST258 is a high-risk clone responsible for 80% of KPC-producing K. pneumoniae outbreaks in the United States [8]. However, other mechanisms of KPC dissemination have been described, for example horizontal transfer of mobile genetic elements [9]. The KPC-coding gene, blaKPC, is usually found within a Tn4401 transposon, a mobile genetic element derived from the Tn3 transposon family that facilitates its spread [8, 10]. However, the blaKPC gene has also been found in non Tn4401 elements (NTE). The first NTE was described in China in 2007 [11] and additional NTEs with different structures were later described in countries like Argentina [12], Colombia [9], Chile [13], and Brazil [14,15,16]. The blaKPC gene is usually carried on plasmids of different incompatibility groups (Inc). A recent meta-analysis made with 435 KPC-bearing plasmids, showed that the most frequent incompatibilty group was IncN [4]. These elements have contributed to the dissemination of KPC within K. pneumoniae and other bacterial species.
The first KPC-producing strain in Chile was detected in 2012 in a K. pneumoniae isolated from a patient traveling from Italy [17]. Since then, KPC has been found in several species of Enterobacteriaceae throughout the country [13]. Surprisingly, the first KPC-producing strain in our University Hospital (Clinical Hospital of Red de Salud UC-CHRISTUS) was detected in 2015 in a P. aeruginosa strain recovered from an adult patient at the intensive care unit. Shortly after the detection of this first KPC-producing P. aeruginosa, KPC was subsequently found in several species of Enterobacteriaceae in various hospital units, in a relatively short period (2 months) [7]; the event fulfilled the classical outbreak definition of The World Health Organization [18]. This observation led us to hypothesize that the blaKPC gene might have been transferred from the index KPC-P. aeruginosa to Enterobacteriaceae through horizontal gene transfer. Therefore, we aimed to elucidate the possible mobile genetic elements harboring KPC and possible mechanisms of genetic transfer and dissemination of the blaKPC gene among bacterial isolates associated with this multi-species outbreak.
Results
Isolates included in the outbreak analysis
A detailed molecular analysis of 3 P. aeruginosa and 10 Enterobacteriaceae associated with the outbreak, obtained from six different patients was performed. The index KPC-P. aeruginosa (Pae-1) was isolated from P1 in the ICU (Table 1) and it was also found to carry blaVIM (Table 2). The first KPC-producing K. pneumoniae (Kpn-3) was isolated almost 20 days later from P3 in the step-down unit. One month later, three KPC-producing isolates were obtained from the same patient (P3) in the same unit: K. pneumoniae (Kpn-4), E. coli (Eco-5) and E. cloacae (Ecl-6). Eleven days later three additional KPC-producing E. coli (Eco-7, Eco-8) and one E. cloacae (Ecl-9) were again recovered from P3. At the same time of isolation of the first KPC-producing K. pneumoniae, a new KPC-producing K. pneumoniae (Kpn-10) was recovered from P4 in the surgical unit, and two KPC-producing isolates of C. freundii (Cfr-11) and one E. cloacae (Ecl-12) respectively, were recovered from P5 in the pediatric ICU. Of note, the two isolates from the pediatric care unit also carried the blaVIM gene (Table 2). Two additional P. aeruginosa isolates were included: one of them was the following consecutive VIM-positive P. aeruginosa (Pae-2) isolated from P2 in the emergency department 24 days after the index case, and the second was Pae-13, isolated more than 2 months after the index case in the same ICU, but from a different patient (P6) (Tables 1 and 2). All isolates carried blaKPC-2, except for Pae-2, that only harbored blaVIM. All P. aeruginosa isolates carried the blaVIM-2 metallo-beta-lactamase. In contrast, Cfr-11 and Ecl-12 carried blaVIM-1 (Table 2).
Molecular epidemiology of the isolates
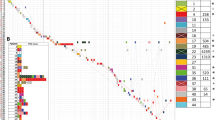
PFGE analysis determined that isolates Pae-1 and Pae-13 were clonal (Fig. 1). Of note, those isolates were recovered from different patients and time points, but within the same ICU. All E. coli isolates were recovered from the same patient (P3), but only Eco-7 and Eco-8 were clonal. Eco-5, obtained 11 days before in a different hospital unit was a different strain. Similar to the case with Eco-5, E. cloacae isolates were not clonally related, despite two of them were recovered from the same patient (P3) within 11 days (Ecl-6 and Ecl-9), but in two different units. K. pneumoniae isolates were not clonal despite two of them, Kpn-3 and Kpn-4 were recovered from the same patient (P3). The unprocessed gel photographs are shown in Supplementary Fig. S1.
Dendrograms obtained from the analysis of PFGE patterns. Panels correspond to E. coli (a), E. cloacae (b), K. pneumoniae (c), and P. aeruginosa (d) isolates. Dendrograms were constructed using the Dice coefficient and unweighted pair group method with arithmetic mean (UPGMA). The 95% similarity cut-off is indicated with a dashed line. A standard ATCC strain of each species was included in every analysis. Pulsotypes for each isolate are indicated in the right column. Lanes that were non-adjacent in the original gel were cropped to be positioned according to dendrogram order. The unprocessed gel photographs are shown in Supplementary Fig. S1
In terms of our MLST results, all three E. coli belonged to the ST378. K. pneumoniae isolates were ST11 (Kpn-3 and Kpn-4) and ST25 (Kpn-10). Both E. cloacae recovered from P3 were ST45, whereas Ecl-12 was ST114. The KPC-harboring P. aeruginosa isolates isolated from the ICU were ST654 and Pae-2 was ST282 (Table 2).
Plasmid analysis
P. aeruginosa isolates (Pae-1, Pae-2 and Pae-13) did not carry plasmids of any of the incompatibility groups most frequently found among Pseudomonas species, that were sought through PCR [19]. All but one (Ecl-12) enterobacterial isolates carried an IncN type plasmid (Table 2). Also, all isolates recovered from P3 except for the first K. pneumoniae isolate Kpn-3, carried a plasmid of the IncA/C type. The E. cloacae isolate Ecl-6 recovered from this patient additionally carried plasmids of the IncF1A and IncF1B types. Plasmids were extracted and visualized in a 0.75% agarose gel (Fig. 2). Typical patterns of relaxed, supercoiled and linear plasmid forms were observed in most isolates. Isolate Ecl-6 carried several plasmids, in accordance to PCR results. Although no plasmids were amplified in isolate Ecl-12 through PCR using a set of primers directed to the plasmids usually found in Enterobacteriaceae, it exhibited a band of less than 4 Kb (Fig. 2).
Genetic environment of bla KPC gene and plasmid
WGS was performed in 7/13 isolates, including KPC-P. aeruginosa Pae-13 and 6 isolates from P3: Kpn-3, Ecl-9, Eco-8, Eco-5, Eco-7, and Ecl-6.
The blaKPC-containing contig of Pae-13 was 35,034 bp long (Genbank accession N° MT949191) and it aligned to a 43,660 bp plasmid (pPA2047) isolated in Argentina (Genbank accession N° MN082782, November 2019). Contigs were then assembled using plasmid pPA2047 as the reference sequence and the plasmid depicted in Fig. 3a was obtained. Analysis of the genetic environment showed that Pae-13 harbored blaKPC embedded in an intact Tn4401 transposon of the b isoform (Fig. 3a). This plasmid did not amplify any of the replicons corresponding to incompatibility groups of Pseudomonas species [19] or Enterobacteriaceae [20]. Moreover, it was non-typeable according to PlasmidFinder software [21].
Genetic environment of blaKPC gene in plasmids from P. aeruginosa (a) and enterobacterial isolates (b) obtained through WGS analysis. Plasmid from P. aeruginosa Pae-13 was obtained through assembly against plasmid pPA2047 and the blaKPC gene embedded in Tn4401 is shown in yellow (a). Plasmids from enterobacterial isolates were obtained through assembly against plasmid pEC881_KPC and the blaKPC gene is embedded in a NTEKPC-IIe element (b)
The size of the blaKPC gene-containing contigs in enterobacterial genomes ranged from 18,252 bp to 42,936 bp (Genbank accession N°: MT949189 for Kpn-3; MT949193 for Ecl-6, Ecl-9 and Eco-5; MT949190 for Eco-8; MT949192 for Eco-7). All of them aligned to a 59,373 bp long plasmid of the IncN type named pEC881_KPC recovered in 2013 from a carbapenem-resistant E. coli in Colombia (Genbank accession N° CP019026.1). Contigs were then assembled using plasmid pEC881_KPC as the reference sequence. The assembly retrieved the plasmid sequence with different identity and completeness levels for all six enterobacterial isolates analyzed (Fig. 3b). The blaKPC gene was not embedded in transposon Tn4401 in any of the analyzed enterobacterial genomes (Fig. 3b). In 4 of the 6 isolates, blaKPC was harbored in a variant of NTEKPC designated NTEKPC-IIe. Upstream of blaKPC gene there was a 570 pb truncated blaTEM-1 gene (deletion of 291 pb) followed by an insertion sequence that was 84% similar to ISEc63, a 4473 bp element that belongs to the Tn3 family. This IS contains, a Tn3 family resolvase, a DDE-transposase (contains 3 acidic amino acids, DDE) and a 50 bp inverted repeat left of ISec63 (IRL, Fig. 3b). The DDE-transposase gene had a single nucleotide deletion of G384 that resulted in a frameshift from amino acid 128, producing a truncated protein in the 4 isolates. Downstream the blaKPC gene there was the remnant of an ISKpn6 gene (ΔISKpn6, Fig. 3b), and the inverted repeat right of Tn4401 (IRR, Fig. 3b), traits that are common among most NTEKPC sequences [10]. The ISec63-like element together with the blaKPC gene plus Tn4401 remnants were inserted in the operon containing the genes required for conjugative transfer of the plasmid (TraI, TraD, VirB11, VirB9, VirB8, VirB5, and VirB4) (pink arrows, Fig. 3b). The region located upstream the ISec63-like element was highly variable among the 6 isolates analyzed: Eco-5, Eco-7, and Ecl-6 lack some genes respective to isolates Kpn-3, Ecl-9, and Eco-8. It could also be stated that Ecl-9 and Eco-8 are different from Kpn-3 (Fig. 3b).
No other IS or transposon structure was found in the plasmid. The plasmid contained other genes coding for proteins involved in antibiotic resistance, eg: Sul1 (sulphonamides), QnrB (quinolones), QacEΔ1 (quaternary ammonium compounds), AadA16 and AacA4 (aminoglycosides) (Fig. 3b). Additionally, it harbored genes involved in DNA repair and metabolism (UmuCD operon, DNA-cytosine methyltransferase, antirestriction protein KlcA and EcoRII).
Discussion
Our results show that KPC-producing P. aeruginosa possessed the blaKPC gene in a different plasmid and transposon structure from that found in Enterobacteriaceae, suggesting that there was no plasmid transfer between them as initially hypothesized.
The multispecies outbreak described herein was most likely driven by horizontal plasmid transfer among Enterobacteriaceae species. Indeed, all enterobacterial isolates studied with WGS harbored essentially the same blaKPC-bearing IncN plasmid. Importantly, this genetic element has been described as a conjugative free mobility plasmid between different species [22]. The fact that most enterobacterial isolates of the same species carrying the IncN plasmid were not clonal further supports the horizontal transfer mechanism.
NTEs have been classified based on the genes adjacent to blaKPC gene: type NTE-I has no insertions respective to the first variant found in China, NTE-II has a partial blaTEM gene upstream blaKPC, and NTE-III has an insertion of tnpR (Tn5563)/IS6100 [10]. The Argentinian and previous Chilean NTEKPC variants were of the type NTEKPC-Ia and were identical to that firstly reported in China. A novel NTEKPC-IId element has been recently described in Brazil, in which there is a truncated blaTEM gene upstream the blaKPC gene, and a truncated ISKpn6 gene followed by relE/parE toxin-antitoxin system downstream the blaKPC gene [16]. The NTEKPC-IIe described here is a novel variant based on the presence of an array of vir genes downstream the blaKPC gene, a region that is usually less variable than the upstream region. A detailed genomic analysis done in Colombia with isolates obtained during KPC emergence showed that the first events responsible for KPC dissemination were horizontal transfer of mobile genetic elements carrying blaKPC-2 in the typical Tn4401 transposon and also in NTEKPC elements, followed by introduction of K. pneumoniae ST258 carrying blaKPC-3 exclusively in Tn4401 and its subsequent clonal dissemination [9]. The events described in Argentina illustrate a similar picture: the first KPC-producing bacteria were of different enterobacterial species and non-ST258 K. pneumoniae carrying blaKPC in NTEs, followed by introduction and clonal dissemination of K. pneumoniae ST258 carrying blaKPC-3 in a typical Tn4401 element [12]. The events described in this work are similar to the beginning of the Colombian and Argentinian KPC epidemics: the blaKPC-2 gene embedded in NTEs disseminates among various enterobacterial species and non-ST258 K. pneumoniae. It would be very interesting to analyze recent Chilean isolates to determine if the switch to the predominant ST258 carrying blaKPC-3 in Tn4401 has occurred.
One of the most remarkable limitations of this work is the lack of experimental evidence about the transferability of the IncN plasmid intra and interspecies. Additionally, the WGS method used provided short reads which make it difficult to obtain a circular complete sequence. This could explain the incomplete sequences obtained for some strains like Eco-5 and Ecl-6. Of note, enterobacterial isolates from patients P4 and P5 lost their KPC genes and were not sequenced. These isolates could have lost their KPC-bearing plasmids, maybe because these plasmids were different and less stable than those of isolates from patient P3, that were stably maintained. In fact, the band of less than 4 Kb of isolate Ecl-12 could correspond to a plasmid belonging to a different incompatibility group. It is possible that interspecies plasmid transfer occurred only in patient P3.
Based on our clonality analysis, only one case of intra-hospital transmission could have occurred: Pae-1 and Pae-13, isolated from P1 and P6 respectively; both isolates were clonal and both patients were hospitalized in the same service, although 2 months apart. Infection control measures such as hand-washing, sterilization of medical devices and contact isolation precautions might have failed in this particular case.
Isolates Pae-1 and Pae-13 were ST654, that is a high-risk clone and has been previously reported in Argentina [23] associated with production of KPC carbapenemase. Sequence types determined for E. coli and C. freundii isolates are not high-risk clones and no previous results were found about these STs associated with carbapenemase production. E. cloacae isolates were of the types ST45 and ST114, that have been previously described in other studies associated with the production of ESBLs and carbapenemases, with ST114 being a high-risk clone [24]. None of the K. pneumoniae isolates recovered belonged to the globally disseminated high-risk clone ST258; two of the isolates were ST11 and one was ST25. However, ST11 is also a clinically relevant genetic lineage, being closely related to ST258 [25]. Although ST25 is less frequent, it is associated with KPC production and it has been previously reported in Chile by the Institute of Public Health (unpublished data). Moreover, ST25 has been described as an hypervirulent clone associated with mucoid phenotype [26].
Conclusions
We describe here a multispecies outbreak of KPC-producing bacteria driven by horizontal gene transfer of blaKPC gene embedded in a novel NTE element, named NTEKPC-IIe. This type of transmission poses a greater challenge to infection control measures often directed against containment of clonal dissemination.
Methods
Clinical isolates
We selected 13 carbapenemase-producing clinical isolates obtained between May and July, 2015, from 6 different patients (P1 – P6) (Table 1). Patients had been admitted to the intensive care unit (ICU), step-down unit, cardiology, surgical and pediatric care units at the Clinical Hospital of Red de Salud UC-CHRISTUS. Isolates species corresponded to K. pneumoniae (Kpn-3, Kpn-4, and Kpn-10), Enterobacter cloacae (Ecl-6, Ecl-9, and Ecl-12), Escherichia coli (Eco-5, Eco-7, and Eco-8), Citrobacter freundii (Cfr-11) and P. aeruginosa (Pae-1, Pae-2, and Pae-13). All included isolates were obtained from surveillance cultures (rectal swabs), as part of an institutional protocol designed to actively screen for carbapenemase-producing bacteria. Such protocol was instituted in 2013 and consisted of monthly rectal swabs performed to all patients hospitalized for more than 5 days in any of the intensive care units of the hospital [27]. Importantly, despite finding many carbapenem-resistant organisms, no carbapenemase-producing bacteria had been recovered before the outbreak described herein [27]. Swabs were plated in chromogenic medium ChromID CARBA (bioMèrieux). Colonies were transferred to Mueller-Hinton agar plates and bacterial species was determined with Matrix-Assisted Laser Desorption/Ionization - Time of Flight (MALDI-TOF) (Bruker-Daltonics, Germany). Carbapenemase production was assessed through the Carba-NP test performed according to CLSI instructions [28].
Detection of carbapenemases genes
Total DNA extraction was performed using the Magna Pure Compact Nucleic acid isolation kit I kit (Roche). The presence of blaKPC and blaVIM genes was determined through PCR using primers listed in Table 3 as previously described [29]. PCR products were purified and sequenced bi-directionally (Macrogen Inc., Korea). The primers used to sequence blaKPC and blaVIM genes are provided in Table 3 [11, 30]. All sequences were corrected and analyzed using Chromas Lite and Clustal Omega online software. To determine the type of KPC and VIM carbapenemases, sequences were compared to the GenBank online database using BLAST software (Basic Local Alignement Search Tool).
Pulsed Field Gel Electrophoresis (PFGE)
Pulsed-field gel electrophoresis (PFGE) of genomic DNA macrorestricted with SpeI or XbaI enzymes was performed to establish a clonal relationship between isolates of the same species, according to the PulseNet protocol of CDC [31]. PFGE conditions were as follows: pulse times ranged from 2 s to 40s for 18 h at 6.0 V/cm at 14 °C. The PFGE profiles obtained were analyzed with GelJ 2.0 software [32] and a dendrogram was constructed using the Dice coefficient (Tolerance 2%) and unweighted pair group method with arithmetic mean (UPGMA). The similarity between band patterns was interpreted according to Tenover criteria [33] setting 95% similarity cut-off values for identifying pulsotypes.
Multi Locus Sequence Typing (MLST)
Amplification and sequencing of 7 housekeeping genes were performed according to Pasteur Institute [34] or PubMLST [35] protocols, and PCR products were sequenced bidirectionally (Macrogen Inc., Korea). Subsequently, the sequences were analyzed using ClustalW Omega online software. The electropherograms were analyzed using Chromas Lite software. Corrected sequences were uploaded to the MLST database to obtain sequence types based on allele combinations.
Plasmid analysis
Identification of plasmid incompatibility groups was performed by multiplex PCR, recognizing incompatibility groups from Enterobacteriaceae and P. aeruginosa as previously described [19, 20].
For plasmid extraction, a single colony of an overnight culture in blood agar plates was resuspended in 2 ml microcentrifuge tubes containing 500 μL distilled water and centrifuged at 3100 g for 10 min. Bacterial pellets were resuspended in 400 μL of cold solution I containing 2 mg/mL lysozyme, 50 mM glucose, 10 mM EDTA, 25 mM Tris-HCl and 0.2 mg/mL RNAse. Samples were stirred vigorously and left at room temperature for 5 min. Eight hundred μL of solution II containing Sodium Hydroxide / Sodium dodecyl sulfate was added, mixed 3 times by inversion, and incubated in ice for 20 min. Six hundred μL of solution III (3 M Sodium Acetate, pH 5.2) was added, vortexed vigorously and incubated for 5 min in ice and then centrifuged at 12,400 g for 15 min. The supernatant (500 μL approx.) was transferred to another tube and mixed with an equal volume of Phenol: Chloroform: Isoamyl alcohol (25: 24: 1), the mixture was centrifuged at 12,400 g for 10 min. Supernatants (500 μL approx.) were transferred to another tube and 2 volumes of cold absolute ethanol were added; it was gently stirred for 2 min and incubated for 35 min at room temperature and centrifuged at 12,400 g for 20 min. Absolute ethanol was removed and 1 volume of cold 70% ethanol was added, subsequently, it was incubated for 5 min and centrifuged for another 5 min at 12,400 g. Finally, ethanol was removed, and the DNA pellet allowed to dry at room temperature. DNA was resuspended in TE (Tris-EDTA) buffer pH 8.0 and stored at − 20 °C. Plasmid DNA was separated on a 0.75% agarose gel electrophoresis for 5 h at 50 V.
Library preparation and DNA sequencing
Unfortunately, when isolates were regrown for genomic DNA extraction and whole-genome sequencing (WGS) isolates from P4 and P5 had lost the plasmid harboring the blaKPC gene (Kpn-10, Cfr-11, and Ecl-12). Isolate Kpn-4 did not lose its KPC gene but its WGS data were low quality, thus it was not included. For this reason, only WGS data of 7 of the 13 original isolates were obtained, and it was performed by Novogene (California, US). A 350 bp insert DNA library was prepared and sequencing was performed in an Illumina Platform PE150. The Q30 obtained was > 90% for all 7 isolates.
Read processing, de novo assembly and annotation of plasmid genomes
Reads were adapter trimmed using Trimmomatic 0.30 with a sliding window quality cutoff of Q15. De novo assembly was performed on samples using plasmidSPAdes as part of the core SPADES version 3.7 package [36]. Genomic annotation of the recovered draft genomes was performed with Prokka tool 1.11 [37]. Annotations were manually reviewed using BLASTP+ against the non-redundant protein NCBI database. Contigs were further aligned against plasmid pEC881_KPC (accession number #CP019026.1) for Enterobacteriaceae and plasmid pPA2047 (accession number MN082782) for P. aeruginosa. Contigs were scaffolded using the MeDuSa open software [38].
Synteny and comparative genomic analyses
Comparisons between individual genomes were performed using BLASTn. Identification of insertions, deletions, and variations in syntenic regions was performed using Easyfig v2.1 [39] using the BLASTn comparison file and gbk files as inputs and calibrating the tBLASTx identity values to a minimum 99% ID for the reference plasmids used. Final visualization and annotations of the aligned contigs ORFs (Open Reading Frames) were made using Geneious vR.10.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request. All data generated or analysed during this study are included in this published article (and its supplementary information files). Genbank accession numbers for deposited data are as follows: MT949189 for Kpn-3; MT949193 for Ecl-6, Ecl-9 and Eco-5; MT949190 for Eco-8; and MT949192 for Eco-7.
Abbreviations
- ATCC:
-
American Type Culture Collection
- CG:
-
Clonal group
- CLSI:
-
Clinical and Laboratory Standards Institute
- EDTA:
-
Ethylenediaminetetraacetic acid
- ICU:
-
Intensive Care Unit
- IRL:
-
Inverrted Repeat Left
- IRR:
-
Inverted Repeat Right
- MALDI-TOF:
-
Matrix Assisted Laser Desorption - Time of Flight Mass Spectrometry
- MLST:
-
Multi-Locus Sequence Typing
- NTE:
-
Non-Tn4401 Element
- ORF:
-
Open Reading Frame
- PFGE:
-
Pulsed-field gel electrophoresis
- ST:
-
Strain Type
- UPGMA:
-
Unweighted Pair Group Method with Arithmetic mean
- WGS:
-
Whole Genome Sequencing
- WT:
-
Wild type
References
Bonomo RA, Burd EM, Conly J, Limbago BM, Poirel L, Segre JA, et al. Carbapenemase-producing organisms: a global scourge. Clin Infect Dis. 2018;66(8):1290–7. https://doi.org/10.1093/cid/cix893.
Wang Q, Zhang Y, Yao X, Xian H, Liu Y, Li H, et al. Risk factors and clinical outcomes for carbapenem-resistant Enterobacteriaceae nosocomial infections. Eur J Clin Microbiol Infect Dis. 2016;35(10):1679–89. https://doi.org/10.1007/s10096-016-2710-0.
Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, et al. Novel Carbapenem-hydrolyzing β-lactamase, KPC-1, from a Carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2001;45(4):1151–61. https://doi.org/10.1128/AAC.45.4.1151-1161.2001.
Brandt C, Viehweger A, Singh A, Pletz MW, Wibberg D, Kalinowski J, et al. Assessing genetic diversity and similarity of 435 KPC-carrying plasmids. Sci Rep. 2019;9(1):11223. https://doi.org/10.1038/s41598-019-47758-5.
Bassetti M, Peghin M. How to manage KPC infections. Ther Adv Infect Dis. 2020;7:204993612091204. https://doi.org/10.1177/2049936120912049.
Villegas MV, Lolans K, Correa A, Kattan JN, Lopez JA, Quinn JP. First identification of Pseudomonas aeruginosa isolates producing a KPC-type Carbapenem-hydrolyzing β-lactamase. Antimicrob Agents Chemother. 2007;51(4):1553–5. https://doi.org/10.1128/AAC.01405-06.
Enberg M, Puente M, Wozniak A, Castillo C, Villagra N, Labarca J, et al. Carbapenemases in Pseudomonas aeruginosa with decreased susceptibility to carbapenems after a decade: from VIM to KPC. Rev Chil Infectol. 2020;37:389–94.
Kitchel B, Rasheed JK, Patel JB, Srinivasan A, Navon-Venezia S, Carmeli Y, et al. Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob Agents Chemother. 2009;53(8):3365–70. https://doi.org/10.1128/AAC.00126-09.
Rojas LJ, Weinstock GM, De La Cadena E, Diaz L, Rios R, Hanson BM, et al. An Analysis of the Epidemic of Klebsiella pneumoniae Carbapenemase-Producing K. pneumoniae: Convergence of Two Evolutionary Mechanisms Creates the “Perfect Storm”. J Infect Dis. 2018;217(1):82–92. https://doi.org/10.1093/infdis/jix524.
Chen L, Mathema B, Chavda KD, DeLeo FR, Bonomo RA, Kreiswirth BN. Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol. 2014;22(12):686–96. https://doi.org/10.1016/j.tim.2014.09.003.
Shen P, Wei Z, Jiang Y, Du X, Ji S, Yu Y, et al. Novel genetic environment of the Carbapenem-hydrolyzing β-lactamase KPC-2 among Enterobacteriaceae in China. Antimicrob Agents Chemother. 2009;53(10):4333–8. https://doi.org/10.1128/AAC.00260-09.
Gomez SA, Pasteran FG, Faccone D, Tijet N, Rapoport M, Lucero C, et al. Clonal dissemination of Klebsiella pneumoniae ST258 harbouring KPC-2 in Argentina. Clin Microbiol Infect. 2011;17(10):1520–4. https://doi.org/10.1111/j.1469-0691.2011.03600.x.
Barría-Loaiza C, Pincheira A, Quezada M, Vera A, Valenzuela P, Domínguez M, et al. Molecular typing and genetic environment of the Bla KPC gene in Chilean isolates of Klebsiella pneumoniae. J Glob Antimicrob Resist. 2016;4:28–34. https://doi.org/10.1016/j.jgar.2016.01.001.
de Souza RC, Dabul ANG, dos Santos Boralli CM, Zuvanov L, da Cunha Camargo IL. Dissemination of blaKPC-2 in an NTEKPC by an IncX5 plasmid. Plasmid. 2019;106:102446. https://doi.org/10.1016/j.plasmid.2019.102446.
Cerdeira LT, Lam MMC, Wyres KL, Wick RR, Judd LM, Lopes R, et al. Small IncQ1 and col-like plasmids harboring bla KPC-2 and non-Tn 4401 elements (NTE KPC -IId) in high-risk lineages of Klebsiella pneumoniae CG258. Antimicrob Agents Chemother. 2019;63(3). https://doi.org/10.1128/AAC.02140-18.
de Lima GJ, Scavuzzi AML, Beltrão EMB, Firmo EF, de Oliveira ÉM, de Oliveira SR, et al. Identification of plasmid IncQ1 and NTEKPC-IId harboring Bla KPC-2 in isolates from Klebsiella pneumoniae infections in patients from Recife-PE, Brazil. Rev Soc Bras Med Trop. 2020;53. https://doi.org/10.1590/0037-8682-0526-2019.
Cifuentes M, García P, San Martín P, Silva F, Zúñiga J, Reyes S, et al. Primer caso de detección de blaKpc en Chile: desde Italia a un hospital público de Santiago. Rev Chil Infectol. 2012;29(2):224–8. https://doi.org/10.4067/S0716-10182012000200018.
World Health Organization. Environment, Climate Change and Health. Disease outbreaks. https://www.who.int/environmental_health_emergencies/disease_outbreaks/en/. Accessed Aug 2020.
Krasowiak R, Smalla K, Sokolov S, Kosheleva I, Sevastyanovich Y, Titok M, et al. PCR primers for detection and characterisation of IncP-9 plasmids. FEMS Microbiol Ecol. 2002;42(2):217–25. https://doi.org/10.1111/j.1574-6941.2002.tb01011.x.
Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005;63(3):219–28. https://doi.org/10.1016/j.mimet.2005.03.018.
Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, et al. In Silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58(7):3895–903. https://doi.org/10.1128/AAC.02412-14.
Pitout JDD, Nordmann P, Poirel L. Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob Agents Chemother. 2015;59(10):5873–84. https://doi.org/10.1128/AAC.01019-15.
Pasteran F, Faccone D, Gomez S, De Bunder S, Spinelli F, Rapoport M, et al. Detection of an international multiresistant clone belonging to sequence type 654 involved in the dissemination of KPC-producing Pseudomonas aeruginosa in Argentina. J Antimicrob Chemother. 2012;67(5):1291–3. https://doi.org/10.1093/jac/dks032.
Izdebski R, Baraniak A, Herda M, Fiett J, Bonten MJM, Carmeli Y, et al. MLST reveals potentially high-risk international clones of Enterobacter cloacae*. J Antimicrob Chemother. 2015;70(1):48–56. https://doi.org/10.1093/jac/dku359.
Chen L, Mathema B, Pitout JDD, DeLeo FR, Kreiswirth BN. Epidemic Klebsiella pneumoniae ST258 is a hybrid strain. MBio. 2014;5(3):e01355–14. https://doi.org/10.1128/mBio.01355-14.
Li J, Huang Z-Y, Yu T, Tao X-Y, Hu Y-M, Wang H-C, et al. Isolation and characterization of a sequence type 25 carbapenem-resistant hypervirulent Klebsiella pneumoniae from the mid-south region of China. BMC Microbiol. 2019;19(1):219. https://doi.org/10.1186/s12866-019-1593-5.
Gutiérrez C, Labarca J, Román JC, Sanhueza F, Moraga M, Wozniak A, et al. Vigilancia de enterobacterias productoras de carbapenemasas en cultivos rectales en un hospital universitario de Santiago, Chile. Rev Chil Infectol. 2013;30(1):103–6. https://doi.org/10.4067/S0716-10182013000100019.
Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing. M02-A12 ed. CLSI document; 2015.
Wozniak A, Villagra NA, Undabarrena A, Gallardo N, Keller N, Moraga M, et al. Porin alterations present in non-carbapenemase-producing Enterobacteriaceae with high and intermediate levels of carbapenem resistance in Chile. J Med Microbiol. 2012;61(9):1270–9. https://doi.org/10.1099/jmm.0.045799-0.
Beriş FŞ, Akyildiz E, Özad Düzgün A, Say Coşkun US, Sandalli C, Çopur ÇA. A novel Integron gene cassette harboring VIM-38 Metallo-β-lactamase in a clinical Pseudomonas aeruginosa isolate. Ann Lab Med. 2016;36(6):611–3. https://doi.org/10.3343/alm.2016.36.6.611.
Centers for Disease Control and Prevention. PulseNet Methods & Protocols. https://www.cdc.gov/pulsenet/pathogens/protocols.html. Accessed Jul 2019.
Heras J, Domínguez C, Mata E, Pascual V, Lozano C, Torres C, et al. GelJ – a tool for analyzing DNA fingerprint gel images. BMC Bioinformatics. 2015;16(1):270. https://doi.org/10.1186/s12859-015-0703-0.
Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33(9):2233–9. https://doi.org/10.1128/JCM.33.9.2233-2239.1995.
Institut Pasteur MLST and whole genome MLST Databases. http://bigsdb.pasteur.fr/. Accessed Nov 2018.
Public Databases for Molecular Typing and Microbial Genome Diversity. https://pubmlst.org/. Accessed Nov 2018.
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–77. https://doi.org/10.1089/cmb.2012.0021.
Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–9. https://doi.org/10.1093/bioinformatics/btu153.
Bosi E, Donati B, Galardini M, Brunetti S, Sagot M-F, Lió P, et al. MeDuSa: a multi-draft based scaffolder. Bioinformatics. 2015;31(15):2443–51. https://doi.org/10.1093/bioinformatics/btv171.
Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27(7):1009–10. https://doi.org/10.1093/bioinformatics/btr039.
Acknowledgements
We thank the staff of the Microbiology Laboratory of the Red de Salud UC-CHRISTUS for their help in the technical aspects of this work. We also thank the Millennium Science Initiative of the Ministry of Economy, Development and Tourism, Government of Chile.
Funding
This work was supported by research funds from SENTRY (Antimicrobial Resistance Surveillance Program), the Red de Salud UC-Christus and the Department of Clinical Laboratories at the School of Medicine of Pontificia Universidad Católica de Chile. The Red de Salud UC-CHRISTUS funded the isolation, identification, carbaNP test, PCR detection of carbapenemase genes, and PFGE analysis. SENTRY funded the WGS analysis (Novogene Inc), MLST and plasmid analysis. The Department of Clinical Laboratories at the School of Medicine of Pontificia Universidad Católica de Chile funded the expenses required for Bioinformatic analysis performed at Universidad del Desarrollo.
Author information
Authors and Affiliations
Contributions
AW: acquisition of data, analysis and interpretation of data, writing and revision of manuscript. CF: acquisition of data, analysis and interpretation of data; writing of manuscript. FM: acquisition and analysis of data, writing of manuscript. PG: design of the study, analysis and interpretation of data. CC: acquisition of data. LR: analysis and interpretation of data. JMM: interpretation of data, writing and revision of manuscript. PCG: conception and design of the study, interpretation of data; critical revision of manuscript and final approval of the version to be submitted. All authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All procedures in this work were approved by the Ethics Committee of the Pontificia Universidad Católica de Chile.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Figure S1.
Unproccessed Pfge Gels.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Wozniak, A., Figueroa, C., Moya-Flores, F. et al. A multispecies outbreak of carbapenem-resistant bacteria harboring the blaKPC gene in a non-classical transposon element. BMC Microbiol 21, 107 (2021). https://doi.org/10.1186/s12866-021-02169-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12866-021-02169-3