Abstract
Background
Infections caused by Shewanella spp. have been increasingly reported worldwide. The advances in genomic sciences have enabled better understanding about the taxonomy and epidemiology of this agent. However, the scarcity of DNA sequencing data is still an obstacle for understanding the genus and its association with infections in humans and animals.
Results
In this study, we report the first isolation and whole-genome sequencing of a Shewanella algae strain from a swine farm in Brazil using the boot sock method, as well as the resistance profile of this strain to antimicrobials. The isolate was first identified as Shewanella putrefaciens, but after whole-genome sequencing it showed greater similarity with Shewanella algae. The strain showed resistance to 46.7% of the antimicrobials tested, and 26 resistance genes were identified in the genome.
Conclusions
This report supports research made with Shewanella spp. and gives a step forward for understanding its taxonomy and epidemiology. It also highlights the risk of emerging pathogens with high resistance to antimicrobial formulas that are important to public health.
Similar content being viewed by others
Background
Shewanella spp. are Gram-negative bacteria mostly found in marine environments. Although infections can be considered rare, the number of reports have increased in recent years [1,2,3,4,5,6,7].. The species reported most frequently in the literature are Shewanella algae and Shewanella putrefaciens [8].
These bacteria were first isolated as part of the genus Achromobacter in 1931 and characterized as Achromobacter putrefaciens [9]. Later, due to advances in molecular biology, Shewanella was assigned as a new genus in the family Vibrionaceae [10]. More recently it is recognized as part of the family Shewanellaceae [11]. Due to its great genomic diversity, some strains can be classified under this group based on genetic data, even though they are phenotypically different [8].
The genus was recently revised by Thorell et al. (2019) [12] based on the genome sequences of strains described in previous studies. More than 48% of the strains (64 out of 131) were reassigned to different species or sub-species after the analysis. This reinforces the importance of research involving isolation and genomic characterization to have a better understanding of the bacteria and their epidemiology.
Shewanella spp. are believed to be susceptible to most antibiotics used in human medicine, such as third-generation cephalosporins, gentamicin and ciprofloxacin [7]. However, recent studies have shown an increasing resistance of new isolates [1, 13, 14]. This resistance, in addition to the increasing number of infections, underscores the importance of research focusing on these bacteria and how to prevent new infections.
In this study, we report the first isolation of a Shewanella algae strain from a swine farm in Brazil using the boot sock method. The isolation happened unexpectedly through a protocol designed for Salmonella spp.. We also report the whole-genome sequence and describe the antimicrobial susceptibility profile in order to improve the understanding of the genus.
Results and discussion
In this study, a Shewanella algae strain was isolated from a pen’s floor of a swine farm in Brazil. To the best of our knowledge, this is the first report of isolation in a swine production facility.
Infectious caused by Shewanella spp. have been increasingly reported in humans. Since this microorganism is widely found in marine environments, many human cases have been linked to seawater exposure prior to the infection. Shewanella spp. are considered opportunistic bacteria, therefore, infections frequently occur in individuals who are immunocompromised or with pre-existing conditions [15]. Although the origin of the isolated strain remains unknown, the presence of this resistant zoonotic microorganism at such a different environment may represent a threat to the employees that may be exposed to it daily.
Shewanella algae may grow in culture media for Enterobacteriaceae and can be misidentified as Shewanella putrefaciens
The strain was isolated after incubation in Tetrathionate broth and recovered from Hektoen agar. The selected colonies were submitted to Triple Sugar Iron and Lysine Iron Agar tests and became suggestive for Salmonella spp.. However, this hypothesis was discarded since they were oxidase-positive. Biochemical identification of the isolate was carried out using the API 20E kit (bioMérieux®, France). As a result, the strain was identified as Shewanella putrefaciens group, with an excellent identification (99.9%), which was later characterized as Shewanella algae by whole-genome sequencing.
Shewanella algae have some biochemical traits that differ from Shewanella putrefaciens, such as nitrite reduction, the ability to grow in higher temperatures and under higher concentrations of sodium chloride (NaCl). The colonies have a mucoid aspect, and they are hemolytic in blood agar. However, strains can be misidentified as Shewanella putrefaciens because many commercial identification systems, such as the API 20E (bioMérieux®, France) and API 20NE (bioMérieux®, France), do not contain S. algae in their database [16]. Botelho-Nevers et al. (2005) [17] also misidentified a S. algae strain as S. putrefaciens after biochemical identification. These findings highlight the importance of further investigation towards correct identification of the strains.
In addition, the results show that Shewanella algae has the ability to grow in culture media designed for the isolation of Enterobacteriaceae, and the morphological aspects of its colonies may incorrectly lead to classifying this bacteria as Salmonella spp. [18]. Therefore, these findings reinforce the necessity of using biochemical, serological, and molecular testing for differentiating Salmonella spp. from other bacteria.
Whole-genome sequencing reveals greater similarity of the strain with Shewanella algae
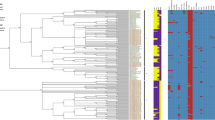
Following identification, the strain was subjected to whole-genome sequencing, and characterized as Shewanella algae. The draft-genome (276 contigs > 200 bases, N50 = 88.103 bases) has 4,944,286 bases, in which 4265 genes were detected with a GC-content (guanine-cytosine content) of 53.15%. The draft-genome of the isolate, named A3/19, was deposited on GenBank under the accession number GCA_014444625.1. When the isolate’s draft-genome was compared to 21 other species of Shewanella retrieved from the GenBank database, it showed an ANI > 98% to two species of Shewanella algae against ANI < 84% of similarity to two Shewanella putrefaciens genomes (Fig. 1). Since ANI > 95% is a robust indicator of two samples belonging to the same species [19] it can be concluded that this strain is a Shewanella algae species. A phylogenetic analysis based on 16S ribosomal gene sequence also indicated that the isolate A3/19 is, indeed, Shewanella algae (Fig. 2).
Average Nucleotide Identity (ANI) of the isolated strain to 21 Shewanella species. A heatmap and a hierarchical clustering are shown. Pairs of genomes with ANI > 95% can be considered as from the same species. Legend: NGS_9_1_Montagem2: S. algae: SAMN15541815; S. algae: NZ_CP046378.1; S. algae_2: NZ_LN10019.1; S. amazonensis: NC_008700.1; S. baltica: NC_016901.1; S. bicestrii: NZ_CP022358.1; S. denitrificans: NC_007954.1; S. donghaensis: NZ_CP041783.1; S. frigidimarina: NC_008345.1; S. halifaxensis: NC_010334.1; S. japonica: NZ_CP020472.1; S. livingstonensis: NZ_ CP034015.1; S. loihica: NC_009092.1; S. marisflavi: NZ_CP022272.1; S. maritima: NZ_CP036200.1; S. pealeana: NC_009901.1; S. piezotolerans: NC_011566.1; S. polaris: NZ_CP041036.1; S. psychrophila: NZ_CP014782.1; S. putrefaciens: NC_009438.1; S. putrefaciens_2: NC_017566.1; S. sediminis: NC_009831.1; S. violacea: NC_014012.1; S. woodyi: NC_010506.1
Antimicrobial susceptibility test and detection of resistance genes
The isolated strain was resistant to 46.7% of the antibiotics tested (7 out of the 15 – POLISENSIDISC, DME®, Brazil) as shown in Table 1. Shewanella spp. are usually susceptible to Tetracycline, Chloramphenicol, Ciprofloxacin and Amoxicillin [6, 7, 20]. However, our strain was resistant to all of these formulas. In swine production, these antimicrobials have been broadly used as in-feed or therapeutically [21], which can possibly explain the resistance found in this study.
In the genetic characterization, we found 26 genes similar to antimicrobial resistance genes (See Additional file 1), which predicts that this strain could be resistant to 21 antibiotics: Amoxicillin, Ampicillin, Streptomycin, Trimethoprim, Amikacin, Sulfamethoxazole, Chloramphenicol, Ciprofloxacin, Doxycycline, Cephalothin, Piperacillin, Ticarcillin, Tobramycin, Gentamicin, Ampicillin + Clavulanic acid, Amoxicillin + Clavulanic acid, Tigecycline, Imipenem, Florfenicol, Tetracycline and Minocycline. These results, based on antimicrobial susceptibility tests and whole genome sequencing, suggest that this is possibly a multidrug resistant strain.
In addition, a total of eleven genes found in the draft-genome (see Additional file 2) were similar to virulence factors, which suggests that, even though the strain was isolated from an environmental sample, it is a potentially hazardous microorganism that could cause infections in both animals and humans.
Conclusion
This study reports the first isolation and whole-genome sequencing of a Shewanella algae strain from a swine production farm in Brazil. The genetic characterization was important to classify the isolate correctly, to confirm that it is a multidrug-resistant strain, and to alert for the potential of being an infective microorganism. The resistance profile found in this study highlights the risk of the indiscriminate usage of antibiotics in animal production. These findings draw attention to the power of genomic approaches for understanding Shewanella spp. strains in order to improve research made with these bacteria.
Methods
The isolation of Shewanella algae happened unexpectedly, since their colonies can be phenotypically similar to Salmonella spp. in many culture media for Enterobacteriaceae [18]. The aim of this methodology was to correctly identify the strain through whole-genome sequencing and evaluate resistance through antimicrobial sensitivity test.
Sample collection
The samples were collected from a farm in the State of Sao Paulo, Brazil. The pens of weaned pigs were sampled using the boot sock method on the surface of the pen floor. The boot socks were prepared in the laboratory using disposable shoe covers that were sterilized at 121 °C for 15 min. At the farm, two layers of disposable shoe covers were worn by one of our collaborators and dragged onto the surface area of the pen to reach as much area as possible. One layer was used to cover the shoes and decrease contamination, and the other to perform the collection and testing. The samples were kept in a sterile bag, and stored on ice until processing.
Isolation and identification of strains
The samples were subjected to three different protocols for the isolation of Salmonella enterica subsp. enterica, based on the use of different culture media and growth conditions. A schematic representation of the methodology used for the isolation of the colonies is described in Fig. 3.
The enrichment step was performed twice with three different broths: Tetrathionate (TT) (Neogen®, United States), Rappaport Vassiliadis Soya (RVS) (KASVI®, Brazil) and Selenite Cystine (SC) (KASVI®, Brazil). First, samples were inoculated in glass bottles containing 225 mL of each broth and incubated at the recommended temperatures for 18–24 h. After incubation, a 1 mL aliquot was inoculated into 9 mL of broth and again incubated for 18–24 h. The samples were plated onto six different media: Hektoen (HE) (KASVI®, Brazil), Salmonella-Shigella (SS) (KASVI®, Brazil), MacConkey (MC) (KASVI®, Brazil), Brilliant-Green (BG) (KASVI®, Brazil), Xylose Lysine Deoxycholate (XLD) (KASVI®, Brazil) and Xylose Lysine Tergitol-4 (XLT-4) (KASVI®, Brazil). All media were prepared following the manufacturer’s recommendations and incubated at the suggested temperatures.
The broths used in this protocol were supplemented with a solution of Novobiocin (INLAB®, Brazil) (40 μg/mL) as suggested by Pessôa and Peixoto (1971) [22] and ISO 6579 (NBR, 2014) [23] in order to decrease competition with Proteus spp..
The suspected colonies were subjected to testing on Lysine Iron Agar (KASVI®, Brazil) and Triple Sugar Iron Agar (KASVI®, Brazil). This procedure is used for differentiating bacteria of the family Enterobacteriaceae.
The final identification was performed with API 20E (bioMérieux®, France) kit, following all manufacturer’s instructions. This biochemical system allows the identification of Enterobacteriaceae and other non-fastidious Gram-negative bacteria.
Antibiogram
The sensitivity to antibiotics was tested using the POLISENSIDISC 15 (DME®, Brazil), following the product’s instructions. This system tests 15 antibiotics: Amikacin, Amoxicillin/Clavulanic Acid, Ampicillin, Aztreonam, Cefazolin, Cefepime, Cefoxitin, Ceftadizime, Ceftriaxone, Ciprofloxacin, Chloramphenicol, Gentamicin, Meropenem, Trimethroprim/Sulfamethoxazole, Tetracycline.
Whole-genome sequencing and phylogenetic assessment
The sequencing was carried out in an Illumina Miseq platform, using kits and protocols from the manufacturer. Raw data was processed by Fastp [24]. Then, it was input to SPAdes [25] in order to assemble the bacterial genome. Annotation was carried out with NCBI Prokaryotic Genome Annotation Pipeline (PGAP), and the detection of genes related to virulence and antimicrobial resistance was performed with Abricate (Seemann T, Abricate, Github) [26], using the databases “The virulence factor database” - VFDB and ResFinder [23]. The ANI was calculated using PyANI [27].
The sequences of 16S Ribosomal genes were obtained either from Genbank or from our whole genome sequencing data. The alignment was performed with Muscle [28], and the Maximum Likelihood phylogeny was carried out using IQTree [29], selecting the best substitution model with ModelFinder [30] and testing for best branch supports with the ultrafast bootstrap [31]. The resulting tree was visualized using FigTree [32].
Availability of data and materials
The dataset supporting the conclusions of this article is available in the GenBank, [GCA_014444625.1 https://www.ncbi.nlm.nih.gov/assembly/GCA_014444625.1].
Abbreviations
- GC-content:
-
Guanine-Cytosine content
- TT:
-
Tetrathionate
- RVS:
-
Rappaport Vassiliadis Soya
- SC:
-
Selenite Cystine
- HE:
-
Hektoen
- SS:
-
Salmonella-Shigella
- MC:
-
MacConkey
- BG:
-
Brilliant-Green (BG)
- XLD:
-
Xylose Lysine Deoxycholate
- XLT-4:
-
Xylose Lysine Tergitol-4
- S. algae :
-
Shewanella algae
- S. putrefaciens :
-
Shewanella putrefaciens
- S. amazonensis :
-
Shewanella amazonensis
- S. baltica :
-
Shewanella baltica
- S. bicestrii :
-
Shewanella bicestrii
- S. denitrificans :
-
Shewanella denitrificans
- S. donghaensis :
-
Shewanella donghaensis
- S. frigidimarina :
-
Shewanella frigidimarina
- S. halifaxensis :
-
Shewanella halifaxensis
- S. japonica :
-
Shewanella japonica
- S. livingstonensis :
-
Shewanella livingstonensis
- S. loihica :
-
Shewanella loihica
- S. marisflavi :
-
Shewanella marisflavi
- S. maritima :
-
Shewanella maritima
- S. pealeana :
-
Shewanella pealeana
- S. piezotolerans :
-
Shewanella piezotolerans
- S. polaris :
-
Shewanella polaris
- S. psychrophila :
-
Shewanella psychrophila
- S. sediminis :
-
Shewanella sediminis
- S. violacea :
-
Shewanella violacea
- S. woodyi :
-
Shewanella woodyi
- rDNA:
-
ribossomal DNA
References
Maqsood Ali A, Noorulamin M, Arif S. Isolation of Shewanella putrefaciens in an elderly man with subacute intestinal obstruction & appendicitis. IDCases. 2017;9(June):45–6.
Badami A, Shimonovich S, Kolpon S, Rubinshteyn V. Shewanella putrefaciens: a rare anaerobic bacterium mimicking necrotizing soft tissue infection. Surg Infect Case Rep. 2017;2:88–91.
López Aperador C, Bosh Benitez-Parodi E, Chamorro Buchelli I, Guerra Rodriguez R, Auyanet Saavedra I, Toledo Gonzalez A. Peritonitis por Shewanella putrefaciens: a propósito de un caso. Rev la Soc Española Nefrol. 2016;36(4):444–5.
Constant J, Chernev I, Gomez E. Shewanella putrefaciens infective endocarditis. Brazilian J Infect Dis. 2014;18(6):686–8.
Pagani L, Lang A, Vedovelli C, Moling O, Rimenti G, Pristera R, et al. Soft tissue infection and bacteremia caused by Shewanella putrefaciens. J Clin Microbiol. 2003;41(5):2240–1.
Patel R, Abraham A, Thomas J, Zhi W, Ahmed S, Verley J. A rare case of pneumonia caused by Shewanella putrefaciens. Case Rep Med. 2012;2012:3–6.
Vignier N, Barreau M, Olive C, Baubion E, Théodose R, Hochedez P, et al. Human Infection with Shewanella putrefaciens and S . algae : Report of 16 Cases in Martinique and Review of the Literature. Am J Trop Med Hyg. 2013;89(1):151–6.
Hau HH, Gralnick JA. Ecology and biotechnology of the genus Shewanella. Annu Rev Microbiol. 2007;61:237–58.
Derby HA, Hammer BW. Bacteriology of butter bacteriological studies on surface taint butter. Iowa Agric Exp Stn Res Bull. 1931;145:387–416.
MacDonell MT, Colwell RR. Phylogeny of the vibrionaceae, and recommendation for two new genera, listonella and shewanella. Syst Appl Microbiol. 1985;6:171–82.
Ivanova EP, Flavier S, Christen R. Phylogenetic relationships among marine Alteromonas-like proteobacteria: emended description of the family Alteromonadaceae and proposal of Pseudoalteromonadaceae fam. Nov., Colwelliaceae fam. Nov., Shewanellaceae fam., Moritellaceae fam. Nov., Ferrimon. Int J Syst Evol Microbiol. 2004;54:1773–88.
Thorell K, Meier-kolthoff JP, Sjöling Å, Martín-rodríguez AJ, Bowman JP. Whole-genome sequencing redefines Shewanella taxonomy. Front Microbiol. 2019;10(August):1–14.
Lee Y, Tung K, Cheng J, Wu Z, Chen S. Genomic characterization of carbapenem-resistant Shewanella algae isolated from Asian hard clam ( Meretrix lusoria ). Aquaculture. 2019;500(October 2018):300–4.
Wu Z, Huang Y, Chao W, Ho S, Cheng J, Liu P. Reversal of carbapenem-resistance in Shewanella algae by CRISPR / Cas9 genome editing. J Adv Res. 2019;18:61–9.
Tsai MS, You HL, Tang YF, Liu JW. Shewanella soft tissue infection: case report and literature review. Int J Infect Dis. 2008;12(6):119–24.
Holt HM, Gahrn-Hansen B, Bruun B. Shewanella algae and Shewanella putrefaciens: clinical and microbiological characteristics. Clin Microbiol Infect. 2005;11(5):347–52. Available from:. https://doi.org/10.1111/j.1469-0691.2005.01108.x.
Botelho-Nevers E, Gouriet F, Rovery C, Paris P, Roux V, Raoult D, et al. First case of osteomyelitis due to Shewanella algae. J Clin Microbiol. 2005;43(10):5388–90.
Wisplinghoff H. Pseudomonas spp., Acinetobacter spp. and Miscellaneous Gram-negative bacilli. In: Cohen J, Opal SM, Powderly WG, editors. Infectious Diseases. 4th Editio: Elsevier Ltd; 2017. p. 1579–99.
Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun. 2018;9(5114):1–8.
Mohan N, Sharma S, Padhi TR, Basu S, Das TP. Traumatic endophthalmitis caused by Shewanella putrefaciens associated with an open globe fishhook injury. Eye. 2014;28:235.
Lekagul A, Tangcharoensathien V, Yeung S. Patterns of antibiotic use in global pig production: a systematic review. Vet Anim Sci. 2019;7(April):100058.
Vital Álvares Pessôa G, Sandoval Peixoto E. Novobiocin-Selenite broth. A medium of improved selectivity for the isolation of Salmonella from faeces. Rev Inst Adolfo Lutz. 1971;31:1–3.
ABNT. NBR ISO 6579: Microbiology of food and animal feeding stuffs - Horizontal method for the detection of Salmonella spp; 2014. p. 1–45.
Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(9):884–90.
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–77.
Seemann T. ABRicate. Github; 2019.
Pritchard L, Glover RH, Humphris S, Elphinstone JG, Toth IK. Genomics and taxonomy in diagnostics for food security: soft-rotting enterobacterial plant pathogens. Anal Methods. 2016;8:12–24.
Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–7.
Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32(1):268–74.
Kalyaanamoorthy S, Minh BQ, Wong TKF, Von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14(6):587–9.
Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UFBoot2: improving the ultrafast bootstrap approximation. Molecular biology and evolution. Mol Biol Evol. 2018;35(2):518–22.
Rambaut A. Figtree [Internet]. 2009. Available from: http://tree.bio.ed.ac.uk/software/figtree/. Accessed 4 Sept 2020.
Acknowledgments
We thank FAPESP and CAPES for supporting our research, and Ashli Moore for proofreading this manuscript.
Funding
This study was funded by the Sao Paulo Research Foundation (FAPESP) (Grant n. 2017/22184–7 and 2019/19473–2). This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brasil (CAPES) – Finance Code 001. The funding agencies had no role in the design of study and collection of samples, analysis and interpretation of data, and in writing the manuscript.
Author information
Authors and Affiliations
Contributions
VBAA, ACNV, VLAR and AMCV were responsible for the isolation and identification of the strain prior to whole-genome sequencing. VBAA and EDM performed the collection of samples. EC, PAM and VLAR performed all the genetic characterization and analysis of data. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The Ethics Committee for Animal Utilization of the Universidade de Sao Paulo approved this study prior to the beginning of experiments (n. 9741230818).
The samples were collected at a third-party farm under the consent to participate in our study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1.
Antimicrobial resistance genes of the isolate A3/19. The list of 26 resistance genes are provided in this file with all the important information related to them.
Additional file 2.
Virulence factors genes of the isolate A3/19. The list of 11 virulence genes are provided in this file with all the important information related to them.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Alves, V.B.d.A., Carvalho, E., Madureira, P.A. et al. First isolation and whole-genome sequencing of a Shewanella algae strain from a swine farm in Brazil. BMC Microbiol 20, 360 (2020). https://doi.org/10.1186/s12866-020-02040-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12866-020-02040-x