Abstract
Background
Wolbachia are the most widely spread endosymbiotic bacteria, present in a wide variety of insects and two families of nematodes. As of now, however, relatively little genomic data has been available. The Wolbachia symbiont can be parasitic, as described for many arthropod systems, an obligate mutualist, as in filarial nematodes or a combination of both in some organisms. They are currently classified into 16 monophyletic lineage groups (“supergroups”). Although the nature of these symbioses remains largely unknown, expanded Wolbachia genomic data will contribute to understanding their diverse symbiotic mechanisms and evolution.
Results
This report focuses on Wolbachia infections in three pseudoscorpion species infected by two distinct groups of Wolbachia strains, based upon multi-locus phylogenies. Geogarypus minor harbours wGmin and Chthonius ischnocheles harbours wCisc, both closely related to supergroup H, while Atemnus politus harbours wApol, a member of a novel supergroup S along with Wolbachia from the pseudoscorpion Cordylochernes scorpioides (wCsco). Wolbachia supergroup S is most closely related to Wolbachia supergroups C and F. Using target enrichment by hybridization with Wolbachia-specific biotinylated probes to capture large fragments of Wolbachia DNA, we produced two draft genomes of wApol. Annotation of wApol highlights presence of a biotin operon, which is incomplete in many sequenced Wolbachia genomes.
Conclusions
The present study highlights at least two symbiont acquisition events among pseudoscorpion species. Phylogenomic analysis indicates that the Wolbachia from Atemnus politus (wApol), forms a separate supergroup (“S”) with the Wolbachia from Cordylochernes scorpioides (wCsco). Interestingly, the biotin operon, present in wApol, appears to have been horizontally transferred multiple times along Wolbachia evolutionary history.
Similar content being viewed by others
Background
Wolbachia are endosymbiotic alpha-proteobacteria infecting a broad range of arthropods and nematodes [1, 2]. The bacteria of this genus are considered to be the most widely spread symbionts in the animal world, perhaps infecting half of insect species [3,4,5]. Wolbachia are maternally inherited and can induce variable phenotypes in their hosts through mutualism or parasitism [6,7,8,9]. Wolbachia are genetically diverse, as are the interactions with their hosts [10, 11]. Currently, there is a general consensus to classify them in monophyletic lineage groups (“supergroups” A to R). Wolbachia belonging to supergroups C, D and J exclusively infect filarial nematodes (Onchocercidae) [12,13,14]. Supergroup L exclusively contains plant parasitic nematodes (Pratylenchidae) [15, 16]. Supergroup F Wolbachia is, so far, the only clade composed by some strains infecting arthropods and some infecting filarial nematodes [17, 18]. All other described supergroups exclusively infect arthropods [19,20,21,22,23]. The proposed supergroup G members [24] and R [25] are now considered to be part of supergroup B [26] and A [27], respectively, and are no longer considered as separate supergroups.
In the last few years, the number of published Wolbachia genomes has increased and currently 48 draft and 21 complete genomes of Wolbachia are available at the NCBI database. However, these data are not a good representation of the entirety of Wolbachia diversity. Indeed, among the 21 complete genomes, 15 are symbionts of insects belonging to either supergroup A or B (including 7 strains from species of Drosophila), 2 are symbionts of insects belonging to other supergroups (F and E) and 4 are symbionts of nematodes (supergroups L, C or D). Wolbachia has been identified in other arthropods, such as isopods [28, 29] and arachnids [24]. Limited genomic data for isopods are available, for example a draft genome of wCon, infecting Cylisticus convexus and a draft genome of wVulC infecting Armadillidium vulgare [30]. No genomic data is available for arachnid symbionts while several studies identified Wolbachia in spiders [24, 25, 31], mites [20] or scorpions [32].
In the current study, we focused on the Wolbachia symbionts of pseudoscorpions. Pseudoscorpions inhabit wooded areas associated with rotting vegetation. Unlike most arthropods, they are viviparous with embryos obtaining nutrients from the maternal reproductive tract [33]. The presence of Wolbachia was described for the first time in pseudoscorpions in 2005 in Cordylochernes scorpioides [34]. It has been reported that the endosymbiont is responsible for the “male killing” phenotype in C. scorpioides, associated with a high rate of spontaneous abortion.
We examined the prevalence of Wolbachia infections in pseudoscorpion population samples from an area in Montpellier (France) where three different species of pseudoscorpions were collected and determined to be positive for Wolbachia infection. The molecular analyses phylogenetically identified two different Wolbachia groups, of which one strain is divergent from currently accepted Wolbachia supergroups. We used Wolbachia DNA enrichment capture [35, 36] to ensnare Wolbachia DNA for genomic sequencing from this Wolbachia strain, wApo, infecting Atemnus politus.
Results
Identification and evolution of Pseudoscorpions
In all, 94 pseudoscorpion specimens were collected from an area of Montpellier (France). Sixty specimens were morphologically identified as Atemnus politus (Simon, 1878), 24 specimens belong to the species Geogarypus minor (Koch, 1873) and 10 specimens belong to the species Chthonius ischnocheles (Hermann, 1804). The specimen identification is consistent with the analysis of the mitochondrial cytochrome oxidase I gene marker (COI). The COI sequences of the specimens belonging to Geogarypus minor are identical to each other and to the sequence of Geogarypus nigrimanus (JN018180 specimen voucher MNHN-JAB62). The species Geogarypus nigrimanus has been recently synonymized with Geogarypus minor [37]. Regarding the produced COI sequences of the specimens belonging to Chthonius ischnocheles species, the specimens IV3–1, Q3–1, IV1–1 and Q4 are between 99.8–100% identical and match the sequence of Chthonius ischnocheles available at the NCBI database (JN018172 specimen voucher MNHN-JAB62). The existence of cryptic species of Chthonius ischnocheles has been documented [38] and the specimens IV-J5 and IV-S1 form a paraphyletic group with the other Chthonius ischnocheles (Fig. 1).
Phylogenetic tree of pseudoscorpions based on the COI (mitochondrial cytochrome oxidase I) gene. The total length of datasets is 659 bp. The topology was inferred using Maximum Likelihood (ML) inference using IQTREE [39]. The Best-fit model calculated using ModelFinder according to BIC index was TVM + R7. Nodes are associated with Bootstrap values based on 1000 replicates, only bootstrap values superior to 70 are indicated
The COI sequences of the specimens identified as Atemnus politus present more intraspecific variability: calculated pairwise similarities range between 97.7–99.8%. The blastn analysis [40] does not provide identification of strongly similar sequences in databases; indeed no COI sequences from species related to the Atemnus genus have been deposited in the NCBI database. The most similar COI sequence in the database belongs to Atemnidae sp. JA-2011 voucher (JN018203), showing only 79.74% similarity. The COI-based phylogeny indicates that all the specimens of Atemnus politus form a clade which is a sister group of the clade composed of Paratemnoides sumatranus and Oratemnus curtus species, both representatives of the Atemnidae family (Fig. 1). The COI analysis shows all specimens of Chthonius ischnocheles form a sister group of other species belonging to Chthonius species (C. tetrachelatus, C. dacnodes) (Fig. 1). To date, the only species of pseudoscorpion described as a Wolbachia host, Cordylochernes scorpioides, belongs to the Chernetinae and is a sister group to the Atemnidae family (including Atemnus politus) (Fig. 1).
Prevalence of Wolbachia infection
Wolbachia was detected by PCR amplification of wsp and gatB markers (see Methods) in the three studied pseudoscorpion species and in accordance with the generally used nomenclature of Wolbachia strains, we have named these new strains according to their hosts: wApol for bacteria infecting Atemnus politus, wGmin for bacteria infecting Geogarypus minor and wCisc for bacteria infecting Chthonius ischnocheles.
The A. politus specimens have the highest prevalence of Wolbachia infection with 43.3% positive samples (26 positive of 60 specimens examined). 16.7% of G. minor individuals appear to be infected (4 positive of 24 specimens examined), while only 10% of the C. ischnocheles (1 positive of 10 specimens examined) show PCR amplification of the Wolbachia markers (Table S1).
The wApol draft genome and annotation
Two draft genomes were produced for wApol Wolbachia from Atemnus politus (specimens K5 and K3) using target capture enrichment [35, 36]. For Illumina sequencing, an enrichment of 45-fold was observed (0.4% of the reads mapped to the draft genome without enrichment against 18% with) and 50-fold enrichment was observed with PacBio sequencing (0.8% of the reads mapped to the draft genome without enrichment as opposed to 40.9% with). Unfortunately, the amounts of DNA available for the two specimens were low, and thus the samples were processed simultaneously using individual barcoded adaptors (see Methods) and for PacBio sequencing, relatively few total reads were produced (Table 1). The hydrid de novo assembly allowed production of two draft genomes. For specimen K5, a 373 contig draft genome of 1,445,964 bp was obtained, with an average G + C content of 35.6% (largest contig, 25,286 bp, N50 = 5741 bp, average sequencing depth of 420X). For specimen K3, a 200 contig draft genome of 1,404,177 bp was obtained, with an average G + C content of 35.49% (largest contig, 40,755 bp, N50 = 10,346 bp, average sequencing depth = 205X) (Table 1).
Among 221 single-copy orthologue genes conserved among proteobacteria (BUSCOs database), 155 are present in the wApol K5 draft genome and 151 in wApol K3, suggesting 70.2 and 69% complete BUSCOs, respectively. This percentage can be used to assess the level of completeness of genomes (see Methods). By comparison, the Wolbachia from Drosophila melanogaster, wMel, has a higher level of BUSCOs in the current analysis with 180 genes (81.4%) and Wolbachia from Pratylenchus penetrans, wPpe, has a low level with 161 genes (72.9%) (Table S2). The low level of BUSCOs observed for wApol suggest that either the wApol draft is incomplete or this genome is highly degraded as has been described for Wolbachia from Onchocerca ochengi (74.7% complete BUSCOs) [44].
The Average Nucleotide Identity (ANI) calculation (see Methods) indicates that the two wApol drafts have 99% identity and they are most similar to wCle with 87% identity (Fig. 2). According to the analysis of available complete genomes, strains which are representative of the same supergroup share more than 94% identity; for example, 99% for wOo and wOv (from supergroup C), 94% for wPip and wTpre (from supergroup B) or 96% for wMel and wCau (from supergroup A).
Summary of ANI calculations for Wolbachia genomes. The Average Nucleotide Identity (ANI) between the wApol draft genomes and 13 complete genomes of Wolbachia evaluated using the ANI Calculator [45]
One thousand seven hundred forty-six coding sequences (CDS) were annotated using RAST from wApol K5 and 2215 CDS from wApol K3. The high number of CDSs might be explained by presence of multiple copies of same genes due to incompleteness of the draft genomes. The two draft genomes contain a low number of transposable elements (55 mobile elements, 16 group II intron-associated genes, 52 ISs for K5; 35 mobile elements, 9 group II intron-associated genes, 36 ISs for K3) compared to other Wolbachia strains infecting insects (with the exception of wTpre), but a high number compared to Wolbachia infecting filarial nematodes (Table S3). Numerous phage-like genes were observed in the wApol K5 draft genome, with 54 CDS (coding sequences) detected on 37 different contigs with 9 annotated as prophage sequences (Table S4). Fewer phage-like genes were detected in the wApol K3 draft genome, with 20 CDS detected on 12 different contigs and only one annoted as a prophage sequence (Table S4). The number of phage-like genes is highly variable among the studied Wolbachia genomes and the number observed in wApol is close to wMel (with 44 CDS) or wCle (with 20 CDS). However, the variability in transposable element and phage numbers observed between the two wApol draft genomes suggests that the genome sequences are incomplete.
Although the wApol drafts are likely not complete, functional annotation of genes by KAAS highlights that the biotin pathway is present, while it is absent in most other studied Wolbachia (Table S5). Only wCle (supergroup F), wNfla, wNleu (supergroup A), wLug, wStri and wVulC (supergroup B) have a complete biotin pathway. Only bioC is not detected in the wApol K5 draft, but the operon is detected on two contigs: the genes bioA and bioD on contig #258 and the genes bioH, bioF and bioB on contig #172 (Fig. 3a). In all studied Wolbachia which contain the operon, the bioC gene is present between the bioD and bioH genes and the absence might be due to the fragmentation of the wApol K5 draft sequence (Fig. 3a). For the wApol K3 draft genome, only one contig (#148) containing bioH, bioF and bioB was detected. The organization of the biotin operon appears to be conserved between all Wolbachia genomes which contain the complete pathway (Fig. 3a). The phylogenies of the biotin genes (containing bioA, bioB, bioH, bioD and bioF) shows that the biotin operon of wApol K5 is most closely related to the operon of wCle (Fig. 3b).
Organization and molecular analysis of the biotin operons of Wolbachia. a Organization of the biotin operon. The operons among the Wolbachia and four outgroups were identified and then aligned using MAFFT [46]. b Phylogeny of the biotin operon (concatenation of contig containing bioA and bioD with the contig containing bioH, bioF and bioB). The length of datasets is 4603 bp. The topology was inferred using Maximum Likelihood (ML) inference using IQ-TREE [39]. The Best-fit model calculated using ModelFinder according to AIC index was TVM + I + G4. Nodes are associated with Bootstrap values based on 1000 replicates. The Wolbachia supergroups are indicated by the color: purple for supergroup F; dark blue for supergroup A; orange for supergroup A and green for supergroup S
Multi-locus phylogenies and phylogenomics
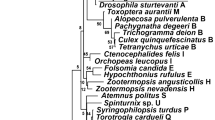
The analysis of Wolbachia in Geogarypus minor, wGmin, was more complex. Two genes were sequenced for wGmin, coxA and ftsZ, and only nested PCR provided clean sequences to enable phylogenetic comparisons. The Wolbachia phylogeny, based on the two genes, identified two different sub-groups of Wolbachia in our samples (Fig. 4a). The Wolbachia infecting Chthonius ischnocheles, wCisc, and Geogarypus minor, wGmin, group together and appear closely related to supergoup H Wolbachia infecting Zootermopsis termite species (wZneg and wZang). The Wolbachia from Atemnus politus, wApol, and the Wolbachia from Cordylochernes scorpioides, wCsco, form a clade which is divergent from known supergroups. This clade is a sister group to supergroup C Wolbachia, which contains the symbiont present in filarial nematodes (Onchocerca spp. and Dirofilaria immitis). The phylogeny based on six genes (16S rDNA, ftsZ, dnaA, gatB, fbpA and coxA) confirms the topology of wApol and wCisc (wGmin and wCsco not included) (Fig. 4b). Our analysis shows the same topology for wApol sequences from the two draft genomes (K5 and K3) as well as sequences of wApol K5 amplified by PCR, confirming that this topology is not an artifact of draft missassembly. In addition, to verify the accuracy of the Wolbachia presence in Geogarypus minor, the coxA gene was sequenced from another specimen (III-J5) derived from an independent DNA extraction, and PCR amplification and the sequences were identical to another specimen III-T3 (accession number MT273088).
Unrooted phylogenetic trees of Wolbachia based on 2 and 6 markers by Maximum Likelihood. a Analysis based on concatenation of ftsZ and coxA; the total length of datasets is 1217 bp. The topology was inferred using Maximum Likelihood (ML) inference using IQ-TREE [39]. The Best-fit model calculated using ModelFinder according to BIC index was TIM3 + I + G4. b Analysis based on concatenation of 16S rDNA, dnaA, ftsZ, coxA, fbpA and gatB; the total length of datasets is 3481 bp. The Best-fit model calculated using ModelFinder according to BIC index was GTR + I + G4. Nodes are associated with Bootstrap values based on 1000 replicates, only bootstrap value superior to 70 are indicated. The Wolbachia supergroups (A–S) are indicated. The bolded names indicate data produced in this study and in the case of wApol, the asterisk (*) indicates sequences produced by PCR amplification
The Wolbachia from Cordylochernes scorpioides had been previously studied using a set of genes not routinely sequenced for Wolbachia evolutionary studies (groEL, fabK, nuoG, NADH dehydrogenase I subunit F, aspS, gltA, coxA, ftsZ, wsp, orpB, nuoD, isocitrate dehydrogenase gene, TPR domain-containing protein gene) [47]. We performed a multi-locus phylogeny based on these 13 genes, including wCsco and sequences isolated from available complete or draft genomes of Wolbachia, as well as those from the wApol produced draft genomes. The observed phylogeny shows that wCsco and wApol cluster together to form a separate clade, a closely related sister group to supergroup C, as observed with the phylogeny based on only two markers (Fig. 4a). The two pseudoscorpions contain closely related Wolbachia strains, with 89.3% identity among these 13 genetic markers (Figure S1).
Two complete phylogenomic analyses were performed: one including 17 Wolbachia genomes representing 6 different supergroups and one including 38 Wolbachia genomes representing 7 different supergroups and including additional draft genome sequences. Three hundred twenty single-copy orthologues were identified among the 17 Wolbachia genomes, while 166 were identified among the 38 Wolbachia genomes. The Maximum Likelihood phylogeny based on the reduced matrix (17 genomes, 320 orthologues, 89,847 amino-acid sites) (Fig. 5a) and the one based on the larger matrix (38 Wolbachia, 167 orthologues, 40,488 amino-acid sites) (Fig. 5b) indicates the wApol group does not cluster with any other Wolbachia group, suggesting it evolved from an independent separate speciation event. We created supplementary phylogenies based on 16S rRNA and ftsZ and have included numerous Wolbachia species to be sure that wApol and wCsco are not grouped with previously described Wolbachia supergroups (Figure S2 and S3).
Phylogenomic analyses of Wolbachia. a Analysis based on concatenation of 320 single-copy orthogroups representing an 89,847 amino-acid matrix. The topology was inferred using Maximum Likelihood (ML) inference using IQTREE [39]. The Best-fit model calculated using ModelFinder according to BIC index was JTT + F + I + G4. b Analysis based on concatenation of 167 single-copy orthogroups representing 40,488 amino-acid matrix. The Best-fit model calculated using ModelFinder according to BIC index was JTT + F + I + G4. Nodes are associated with Bootstrap values based on 1000 replicates, only bootstrap value superior to 70 are indicated. The Wolbachia supergroups (A–S) are indicated
Based upon the phylogenomics analyses, wApol (supergroup S) is closely related to supergroup C (symbionts within the filarial nematodes Onchocerca spp. and Dirofilaria immitis) and supergroup F (including both filarial and arthropods symbionts).
Discussion
Diversity of Wolbachia in pseudoscorpions
This study highlights diversity of Wolbachia harboured by pseudoscorpions. The studied specimens harbouring distinct Wolbachia represent different families, not closely related, within pseudoscorpions. The small number of pseudoscorpion species studied (3 among 3300 described species) represent only small snapshot of the diversity within this group (3 families among 27 described), suggesting that the heterogeneity of Wolbachia among pseudoscorpion species may likely be an underestimate. The species Geogarypus minor, a representative of the Geogarypidae family and the species Chthonius ischnocheles, a representative of the Chthoniidae family, contain Wolbachia closely related to each other and related to supergroup H, as represented by termite Wolbachia. The species Atemnus politus, as a representative of the Atemnidae family, contains Wolbachia closely related to the Wolbachia from the previously described pseudoscorpion Cordylochernes scorpioides and forms a clade as a sister group to supergroup C (infecting exclusively filarial nematodes) and supergroup F (infecting filarial nematodes and arthropods). Within the evolutionary history of pseudoscorpions, at least two events of horizontal transfers of Wolbachia infection have apparantly occurred. More detailed, biodiversity analysis of this group will be required to fully detail the evolutionary dynamics of their Wolbachia.
New supergroup S
The Wolbachia harboured by Atemnus politus (wApol) and Cordylochernes scorpioides (wCsco) form a clade divergent from those Wolbachia supergroups previously described (Figure S2 and S3). The analysis of the wApol draft genome using phylogenomic analysis (Fig. 5), or ANI value (Fig. 2) point out a divergent Wolbachia supergroup. It is important to distinguish the notion of “supergroup” from the notion of “species” (which remains problematic for bacteria [48]). The “supergroup” naming only describes the different evolutionary lineages (or clades) of Wolbachia, and their limits remain arbitrary (as far as it being a monophyletic group).
In the past, two Wolbachia supergroups had been designated and then revised based on further phylogenetic analysis. Supergoup G had been described as a clade of Wolbachia from spider Diaea species based on wsp marker analysis [49]. However, it was later demonstrated that the analysis based upon this marker sequence led to an incorrect conclusion due to recombination events between Wolbachia from supergroups A and B [26]. In the case of supergroup R, four genes (ftsZ, coxA, groEL, and 16S) were analyzed to study Wolbachia from cave spiders (Telema spp.) [25]. From among these four genes, only the groEL marker gave a different topology, excluding these Wolbachia from supergroup A. Upon reanalysis, the differing topology conclusion appears to be due to a G + C composition bias of the groEL gene, causing an incorrect phylogenetic analysis when based on nucleotide sequences [27]. Being aware of these, we created supplementary phylogenies based on 16S rRNA and ftsZ and have included numerous Wolbachia species to be sure that wApol and wCsco are not grouped with previously described Wolbachia supergroups (Figure S2 and S3).
To ensure that the draft genomes of wApol did not create an artificial clade, we further sequenced PCR amplificons from six previously studied marker genes from an independent specimen from the same set of samples as K5 and verified that the data provided the same topology. Based upon this confirmation, we propose that Wolbachia from Atemnus politus and Cordylochernes scorpioides constitute representatives of a new supergroup. To avoid any confusion, we propose not to reuse the previous supergroup R [27] designation and have assigned Wolbachia from the pseudoscorpion Atemnus politus and Cordylochernes scorpioides to a novel supergroup S.
wApol draft genomes and limitation of de novo assembly based on the enrichment method
Two draft genomes, wApol K3 (1,404,177 bp long) and wApol K5 (1,445,964 bp long) were produced, as representatives of supergroup S. These represent an average size for a Wolbachia genome from arthropods with the smallest complete Wolbachia genome being the symbiont from Trichogramma pretiosum (wTpre) which is 1,133,809 bp long [50] and the largest being the symbiont from Folsomia candida (wFol) which is 1,801,626 bp long [11]. The genomes of Wolbachia from filarial nematodes are smaller, between 957,990 bp and 1,080,084 bp [44, 51]. However, the two wApol draft genomes are fragmented, being in 200 contigs and 373 contigs, respectively. Relatively few PacBio reads were obtained due to limited material. Despite an efficient probe hybridization enrichment, the low DNA input required pooling of barcoded samples. Based upon BUSCO, assessment of genome assembly indicates that the draft genomes wApol are probably incomplete. Despite its average size, the percentage of missing single-copy orthologs conserved among proteobacteria and wApol (or missing BUSCOs) (around 22% for K3 and 22.6% for K5) is close to the percentage observed for smaller genomes as wOo (22.6%; 957,990 bp), wOv (23.1; 960,618 bp) or wPpe (21.7%; 975,127 bp). Further, the frequency of transposable elements (Group II intron-associated genes, mobile elements and IS) varies between the two drafts and lower than usually observed for Wolbachia from arthropods (Table S3). In addition, the long repetitive elements in a genome can limit the efficiency of de novo assembly following hybridization capture protocols, as shown with the de novo assembly of wAlbB (Wolbachia from Aedes albopictus), which contains a large numbers of long repetitive Group II intron-associated genes [36].
Since the first observation in Wolbachia from Culex pipiens, the presence of WO bacteriophage in Wolbachia has been well documented [52,53,54,55]. Not all Wolbachia genomes have intact prophage regions; only vestiges of prophage DNA were detected in Wolbachia genomes infecting nematodes belonging to D or L supergoups [15, 44, 51]. In the two wApol draft genomes, we identified several coding sequences annoted as phage-like proteins, such as structural phage proteins (tail, portal phage, capsid genes) (Table S4). Although, the fragmented nature of the draft genomes limits interpretation, wApol has clearly been associated with WO bacteriophage, unlike supergroup C Wolbachia in which no structural phage proteins are found (Table S4) [44]. In addition, the presence of a prophage region appears to be a limitation for de novo genome assembly using the Wolbachia genome enrichment method, perhaps due to missing probes based upon the original design or the repetitive pattern of this region, a similar observation to that of the wAlbB genome assembly, which also failed to assemble the WO region [36].
Rare presence of biotin pathway in Wolbachia
Although it is difficult to conclude about the absence of a particular gene because the wApol draft genomes are likely incomplete, our study highlights the biotin pathway as being more complete in wApol than in numerous studied Wolbachia. Until recently, only wCle (Wolbachia from bedbug Cimex lectularius) contains a complete pathway for biotin (vitamin B7) [56]. More recently, a complete pathway for biotin was identified in wNfla and wNleu (supergroup A) [57], as well as in wLug and wStri (supergroup B) [58]. Interestingly, wCle provisions biotin (but not thiamin), which significantly contributes to the fitness of host bedbug [56]. In planthoppers, wLug and wStri Wolbachia appear to increase the fecundity of their hosts, which may be related to a beneficial effect of Wolbachia-synthesized biotin and riboflavin [58]. It is tempting to speculate that this pathway in wApol may also be important in the association between Wolbachia and its pseudoscorpion hosts. From this analysis, we also identified the complete biotin pathway for Wolbachia from the isopod Armadillidium vulgare wVulC genome. The presence of the biotin operon among the Wolbachia appears rare, but when present, its physical organization appears conserved among them (Fig. 3). Phylogenetic analysis of the biotin gene cluster supports the suggestion that the biotin operons were acquired by lateral transfer from endosymbiotic bacteria, such as Cardinium species, as previously suggested [57, 58]. The lack of congruence between the phylogeny of the biotin operons and Wolbachia phylogeny suggests multiple independent transfers of the operon within Wolbachia evolutionary history.
Conclusion
Our results emphasize the diversity of Wolbachia among the pseudoscorpion family. We identified infection with two different groups of Wolbachia, suggesting their independent evolutionary inheritance, likely via host-switching. Horizontal transmission of Wolbachia among insects has been previously documented [31, 59, 60] but this is the first time that a group of pseudoscorpions has been similarly analyzed. The Wolbachia symbiont from Atemnus politus, wApol is divergent from previous described supergroups and we propose a novel clade composed of wApol and wCsco, named supergroup S. The multi-locus phylogenies and phylogenomics indicate that the supergroup S is closely related to the supergroup F (which contains Wolbachia symbionts of arthropods and filarial nematodes) and supergroup C (comprised exclusively Wolbachia of filarial nematodes).
We produced two draft genomes of wApol, one of 200 contigs (1,404,177 bp; K3) and the second of 373 contigs (1,445,964 bp; K5) using target enrichment capture. The genome sequences remain incomplete due to the low input of DNA, limiting the PacBio long read sequencing and due to the presence of large repetitive motifs and prophage region(s), previously demonstrated to interfere with complete genome assembly based on the Wolbachia genome capture enrichment.
Interestingly, the annotation of the wApol draft genome contains the biotin pathway, as opposed to its lack in most of other Wolbachia, while speculative, it might be suggested that wApol may provision biotin to their pseudoscorpion hosts, similar to wCle in its bedbug host or wStri and wLug in planthoppers. The nature of the pseudoscorpion-Wolbachia biology emphasizes the amazing complexity and evolutionary trajectories of these ubiquitous symbionts and provides the background for future comparative studies.
Methods
Source of material, DNA extraction and characterization
Specimens were caught in the field using classical soil microfauna recovery methods using a Berlese–Tullgren funnel [49, 61]. The specimens were collected at Montpellier in 2017 (43°37′52.0″N 3°52′04.6″E).
DNA from specimens was extracted using the Monarch® Genomic DNA Purification Kit following the recommended protocol for extraction from tissues (New England Biolabs (NEB), USA) with overnight incubation at 56 °C with proteinase K. The quality of the extraction was verified by a PCR amplification of the host COI gene (Table S6). A total of 20 sequences were deposited in the GenBank Data Library: MN923050-MN923069 (Table S7).
Detection and molecular characterization of Wolbachia symbionts
The determination of Wolbachia infection in populations was determined by a series of individual specific PCRs (Table S1). The presence of Wolbachia was initially screened using PCR amplification of the Wolbachia surface protein (wsp) gene. The presence of Wolbachia was further confirmed using PCR amplification of the gatB gene for all specimens which tested positive for wsp amplification or for those giving dubious results, as well as a randomly selected set of specimens which were negative for wsp amplification.
The molecular characterization of Wolbachia was determined by PCR amplification of six genes (16S rDNA gene, dnaA, coxA, fbpA, gatB and ftsZ) as described in Lefoulon et al. [14] (Table S6). For some specimens, nested PCR amplification was necessary to obtain sufficient PCR product. A total of 14 sequences were deposited in the GenBank Data Library: MN931247 to MN931248, MN931689 to MN931700 and MT273088 (Table S8).
Next-generation sequencing of Atemnus politus
Atemnus politus specimens (K5 and K3) were collected from the same location, but at different times within the same year (respectively 5 April and 22 February 2017). The extracted DNAs were low (around 350 ng total based on Nanodrop™ quantification Thermo Scientific, USA)) and fragmented (average size around 6000 bp). In order to attain an amount of DNA necessary for the enrichment method, the DNA of the different specimens was individually barcoded and processed simultaneously, using SeqCap® barcoded adaptors (Roche NimbleGen, USA) (Table 1). Samples were not pooled to avoid concerns of potential Wolbachia strain differences. Four different libraries preparations were processed: one Illumina and one PacBio library using DNA enrichment and Illumina and PacBio libraries without enrichment, to determine enrichment efficiency (Table 1). The enrichment method has been described by Lefoulon et al. [36] and it is based on the use of biotinylated probes to capture Wolbachia DNA (probes designed by Roche NimbleGen).
For PacBio sequencing (Pacific Biosciences, USA), the Large Enriched Fragment Targeted Sequencing (LEFT-SEQ) method, as previously described, was utilized [36] without the DNA fragmentation step. The PacBio library without enrichment was produced using the SMRTbell® Express Template Prep Kit 2.0 for Low DNA input, using the barcoded Overhang Adapter Kit- 8B (PacBio). The enriched PacBio library was sequenced using 2 SMRT cells with the PacBio RS II system and the library without enrichment used 1 SMRT cell, all on the PacBio Sequel I system.
For Illumina sequencing, a modified protocol was performed, which eliminated the last steps designed for the PacBio library protocol (end repair, ligation of PacBio adaptor and purification) and in which DNA was fragmented using NEBNext® Ultra™ II FS DNA Library Prep Kit (NEB) at 20 °C for 30 min, resulting in DNA fragments with an average size of 350 bp. We used 100 ng of DNA per sample for each capture and used SeqCap barcoded adaptors (Roche Nimblegen) to process simultaneous multiple samples. The Illumina library without enrichment was produced using the NEBNext® Ultra™ II FS DNA Library Prep Kit following the manufacturer’s recommendations (NEB). The enriched Illumina library was sequenced on three independent Ilumina MiSeq runs: one mate-pair 150 bp read and two mate-pair 300 bp reads. The unenriched Illumina library was sequenced with one, single-end, 150 bp NextSeq run. All sequencing was performed at NEB.
De novo assembly pipeline
Illumina reads were filtered by quality and “cleaned” using the wrapper Trim Galore! (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/), and then merged with PEAR [62]. PacBio circular consensus sequences (CCS) were generated using SMRT® pipe RS_ReadsOfInsert Protocol (PacBio) with a minimum 3 full passes and minimum predicted accuracy superior at 90. The adaptors were removed by trimming off the first and last 65 bp of the reads, any reads smaller than 20 bp, or reads containing residual adaptor sequences (potential chimeric reads) using seqkt (github.com/lh3/seqtk) (analyses were performed with an in-house shell script).
A first hybrid de novo assembly was done using Unicycler [63]. The potential contigs belonging to Wolbachia were detected by nucleotide similarity using blastn [40] and selected (similarity superior at 80%, bitscore superior at 50). The Illumina reads were mapped against this contig selection using Bowtie2 (with the very sensitive settings) [64] and the PacBio reads using ngmlr (with the PacBio preset settings) [65].
A second hybrid de novo assembly was performed using Unicycler with this new selection of reads. Selection of Wolbachia contigs was performed using blastn a second time (similarity superior at 80%, bitscore superior at 50) and then verified manually. Assembly statistics were calculated using QUAST [41].
Comparative genomic analyses and annotation of wApol
The comparative genomic analyses described below included analyses of 10 available complete genomes of Wolbachia and 4 draft genomes (Table S8): wMel, Wolbachia from Drosophila melanogaster (NC_002978), wCau, Wolbachia from Carposina sasakii (CP041215) and wNfla, Wolbachia from Nomada flava (LYUW00000000) for supergroup A; wPip, Wolbachia from Culex quinquefasciatus (NC_010981), wTpre, Wolbachia from Trichogramma pretiosum (NZ_CM003641), wLug, Wolbachia from Nilaparvata lugens, wStri (MUIY01000000), Wolbachia from Laodelphax striatella (LRUH01000000) and wVulC, Wolbachia from Armadillidium vulgare (ALWU00000000) for supergroup B; wPpe, Wolbachia from Pratylenchus penetrans for supergroup L (NZ_MJMG01000000); wCle, Wolbachia from Cimex lectularius for supergroup F (NZ_AP013028); wFol, Wolbachia from Folsomia candida for supergroup E (NZ_CP015510); wBm, Wolbachia from Brugia malayi (NC_006833) for supergroup D; wOv Wolbachia from Onchocerca volvulus (NZ_HG810405) and wOo, Wolbachia from Onchocerca ochengi (NC_018267) from supergroup C.
The Average Nucleotide Identity (ANI) between the wApol draft genomes and available complete genomes of Wolbachia was performed using the ANI Calculator [45]. The completeness of the draft genome was studied using BUSCO v3 [43]. BUSCO estimates the completeness of genomes analyzing gene content and comparing to selection of near-universal single-copy orthologue genes (here, 221 genes in common among proteobacteria (proteobacteria_odb9)).
The processed drafts were analyzed using RAST pipeline [42]. Transposable elements were identified: insertion sequences (ISs) using ISSAGA [66] and mobile element and group II introns using RAST pipeline. The phage-like proteins were identified using the RAST pipeline. KEGG Orthology (KO) assignment were generated using KAAS (KEGG Automatic Annotation Server [67]. KAAS assigned orthologue genes by blast comparison against KEGG genes database using BBH (bi-directional best hit) method. The same assignment analysis was performed for the wApol draft genome, the set of 14 complete or draft genomes and 1 supplementary draft genome: wNleu, Wolbachia from Nomada eucophthalma (LYUV00000000). The assigned KOs were ordered in 165 different KEGG pathways (Table S5). For the biotin operon, the amino-acid sequences were identified with the KAAS assignments of wApol and other studied Wolbachia genomes and the complete operons were identified using tblastn and then aligned. The nucleotide sequences were aligned using MAFFT [46], concatenated using Seaview [68] and a phylogenetic analysis was performed with Maximum Likelihood inference inference using IQ-TREE [39].
Multi-locus phylogeny and Phylogenomics
Two multi-locus phylogenies were performed. The first phylogeny was based on six markers (16S rDNA, dnaA, ftsZ, coxA, fbpA and gatB), classically used for Wolbachia phylogeny [14, 69]. The produced sequences were analyzed with available sequences extracted from 49 Wolbachia complete or draft genomes and the addition of sequences from Wolbachia from Zootermopsis angusticollis and Zootermopsis nevadensis (Table S8). A supplementary analysis was based on 13 markers (groEL, fabK, nuoG, NADH dehydrogenase I subunit F, aspS, gltA, coxA, ftsZ, wsp, orpB, nuoD, isocitrate dehydrogenase gene, TPR domain-containing protein gene) which includes less Wolbachia strains (because these markers were rarely used for phylogenetic analyses) but included the only Wolbachia known to infect pseudoscorpions, Wolbachia from the pseudoscorpion Cordylochernes scorpioides [34, 47]. This analysis included 14 complete genomes in addition to the wApol draft genome and wCsco Wolbachia from Cordylochernes scorpioides (Table S8).
For phylogenomic analyses, single-copy orthologue genes among a selection of Wolbachia genomes were identified using Orthofinder [70]. Two types of phylogenomics studies were performed: one included only 15 complete genomes and one included 36 complete or draft genomes (Table S8). Differences in completeness of draft genomes can be variable and have a negative effect on the robustness of the analyses, and thus the two separate analyses were performed.
The orthologue sequence alignments were generated by MAFFT [46]. For the multi-locus phylogenies, a supermatrix of these six alignments was generated using Seaview [68], and for phylogenomics, the supermatrix was produced by Orthofinder (implemented as functionality). For the later, the poorly aligned positions of the produced orthologue genes alignments were eliminated using Gblocks [71]. The phylogenetic analyses were performed with Maximum Likelihood inference using IQ-TREE [39]. The most appropriate model of evolution was evaluated by Modelfinder [72] (implemented as functionality of IQ-TREE). The robustness of each node was evaluated by a bootstrap test (1000 replicates). The phylogenetic trees were edited by FigTree (https://github.com/rambaut/figtree/) and Inkscape (https://inkscape.org/).
Availability of data and materials
Data generated are available in GenBank: BioProject PRJNA593570; BioSample SAMN13481355 for Wolbachia endosymbiont strain of Atemnus sp. Specimen K5 (genome: WQMQ00000000); BioSample SAMN14519337 for Wolbachia endosymbiont strain of Atemnus sp. Specimen K3 (genome: JAAXCS000000000). The raw data are available in GenBank as Sequence Read Archive (SRA): SRX7550679 to SRX7550680, SRX8097933 to SRX8097936. In addition, a total of 34 sequences were deposited in the GenBank: MN923050 to MN923069, MN931247 to MN931248, MN931689 to MN931700 and MT273088.
Abbreviations
- bp(s):
-
DNA nucleotide base pair(s)
- CDS:
-
Coding sequences
- G + C:
-
DNA nucleotides Guanine plus Cytosine
- contig:
-
continuous DNA segment
- IS:
-
DNA insertion element
- nt:
-
DNA nucleotide
- PCR:
-
polymerase chain reaction
References
Sironi M, Bandi C, Sacchi L, Di Sacco B, Damiani G, Genchi C. Molecular evidence for a close relative of the arthropod endosymbiont Wolbachia in a filarial worm. Mol Biochem Parasitol. 1995;74(2):223–7.
Werren JH, Zhang W, Guo LR. Evolution and phylogeny of Wolbachia: reproductive parasites of arthropods. Proc Biol Sci. 1995;261(1360):55–63.
Hilgenboecker K, Hammerstein P, Schlattmann P, Telschow A, Werren JH. How many species are infected with Wolbachia?--a statistical analysis of current data. FEMS Microbiol Lett. 2008;281(2):215–20.
Zug R, Hammerstein P. Still a host of hosts for Wolbachia: analysis of recent data suggests that 40% of terrestrial arthropod species are infected. PLoS One. 2012;7(6):e38544.
Sazama EJ, Ouellette SP, Wesner JS. Bacterial Endosymbionts are common among, but not necessarily within, Insect Species. Environ Entomol. 2019;48(1):127–33.
Martin C, Gavotte L. The bacteria Wolbachia in filariae, a biological Russian dolls’ system: new trends in antifilarial treatments. Parasite. 2010;17(2):79–89.
Hoerauf A, Mand S, Volkmann L, Buttner M, Marfo-Debrekyei Y, Taylor M, Adjei O, Buttner DW. Doxycycline in the treatment of human onchocerciasis: kinetics of Wolbachia endobacteria reduction and of inhibition of embryogenesis in female Onchocerca worms. Microbes Infect. 2003;5(4):261–73.
Werren JH, Baldo L, Clark ME. Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol. 2008;6(10):741–51.
Zug R, Hammerstein P. Bad guys turned nice? A critical assessment of Wolbachia mutualisms in arthropod hosts. Biol Rev Camb Philos Soc. 2015;90(1):89–111.
Comandatore F, Cordaux R, Bandi C, Blaxter M, Darby A, Makepeace BL, Montagna M, Sassera D. Supergroup C Wolbachia, mutualist symbionts of filarial nematodes, have a distinct genome structure. Open Biol. 2015;5(12):150099.
Kampfraath AA, Klasson L, Anvar SY, Vossen R, Roelofs D, Kraaijeveld K, Ellers J. Genome expansion of an obligate parthenogenesis-associated Wolbachia poses an exception to the symbiont reduction model. BMC Genomics. 2019;20(1):106.
Bandi C, Anderson TJ, Genchi C, Blaxter ML. Phylogeny of Wolbachia in filarial nematodes. Proc Biol Sci. 1998;265(1413):2407–13.
Casiraghi M, Bain O, Guerrero R, Martin C, Pocacqua V, Gardner SL, Franceschi A, Bandi C. Mapping the presence of Wolbachia pipientis on the phylogeny of filarial nematodes: evidence for symbiont loss during evolution. Int J Parasitol. 2004;34(2):191–203.
Lefoulon E, Bain O, Makepeace BL, d'Haese C, Uni S, Martin C, Gavotte L. Breakdown of coevolution between symbiotic bacteria Wolbachia and their filarial hosts. PeerJ. 2016;4:e1840.
Brown AM, Wasala SK, Howe DK, Peetz AB, Zasada IA, Denver DR. Genomic evidence for plant-parasitic nematodes as the earliest Wolbachia hosts. Sci Rep. 2016;6:34955.
Haegeman A, Vanholme B, Jacob J, Vandekerckhove TT, Claeys M, Borgonie G, Gheysen G. An endosymbiotic bacterium in a plant-parasitic nematode: member of a new Wolbachia supergroup. Int J Parasitol. 2009;39(9):1045–54.
Ferri E, Bain O, Barbuto M, Martin C, Lo N, Uni S, Landmann F, Baccei SG, Guerrero R, de Souza LS, et al. New insights into the evolution of Wolbachia infections in filarial nematodes inferred from a large range of screened species. PLoS One. 2011;6(6):e20843.
Lefoulon E, Gavotte L, Junker K, Barbuto M, Uni S, Landmann F, Laaksonen S, Saari S, Nikander S, de Souza LS, et al. A new type F Wolbachia from Splendidofilariinae (Onchocercidae) supports the recent emergence of this supergroup. Int J Parasitol. 2012;42(11):1025–36.
Bordenstein S, Rosengaus RB. Discovery of a novel Wolbachia super group in Isoptera. Curr Microbiol. 2005;51(6):393–8.
Glowska E, Dragun-Damian A, Dabert M, Gerth M. New Wolbachia supergroups detected in quill mites (Acari: Syringophilidae). Infect Genet Evol. 2015;30:140–6.
Lo N, Casiraghi M, Salati E, Bazzocchi C, Bandi C. How many wolbachia supergroups exist? Mol Biol Evol. 2002;19(3):341–6.
Lo N, Paraskevopoulos C, Bourtzis K, O'Neill SL, Werren JH, Bordenstein SR, Bandi C. Taxonomic status of the intracellular bacterium Wolbachia pipientis. Int J Syst Evol Microbiol. 2007;57(Pt 3):654–7.
Ros VI, Fleming VM, Feil EJ, Breeuwer JA. How diverse is the genus Wolbachia? Multiple-gene sequencing reveals a putatively new Wolbachia supergroup recovered from spider mites (Acari: Tetranychidae). Appl Environ Microbiol. 2009;75(4):1036–43.
Rowley SM, Raven RJ, McGraw EA. Wolbachia pipientis in Australian spiders. Curr Microbiol. 2004;49(3):208–14.
Wang GH, Jia LY, Xiao JH, Huang DW. Discovery of a new Wolbachia supergroup in cave spider species and the lateral transfer of phage WO among distant hosts. Infect Genet Evol. 2016;41:1–7.
Baldo L, Werren JH. Revisiting Wolbachia supergroup typing based on WSP: spurious lineages and discordance with MLST. Curr Microbiol. 2007;55(1):81–7.
Gerth M. Classification of Wolbachia (Alphaproteobacteria, Rickettsiales): no evidence for a distinct supergroup in cave spiders. Infect Genet Evol. 2016;43:378–80.
Almerao MP, Fagundes NJ, de Araujo PB, Verne S, Grandjean F, Bouchon D, Araujo AM. First record of Wolbachia in South American terrestrial isopods: Prevalence and diversity in two species of Balloniscus (Crustacea, Oniscidea). Genet Mol Biol. 2012;35(4 (suppl)):980–9.
Bouchon D, Rigaud T, Juchault P. Evidence for widespread Wolbachia infection in isopod crustaceans: molecular identification and host feminization. Proc Biol Sci. 1998;265(1401):1081–90.
Badawi M, Moumen B, Giraud I, Greve P, Cordaux R: Investigating the Molecular Genetic Basis of Cytoplasmic Sex Determination Caused by Wolbachia Endosymbionts in Terrestrial Isopods. Genes (Basel) 2018, 9(6).
Baldo L, Ayoub NA, Hayashi CY, Russell JA, Stahlhut JK, Werren JH. Insight into the routes of Wolbachia invasion: high levels of horizontal transfer in the spider genus Agelenopsis revealed by Wolbachia strain and mitochondrial DNA diversity. Mol Ecol. 2008;17(2):557–69.
Baldo L, Prendini L, Corthals A, Werren JH. Wolbachia are present in southern african scorpions and cluster with supergroup F. Curr Microbiol. 2007;55(5):367–73.
Weygoldt P. Mating behavior and semen transmission in the pseudoscorpion Withius subruber Simon (Cheliferidae). Zeitschrift fur Tierpsychologie. 1969;26(2):230–5.
Zeh DW, Zeh JA, Bonilla MM. Wolbachia, sex ratio bias and apparent male killing in the harlequin beetle riding pseudoscorpion. Heredity (Edinb). 2005;95(1):41–9.
Geniez S, Foster JM, Kumar S, Moumen B, Leproust E, Hardy O, Guadalupe M, Thomas SJ, Boone B, Hendrickson C, et al. Targeted genome enrichment for efficient purification of endosymbiont DNA from host DNA. Symbiosis. 2012;58(1–3):201–7.
Lefoulon E, Vaisman N, Frydman HM, Sun L, Foster JM, Slatko BE. Large enriched fragment targeted sequencing (LEFT-SEQ) applied to capture of Wolbachia genomes. Sci Rep. 2019;9(1):5939.
Gardini G, Galli L, Zinni M. Redescription of Geogarypus minor, type species of the genus Geogarypus, and description of a new species from Italy (Pseudoscorpiones: Geogarypidae). J Arachnol. 2017;45(3):424–43.
Kotrbova J, Opatova V, Gardini G, Stahlavsky F. Karyotype diversity of pseudoscorpions of the genus Chthonius (Pseudoscorpiones, Chthoniidae) in the Alps. Comp Cytogenet. 2016;10(3):325–45.
Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32(1):268–74.
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:42.
Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29(8):1072–5.
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, et al. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75.
Seppey M, Manni M, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness. Methods Mol Biol. 1962;2019:227–45.
Darby AC, Armstrong SD, Bah GS, Kaur G, Hughes MA, Kay SM, Koldkjaer P, Radford AD, Blaxter ML, Tanya VN, et al. Analysis of gene expression from the Wolbachia genome of a filarial nematode supports both metabolic and defensive roles within the symbiosis. Genome Res. 2012. https://doi.org/10.1242/bio.2012737.
Yoon SH, Ha SM, Lim J, Kwon S, Chun J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek. 2017;110(10):1281–6.
Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30(14):3059–66.
Bordenstein SR, Paraskevopoulos C, Dunning Hotopp JC, Sapountzis P, Lo N, Bandi C, Tettelin H, Werren JH, Bourtzis K. Parasitism and mutualism in Wolbachia: what the phylogenomic trees can and cannot say. Mol Biol Evol. 2009;26(1):231–41.
Chung M, Munro JB, Tettelin H, Dunning Hotopp JC: Using Core Genome Alignments To Assign Bacterial Species. mSystems 2018, 3(6).
Sabu TK, Shiju RT, Vinod K, Nithya S. A comparison of the pitfall trap, Winkler extractor and Berlese funnel for sampling ground-dwelling arthropods in tropical montane cloud forests. J Insect Sci. 2011;11:28.
Lindsey AR, Werren JH, Richards S, Stouthamer R. Comparative Genomics of a Parthenogenesis-Inducing Wolbachia Symbiont. G3 (Bethesda). 2016;6(7):2113–23.
Foster J, Ganatra M, Kamal I, Ware J, Makarova K, Ivanova N, Bhattacharyya A, Kapatral V, Kumar S, Posfai J, et al. The Wolbachia genome of Brugia malayi: endosymbiont evolution within a human pathogenic nematode. PLoS Biol. 2005;3(4):e121.
Fujii Y, Kubo T, Ishikawa H, Sasaki T. Isolation and characterization of the bacteriophage WO from Wolbachia, an arthropod endosymbiont. Biochem Biophys Res Commun. 2004;317(4):1183–8.
Gavotte L, Henri H, Stouthamer R, Charif D, Charlat S, Bouletreau M, Vavre F. A survey of the bacteriophage WO in the endosymbiotic bacteria Wolbachia. Mol Biol Evol. 2007;24(2):427–35.
Masui S, Kuroiwa H, Sasaki T, Inui M, Kuroiwa T, Ishikawa H. Bacteriophage WO and virus-like particles in Wolbachia, an endosymbiont of arthropods. Biochem Biophys Res Commun. 2001;283(5):1099–104.
Wright JD, Sjostrand FS, Portaro JK, Barr AR. The ultrastructure of the rickettsia-like microorganism Wolbachia pipientis and associated virus-like bodies in the mosquito Culex pipiens. J Ultrastruct Res. 1978;63(1):79–85.
Nikoh N, Hosokawa T, Moriyama M, Oshima K, Hattori M, Fukatsu T. Evolutionary origin of insect-Wolbachia nutritional mutualism. Proc Natl Acad Sci U S A. 2014;111(28):10257–62.
Gerth M, Bleidorn C. Comparative genomics provides a timeframe for Wolbachia evolution and exposes a recent biotin synthesis operon transfer. Nat Microbiol. 2016;2:16241.
Ju JF, Bing XL, Zhao DS, Guo Y, Xi Z, Hoffmann AA, Zhang KJ, Huang HJ, Gong JT, Zhang X, et al. Wolbachia supplement biotin and riboflavin to enhance reproduction in planthoppers. ISME J. 2020;14:676–87.
O'Neill SL, Giordano R, Colbert AM, Karr TL, Robertson HM. 16S rRNA phylogenetic analysis of the bacterial endosymbionts associated with cytoplasmic incompatibility in insects. Proc Natl Acad Sci U S A. 1992;89(7):2699–702.
Gerth M, Rothe J, Bleidorn C. Tracing horizontal Wolbachia movements among bees (Anthophila): a combined approach using multilocus sequence typing data and host phylogeny. Mol Ecol. 2013;22(24):6149–62.
Kaydan MB, Benedicty ZK, Kiss B, Szita E. A survey of scale insects in soil samples from Europe (Hemiptera, Coccomorpha). Zookeys. 2016;565:1–28.
Zhang J, Kobert K, Flouri T, Stamatakis A. PEAR: a fast and accurate Illumina paired-end reAd mergeR. Bioinformatics. 2014;30(5):614–20.
Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13(6):e1005595.
Langmead B, Salzberg SL. Fast gapped-read alignment with bowtie 2. Nat Methods. 2012;9(4):357–9.
Sedlazeck FJ, Rescheneder P, Smolka M, Fang H, Nattestad M, von Haeseler A, Schatz MC. Accurate detection of complex structural variations using single-molecule sequencing. Nat Methods. 2018;15(6):461–8.
Varani AM, Siguier P, Gourbeyre E, Charneau V, Chandler M. ISsaga is an ensemble of web-based methods for high throughput identification and semi-automatic annotation of insertion sequences in prokaryotic genomes. Genome Biol. 2011;12(3):R30.
Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35(Web Server issue):W182–5.
Gouy M, Guindon S, Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27(2):221–4.
Baldo L, Dunning Hotopp JC, Jolley KA, Bordenstein SR, Biber SA, Choudhury RR, Hayashi C, Maiden MC, Tettelin H, Werren JH. Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl Environ Microbiol. 2006;72(11):7098–110.
Emms DM, Kelly S. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015;16:157.
Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56(4):564–77.
Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14(6):587–9.
Acknowledgments
We thank Mark Judson from MNHN for the confirmation of the morphological identification of the pseudoscorpion specimens. We thank Andy Gardner, Tom Evans, Rich Roberts, Jim Ellard and Don Comb from New England Biolabs for their support. We thank Laurie Mazzola, Danielle Fuchs and and Kristen Augulewicz from the NEB Sequencing Core.. We also thank anonomous reviewers for helpful and constructive comments.
Funding
Funds provided by New England Biolabs, Inc., Internal Research Funding who approved the design of the study and collection, analysis, interpretation of data and final manuscript.
Author information
Authors and Affiliations
Contributions
E.L., L.G., B.E.S. and C.M. conceived and designed the experiments. E.L., T.C., M.P.S. and F.B. performed the experiments. E.L. and L.G. analyzed the data. L.G. and C.M. contributed materials. E.L., L.G., C.M. and B.E.S wrote the main manuscript text. All authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable, no consent form required.
Competing interests
EL and BS are employed by New England Biolabs, Inc., who provided funding for this project.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Figure S1.
Unrooted phylogenetic trees of Wolbachia based on 13 markers by Maximum Likelihood. Analysis based on concatenation of groEL, fabK, nuoG, NADH dehydrogenase I subunit F, aspS, gltA, coxA, ftsZ, wsp, orpB, nuoD, isocitrate dehydrogenase gene and the TPR domain-containing protein gene. The total length of the dataset is 6461 bp. The topology was inferred using Maximum Likelihood (ML) inference using IQ-TREE [39]. The Best-fit model, calculated using ModelFinder according to BIC index, was GTR + R3. Nodes are associated with Bootstrap values based on 1000 replicates and only bootstrap value superior to 70 are indicated. The Wolbachia supergroups (A–S) are indicated.
Additional file 2: Figure S2.
Phylogeny of Wolbachia based on the 16S ribosomal RNA gene. Analysis based on alignment of 101 16S rRNA sequences of the total length of 445 bp. The topology was inferred using Maximum Likelihood (ML) inference using IQ-TREE [39]. The Best-fit model, calculated using ModelFinder according to BIC index, was K2P + R2. Nodes are associated with Bootstrap values based on 1000 replicates.
Additional file 3: Figure S3.
Phylogeny of Wolbachia based on the ftsZ gene. Analysis based on alignment of 95 ftsZ sequences of the total length of 779 bp. The topology was inferred using Maximum Likelihood (ML) inference using IQ-TREE [39]. The Best-fit model, calculated using ModelFinder according to BIC index, was TIM3 + G4. Nodes are associated with Bootstrap values based on 1000 replicates.
Additional file 4: Table S1.
Wolbachia infections in 3 species of pseudoscorpions caught in the Montpellier (France) area determined by PCR amplification.
Additional file 5: Table S2.
Summary of BUSCOs analyses of Wolbachia complete genomes and wApol.
Additional file 6: Table S3.
List of detected transposable elements.
Additional file 7: Table S4.
List of phage-like genes detectd by RAST pipeline.
Additional file 8: Table S5.
Summary of number of genes assigned to 165 different KEGG pathways from Wolbachia genomes using KASS.
Additional file 9: Table S6.
PCR information.
Additional file 10: Table S7.
NCBI accession of CO1 sequences of pseudoscorpion produced in the study.
Additional file 11: Table S8.
NCBI accession of Wolbachia sequences used in the study and detail of sequences included in molecular analyses. The sequences produced are indicated in bold and blue.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Lefoulon, E., Clark, T., Borveto, F. et al. Pseudoscorpion Wolbachia symbionts: diversity and evidence for a new supergroup S. BMC Microbiol 20, 188 (2020). https://doi.org/10.1186/s12866-020-01863-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12866-020-01863-y