Abstract
Background
Multi-drug resistant forms of Pseudomonas aeruginosa (MDRPA) are a major source of nosocomial infections and when discharged into streams and rivers from hospital wastewater treatment plants (HWWTP) they are known to be able to persist for extended periods. In the city of Manaus (Western Brazilian Amazon), the effluent of three HWWTPs feed into the urban Mindu stream which crosses the city from its rainforest source before draining into the Rio Negro. The stream is routinely used by Manaus residents for bathing and cleaning (of clothes as well as domestic utensils) and, during periods of flooding, can contaminate wells used for drinking water.
Results
16S rRNA metagenomic sequence analysis of 293 cloned PCR fragments, detected an abundance of Pseudomonas aeruginosa (P. aeruginosa) at the stream’s Rio Negro drainage site, but failed to detect it at the stream’s source. An array of antimicrobial resistance profiles and resistance to all 14 tested antimicrobials was detected among P. aeruginosa cultures prepared from wastewater samples taken from water entering and being discharged from a Manaus HWWTP. Just one P. aeruginosa antimicrobial resistance profile, however, was detected from cultures made from Mindu stream isolates. Comparisons made between P. aeruginosa isolates’ genomic DNA restriction enzyme digest fingerprints, failed to determine if any of the P. aeruginosa found in the Mindu stream were of HWWTP origin, but suggested that Mindu stream P. aeruginosa are from diverse origins. Culturing experiments also showed that P. aeruginosa biofilm formation and the extent of biofilm formation produced were both significantly higher in multi drug resistant forms of P. aeruginosa.
Conclusions
Our results show that a diverse range of MDRPA are being discharged in an urban stream from a HWWTP in Manaus and that P. aeruginosa strains with ampicillin and amikacin can persist well within it.
Similar content being viewed by others
Background
Pseudomonas aeruginosa (P. aeruginosa) is a ubiquitous Gram-negative, opportunistic bacterium that is spread in hospitals on the hands of healthcare workers and by contact with improperly cleaned hospital surfaces and equipment on which it can persist over long periods [1, 2]. Pseudomonas aeruginosa can cause, sometimes invasive, infections of burns and surgical wounds that can be life-threating [3, 4]. Pseudomonas aeruginosa has intrinsic multidrug resistance and several strains have acquired resistance to a wide variety of antimicrobials, including 3rd and 4th generation cephalosporins and carbapenems [1, 5–8]. Multi drug resistant Pseudomonas aeruginosa (MDRPA) are thus a significant and growing source of nosocomial infections that have few treatment options and that are especially problematic for immunocompromised patients and intensive care units (ICU) [9–12].
The dissemination of antimicrobial resistant bacteria from hospital wastewater treatment plants (HWWTP) has been a cause for concern for public health professionals [3, 13–22]. Previous studies have identified MDRPA as the primary pathogen found in the discharge of HWWTP, suggesting that the bacterium could have an important role in the spread of antimicrobial resistance, which is a global and increasing healthcare problem [23]. Consistent with this notion, studies investigating the distribution and persistence of P. aeruginosa have shown that the bacterium is widely distributed in the environment and that P. aeruginosa, including MDRPA, discharged from HWWTP can persist in the environment over extended periods [24]. The discharge of MDRPA in hospital effluents is thus increasingly being seen as having an important role in the global spread of antimicrobial resistance and is therefore an important public health concern [24, 25].
In this study, we have conducted a preliminary evaluation of the public health threat that MDRPA pose to individuals that live in contact with HWWTP contaminated waters in the city of Manaus (Western region of the Amazon). Using a range of molecular and culturing techniques we have investigated the diversity, persistence and origin of MDRPA in an urban stream in Manaus. The Mindu stream is used by some of Manaus’s most deprived residents for bathing, basic domestic cleaning (of clothes and cooking utensils) and receives discharge waters from three HWWTP as it crosses the city (See Fig. 1). The stream can also contaminate drinking water wells during times of flooding. Our results suggest that HWWTP are polluting the Mindu stream in Manaus with antibiotic resistant P. aeruginosa and may be putting residents that live in contact with these contaminated waters at risk to nosocomial infections.
Study setting. (a) Manaus city (red point), state of Amazonas (yellow), Brazil (outline). (b) Mindu stream crossing Manaus to its Rio Negro drainage site (Outfall). (c) Mindu stream flows from its source in the Ducke Forest Reserve and crosses Manaus receiving non-treated wastewater and wastewater from the treatment plants of at least three hospitals. Red points show the sites where surface water samples were collected for the preparation of metagenomic libraries. The blue points show the sites where surface water collections were made from the Mindu stream and green squares show hospitals near to Mindu stream
Results and discussion
Bacterial diversity at the Mindu stream´s source and drainage sites
As a preliminary and general assessment of the bacterial ecology of the Mindu stream, two bacterial 16S rRNA gene metagenomic libraries were constructed from bacterial concentrates made from the surface water of: (1) a site close to the stream’s source (the S-Library) and (2) a site close to where the stream drains into the Rio Negro (the outfall site, O-Library) (see Fig. 1). A total of 384 16S rRNA clones were randomly chosen for Sanger sequencing from the two libraries, 192 from the S-Library and 192 from the O-Library. Sequence comparisons made with 156 high-quality sequences obtained from the S-Library and reference sequence deposits maintained at the ribosomal Database Project (RDP) and the National Center for Biotechnology Information (NCBI) allowed for 126 (81,0 %) to be confidently classified to the bacterial genus level, including 23 (18,3 %) from the Pseudomonas genus. Four of these Pseudomonas genus sequences were identical to P. fluorescens (17.4 %), two were identical with P. plecoglossicida (8.7 %), and one identical sequence match (4.3 %) was found for three other Pseudomonas species: P. putida, P. segetis and P. azotoformans. No P. aeruginosa were detected from our S-Library sequencing.
A total of 167 high-quality 16S rRNA gene sequences were obtained from the 192 O-Library clones that were randomly selected for sequencing. Of these high-quality sequences, database similarity matches allowed 134 sequences to be confidently classified to bacterial genus. A very similar proportion of the O-library, 19.6 % (26) as was observed at the S-library (see Fig. 2), were classified by database similarity comparisons as belonging to the Pseudomonas genus. However, in contrast to the S-library, a high proportion of the Pseudomonas sequences from the O-Library, 14 (53.8 %) were identified as P. aeruginosa. Four of the O-Library sequences (15.4 %) were classified as P. fluorescens and two (7.7 %) were classified as P. alcaliphila. Pseudomonas otitidis and P. pseudoalcali sequences were also identified as occurring in the O-library. Hospital effluents are a major source of environmental contamination and contribute substantially to changes in the Pseudomonas communities. Our metagenomic data is therefore in agreement with previous studies that have shown that hospital wastewater discharges can change the structure and composition of the bacterial communities that they are discharged into [16, 26–30].
Antimicrobial resistance profiles of P. aeruginosa from the Mindu stream and a Manaus HWWTP
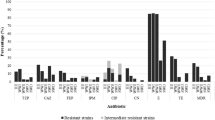
A total 25 P. aeruginosa isolates were tested for drug susceptibility with a panel of 14 antimicrobials (Fig. 3a and b). Nine P. aeruginosa cultures were prepared from bacteria obtained from the raw hospital effluent (RHE) that is fed into the HWWTP for processing and eight were prepared from treated hospital water effluent (THE) samples, which are usually discharged into the Mindu stream. A further eight P. aeruginosa cultures were prepared from Mindu stream surface water samples which were taken from two locations, upstream and downstream of the treatment site (Fig. 1). As can be seen in Fig. 3a, a total of 15 widely variant antimicrobial resistance profiles were found among the HWWTP P. aeruginosa isolates. Overall the isolates from the HWWTP can be seen to display resistance to between one and 12 antimicrobials, with the RHE displaying resistance to an average of 7.4 antimicrobials and those from the THE displaying resistance to an average of 7.6. In stark contrast to what was observed with the P. aeruginosa isolates from the HWWTP, only one resistance profile was detected from the Mindu stream isolates: dual resistance to ampicillin and amikacin. Our results show that P. aeruginosa with antimicrobial profiles matching some of the antimicrobial resistant P. aeruginosa discharged in the THE are able to persist in the Mindu stream and thus are consistent with what would be expected from the observations of others [23, 31, 32]. Our results thus suggest that our studied HWWTP could be polluting the Mindu stream and could be a public health risk to Manaus residents that live in contact with this stream.
Antimicrobial resistant and biofilm formation analysis. Diagram showing (a) the antimicrobial resistance profiles of 25 P. aeruginosa isolates obtained from water samples tested in this study. Isolates are labelled with either RHE, THE or ST to indicate their origin: RHE for raw hospital effluent, THE for treated hospital effluent and ST for stream sample. The susceptibility testing was carried out according to the protocol of the Clinical Laboratory Standards Institute (CLSI, 2012) using 14 different antimicrobials: Levofloxacin (LEV); Norfloxacin (NOR); Ciprofloxacin (CIP); Ceftriaxon (CRO); Cefepime (CPM); Ceftazidima (CAZ); Meropenem (MER); Imipenem (IMP); Amicacina (AMI); Gentamicina (GEN); Ampicilin (AMP); Piperaciline-Tazobactam (PTZ); Aztreonam monobactan (ATM); and Polimixina B-I (POL). Black-squares are used to indicate resistance and white-squares are used to indicate susceptibility. b Frequencies of antimicrobial resistant phenotypes were calculated as percentages of antimicrobial resistance among isolates of same origin. c-d Comparison of biofilm formation in LB broth with or without shaking. Biofilm production expressed as absorbance measurements taken at 600 nm. Comparison between MDRPA versus P. aeruginosa (c); Comparison between HE versus ST isolates (d); e-f Linear regression analysis showing that the number of antimicrobials a MDRPA is resistant to correlates with the amount of biofilm it produces independently of shaking
Genetic characterization of Mindu stream and HWWTP P. aeruginosa
Figure 4 shows pulse field gel electrophoresis (PFGE) fingerprints generated for each of the 25 P. aeruginosa isolates used in the antimicrobial testing (Fig. 3). These fingerprints were generated by running genomic DNA XbaI restriction enzyme digestion products on a PFGE gel. Although clear similarity between these fingerprints are observable between the isolates taken from the HWWTP, there is also clear evidence of high levels of genetic diversity among the RHE and THE bacterial isolates as well as those obtained from the Mindu stream. This approach did not, however, help to resolve the precise origin of antimicrobial resistant P. aeruginosa isolates from the Mindu, but did show that, despite the fact they all have identical antimicrobial resistance profiles, they are not all closely related to one another. These results thus suggest that the P. aeruginosa of the Mindu stream are most probably derived from multiple origins which adds weight to the notion that the P. aeruginosa with resistance to ampicillin and amikacin are at a selective advantage in this stream [23, 31, 32].
The biofilm formation of Mindu stream MDRPA
All 25 above mentioned P. aeruginosa isolates where also assessed for their ability to form biofilms under shaking and non-shaking growth conditions. Biofilm formation was observed to be significantly higher with MDRPA than non-multi-resistant P. aeruginosa (p <0.05) while no differences were seen when the isolates were classified based on their origin (Fig. 3c-d). These data indicate thus that biofilm formation is a characteristic of MDRPA. Consistent with this notion, we also observed a positive association between the number of antimicrobials a P. aeruginosa isolate was resistant to (i.e., the extent of an isolate’s multi-drug resistance) and the extent of biofilm it formed. Our results are thus consistent with other studies that have suggested that multi-drug resistance is associated with biofilm formation [12, 33–36].
Conclusions
A HWWTP in Manaus is discharging MDRPA into the urban Mindu stream, where antimicrobial resistant P. aeruginosa may be at a selective advantage and are certainly able to persist. Many of Manaus’s most vulnerable residents that live in contact the Mindu stream are being exposed to antimicrobial resistant P. aeruginosa and any and all health risks posed by such bacteria. More research is required to determine the extent to which HWWTPs are contributing to the occurrence of antimicrobial resistant P. aeruginosa in the Mindu stream and to determine the public health importance of such bacteria.
Methods
Waste water collections
Figure 1 shows a map of the Brazilian city of Manaus, Amazonas state, with the key study localities indicated. Waste water collections from the Hospital Pronto Socorro 28 de Agosto HWWTP were made twice: once in January 2012, and once in July 2012. Surface water samples from the Mindu stream were collected in July 2012 at locations downstream of where the HWWTPs of Manaus discharge their waste water (as indicated in Figs. 1 and 5). Two samples of HWWTP were collected in raw hospital effluent (RHE) and outflow-treated hospital effluent (THE). All water samples were 250 mL in volume and transported refrigerated for laboratory processing. Samples were processed within six hours of collection at Leonidas and Maria Deane Research Institute, located in Manaus. All samples were filtrated with hydrophilic filters with pore sizes of 0.8 μm (Durapore) and 0.22 μm (Sterifil) (Millipore Corp., Bedford, Mass.) using a vacuum pump to concentrate samples. The bacteria collected on these membranes were used to prepare metagenomic libraries and for P. aeruginosa isolation. P. aeruginosa isolate names reflect when they were collected: samples collected in January all start with the number 1; those collected in July start with the number 2.
Surface water 16S rRNA gene Metagenomic library preparation
Two metagenomic libraries were made by extracting DNA from bacteria collected on membranes prepared from waste water samples collected at the Mindu stream’s source and outfall sites (see Fig. 1). DNA was extracted using the phenol-chloroform method, before being re-suspended in 40 μL of TEN buffer (100 mM Tris-HCl pH 8.5, 100 mM EDTA, 3 M NaCl) [37]. Bacterial 16S rDNA amplicons of ≈ 600 bp were ampified from this extracted DNA using universal 16S rRNA gene-specific primers: forward 5’ CCTACGGGAGGCAGCAG and reverse 5´ CCGTCAATTCCTTTGAGTTT 3´ [30]. The amplicons obtained from each site were ligated into the pGEM plasmid vector (pGEM®-T Easy, Promega) and used to transform into E. coli TOP10 by electroporation. A total of 383 transformed colonies had their 16S rDNA cloned fragments amplified and directly Sanger sequenced using the same primers that were used to amplify them, following an approach similar to one described previously [38]. Generated 16S metagenomic sequences were edited and then taxonomically assigned to genus using both the Bayesian rRNA classifier software from the “Ribosomal Database Project II” (RDP) database [39] and Basic Location Alignment Search Tool (BLAST) software from the National Center of Biotechnology Information (NCBI).
Pseudomonas aeruginosa isolation and antimicrobial susceptibility testing from hospital effluents
Membranes prepared from eight Mindu stream and 17 HWWTP water samples were incubated in 5 mL of LB broth at 37 °C before being plated on to a Pseudomonas Isolation Agar (PIA). The 25 P. aeruginosa isolates were identified by Gram staining and conventional biochemical testing. In brief, using cytochrome oxidase oxidation and fermentation of glucose and lactose; growth in LB Broth at 42 °C; motility; production of H2S; nitrate reduction and subsequent plating onto Pseudomonas Isolation Agar (PIA) for pioverdin production. Antimicrobial susceptibility testing was performed by the agar disk diffusion method against antipseudomonal agents according to Clinical and Laboratory Standards Institute Guidelines, 2012. The following antimicrobials were tested: β-lactamics, including 100ug/10 μg Piperaciline-Tazobactam (PTZ); 10 μg Ampicilin (AMP); 30 μg Aztreonam monobactan (ATM); fluoroquinolones, including 10 μg Norfloxacino (NOR), 5 μg Levofloxacino (LEV), and 5 μg Ciprofloxacin (CIP); 3rd generation cephalosporins, including 30 μg Ceftriaxon (CRO), 30 μg Ceftazidima (CAZ); one 4th generation cephalosporin, 30 μg Cephepime (CPM); aminoglycosides, including 30 μg Amicacin (AMI) and 10 μg Gentamicin (GEN); carbapenens, including 10 μg Imipenem (IMP) and 10 μg Meropenem (MER); and one polymixin B-I 300 U (POL). Isolates resistant to at least three classes of antimicrobials were defined as MDRPA.
Biofilm formation
Biofilm formation was tested in 15 ml polystyrene tubes following the general approach described by [40]. Biofilm formation was assessed culturing P. aeruginosa isolates in 2 ml of LB broth with or without shaking at 37 °C for 24 h. After this, cultures were removed and the adherence pellicle formation was stained with crystal violet for 30 min. After washing in saline phosphate buffer (pH 7.0), the crystals were dissolved in ethanol, and biofilm formation was calculated using optical spectrometry and 600 nm absorbance measurements.
Pulsed-field gel electrophoresis (PFGE)
Bacterial genomic DNA from the 25 P. aeruginosa isolates was fingerprinted by pulsed-field gel electrophoresis (PFGE) following XbaI restriction enzyme digestion [41]. PFGE was ran for 20 h and carried-out using the CHEF-DR III system (Bio-Rad, Melville, NY, USA) and a 50,000-bp λDNA PFGE molecular weight marker (Sigma-Aldrich, St. Louis, MO, USA). The DNA fragments were stained by immersing the gels in 400 ml of 0.5 % TBE buffer containing 100 μl of an ethidium bromide stock solution (10 mg/ml) for 30 min. The gels were photographed, and profiles visualized using the GelDoc® (Bio-Rad, Hercules, CA, USA) photo documentation system. The profiles were analysed and compared with BioNumerics software, version 6.5, using the Dice Coefficient and UPGMA comparison settings with tolerance set to 1 %.
Statistical analysis and data presentation
Data were analysed with GraphPad Prism software version 5. A non-parametric Mann Whitney t-Test was used to compare biofilm formation between MDRPA versus P. aeruginosa and between HE versus ST isolates. Linear regression analysis was used to test for a correlation between the level of biofilm formation (measured by absorbance at 600 nm) and the amount of antimicrobial resistance that a given P. aeruginosa isolate possessed. The results were considered statistically significant when p <0.05.
Abbreviations
- HWWTP:
-
Hospital wastewater treatment plants
- MDRPA:
-
Multi-drug resistant forms of Pseudomonas aeruginosa
- O-Library:
-
Outfall library
- RSE:
-
Raw sewage effluent
- S-Library:
-
Source library
- THE:
-
Treated hospital effluent.
References
Luna RA, Millecker L a, Webb CR, Mason SK, Whaley EM, Starke JR, et al. Molecular epidemiological surveillance of multidrug-resistant Pseudomonas aeruginosa isolates in a pediatric population of patients with cystic fibrosis and determination of risk factors for infection with the houston-1 strain. J Clin Microbiol. 2013;51:1237–40.
Allen JP, Ozer EA, Hauser AR. Different paths to pathogenesis. Trends in Microbiology. 2014;22:168–9.
Bjarnsholt T, Alhede M, Alhede M, Eickhardt-Sørensen SR, Moser C, Kühl M, et al. The in vivo biofilm. Trends Microbiol. 2013;21:466–74.
Cortes JA, Leal AL, Montañez AM, Buitrago G, Castillo JS, Guzman L. Frequency of microorganisms isolated in patients with bacteremia in intensive care units in Colombia and their resistance profiles. Braz J Infect Dis. 2013;17:346–52.
Koutsogiannou M, Drougka E, Liakopoulos A, Jelastopulu E, Petinaki E, Anastassiou ED, et al. Spread of multidrug-resistant Pseudomonas aeruginosa clones in a university hospital. J Clin Microbiol. 2013;51:665–8.
Lambert P a. Mechanism of antibiotic resistance in Pseudomonas aeruginosa. J R Soc Med. 2002;95:S22–6.
Polotto M, Casella T, de Lucca Oliveira MG, Rúbio FG, Nogueira ML, de Almeida MT, et al. Detection of P. aeruginosa harboring bla CTX-M-2, bla GES-1 and bla GES-5, bla IMP-1 and bla SPM-1 causing infections in Brazilian tertiary-care hospital. BMC Infect Dis. 2012;12:176.
Patel SJ, Oliveira AP, Zhou JJ, Alba L, Furuya EY, Weisenberg SA, et al. Risk factors and outcomes of infections caused by extremely drug-resistant gram-negative bacilli in patients hospitalized in intensive care units. Am J Infect Control. 2014;42:626–31.
Jeannot K, Fournier D, Müller E, Cholley P, Plésiat P. Clonal Dissemination of Pseudomonas aeruginosa isolates producing extended-spectrum β-Lactamase SHV-2a. J Clin Microbiol. 2013;51:673–5.
Cornaglia G, Mazzariol a, Lauretti L, Rossolini GM, Fontana R. Hospital outbreak of carbapenem-resistant Pseudomonas aeruginosa producing VIM-1, a novel transferable metallo-beta-lactamase. Clin Infect Dis. 2000;31:1119–25.
Vardakas KZ, Rafailidis PI, Konstantelias A a, Falagas ME. Predictors of mortality in patients with infections due to multi-drug resistant Gram negative bacteria: The study, the patient, the bug or the drug? J Infect. 2013;66:401–14.
Deziel E, Comeau Y, Villemur R. Initiation of Biofilm Formation by Pseudomonas aeruginosa 57RP Correlates with Emergence of Hyperpiliated and Highly Adherent Phenotypic Variants Deficient in Swimming, Swarming, and Twitching Motilities. J Bacteriol. 2001;183:1195–204.
Baquero F, Martínez J-L, Cantón R. Antibiotics and antibiotic resistance in water environments. Curr Opin Biotechnol. 2008;19:260–5.
Diwan V, Tamhankar AJ, Khandal RK, Sen S, Aggarwal M, Marothi Y, et al. Antibiotics and antibiotic-resistant bacteria in waters associated with a hospital in Ujjain, India. BMC Public Health. 2010;10:414.
Kümmerer K, Henninger A. Promoting resistance by the emission of antibiotics from hospitals and households into effluent. Clin Microbiol Infect. 2003;9:1203–14.
Huerta B, Marti E, Gros M, López P, Pompêo M, Armengol J, et al. Exploring the links between antibiotic occurrence, antibiotic resistance, and bacterial communities in water supply reservoirs. Sci Total Environ. 2013;456-457:161–70.
Oberlé K, Capdeville M, Berthe T, Budzinski H, Petit F. Evidence for a complex relationship between antibiotics and antibiotic-resistant Escherichia coli: from medical center patients to a receiving environment. Env Sci Technol. 2012;46:1859–68.
Fuentefria DB, Ferreira a E, Graf T, Corcao G. Pseudomonas aeruginosa: spread of antimicrobial resistance in hospital effluent and surface water. Rev Soc Bras Med Trop. 2008;41:470–3.
Rijal G, Zmuda J, Gore R, Abedin Z, Granato T, Kollias L, et al. Antibiotic resistant bacteria in wastewater processed by the Metropolitan Water Reclamation District of Greater Chicago system. Water Sci Technol. 2009;59:2297–304.
Finley RL, Collignon P, Larsson DGJ, Mcewen S a, Li XZ, Gaze WH, et al. The scourge of antibiotic resistance: The important role of the environment. Clin Infect Dis. 2013;57:704–10.
Rizzo L, Manaia C, Merlin C, Schwartz T, Dagot C, Ploy MC, et al. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: A review. Sci Total Environ. 2013;447:345–60.
Galvin S, Boyle F, Hickey P, Vellinga A, Morris D, Cormican M. Enumeration and characterization of antimicrobial-resistant Escherichia coli bacteria in effluent from municipal, hospital, and secondary treatment facility sources. Appl Environ Microbiol. 2010;76:4772–9.
Romling U, Wingender J, Muller H, Tummler B. A major Pseudomonas aeruginosa clone common to patients and aquatic habitats. Appl Env Microbiol. 1994;60:1734–8.
Kittinger C, Lipp M, Baumert R, Folli B, Koraimann G, Toplitsch D, et al. Antibiotic resistance patterns of Pseudomonas spp. Isolated from the River Danube. Front Microbiol. 2016;7:1–8.
Fuentefria DB, Ferreira AE, Gräf T, Corção G. Spread of Metallo- B -Lactamases: screening reveals the presence of a Bla Spm-1 Gene in hospital sewage in Southern Brazil. Brazilian J Microbiol. 2009;40:82–5.
Filali BK, Taoufik J, Zeroual Y, Dzairi FZ, Talbi M, Blaghen M. Waste water bacterial isolates resistant to heavy metals and antibiotics. Curr Microbiol. 2000;41:151–6.
Casamayor EO, Massana R, Benlloch S, Øvreås L, Díez B, Goddard VJ, et al. Changes in archaeal, bacterial and eukaryal assemblages along a salinity gradient by comparison of genetic fingerprinting methods in a multipond solar saltern. Environ Microbiol. 2002;4:338–48.
Novo A, André S, Viana P, Nunes OC, Manaia CM. Antibiotic resistance, Antimicrobial residues and bacterial community composition in urban wastewater. Water Res. 2013;47:1875–87.
Borneman J, Triplett E. Molecular microbial diversity in soils from eastern Amazonia: Evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl Environ Microbiol. 1997;63:2647–53.
Casamayor EO, Schäfer H, Bañeras L, Pedrós-Alió C, Muyzer G. Identification of and spatio-temporal differences between microbial assemblages from two neighboring sulfurous lakes: Comparison by microscopy and denaturing gradient gel electrophoresis. Appl Environ Microbiol. 2000;66:499–508.
Alonso A, Rojo F, Martínez JL. Environmental and clinical isolates of Pseudomonas aeruginosa show pathogenic and biodegradative properties irrespective of their origin. Environ Microbiol. 1999;1:421–30.
Römling U, Kader A, Sriramulu DD, Simm R, Kronvall G. Worldwide distribution of Pseudomonas aeruginosa clone C strains in the aquatic environment and cystic fibrosis patients. Environ Microbiol. 2005;7:1029–38.
Pratt LA, Kolter R. Genetic analyses of bacterial biofilm formation. Curr Opin Microbiol. 1999;2:598–603.
Weigel LM, Donlan RM, Shin DH, Jensen B, Clark NC, McDougal LK, et al. High-level vancomycin-resistant Staphylococcus aureus isolates associated with a polymicrobial biofilm. Antimicrob Agents Chemother. 2007;51:231–8.
Fricks-Lima J, Hendrickson CM, Allgaier M, Zhuo H, Wiener-Kronish JP, Lynch SV, et al. Differences in biofilm formation and antimicrobial resistance of Pseudomonas aeruginosa isolated from airways of mechanically ventilated patients and cystic fibrosis patients. Int J Antimicrob Agents. 2011;37:309–15.
Donlan RM, Costerton JW, Donlan RM, Costerton JW. Biofilms : Survival mechanisms of clinically relevant microorganisms. Clin Microbiol. 2002;15:167–93.
Maniatis T, Fritsch E, Sambrook J. Molecular cloning: a laboratory manua. Laboratory CSH, editor. 1982.
Crainey J, Wilson M, Post R. Phylogenetically distinct Wolbachia gene and pseudogene sequences obtained from the African onchocerciasis vector Simulium squamosum. Int J Parasitol. 2010;40:569–78.
Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2004;7:203–14.
O’Toole GA, Pratt LA, Watnick PI, Newman DK, Weaver VB, Kolter R. Genetic approaches to study of biofilms. Methods Enzymol. 1999;310:91-109.
Ribot EM, Fair M a, Gautom R, Cameron DN, Hunter SB, Swaminathan B, et al. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis. 2006;3:59–67.
Acknowledgments
This work was supported by grants from the Foundation for the Support of Amazonas State Research – FAPEAM, the National Council for Scientific and Technological Development – CNPq, and Coordination for the Improvement of Higher Education Personnel – CAPES. We thank Dr. Mauricio Sadahyro (National Institute for Research in Amazonia – INPA) for PFGE analysis. We thank Fernanda Rodrigues Fonseca (ILMD) for the outline of hydrological maps from Manaus. We also thank the health staff that helped us in the hospitals and the laboratory technicians of FIOCRUZ Amazonia (Leonidas and Maria Deane Institute – ILMD-FIOCRUZ).
Authors’ contributions
MJTLP, PT, DC, AB, FAP and CBNC performed the experiments. AB contributed with statistical analyses. GP, PAN contributed to this work in data analysis and PPO, JLC, LC and PAN drafted the paper. We declare that all authors have read and approved the final version of this article.
Competing interests
We declare that none of our authors have any actual or potential conflict of interests relating to the publication of this article.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Magalhães, M.J.T.L., Pontes, G., Serra, P.T. et al. Multidrug resistant Pseudomonas aeruginosa survey in a stream receiving effluents from ineffective wastewater hospital plants. BMC Microbiol 16, 193 (2016). https://doi.org/10.1186/s12866-016-0798-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12866-016-0798-0