Abstract
Background
Isabel Island is a Mexican volcanic island primarily composed of basaltic stones. It features a maar known as Laguna Fragatas, which is classified as a meromictic thalassohaline lake. The constant deposition of guano in this maar results in increased levels of phosphorus, nitrogen, and carbon. The aim of this study was to utilize high-quality genomes from the genus Halomonas found in specialized databases as a reference for genome mining of moderately halophilic bacteria isolated from Laguna Fragatas. This research involved genomic comparisons employing phylogenetic, pangenomic, and metabolic-inference approaches.
Results
The Halomonas genus exhibited a large open pangenome, but several genes associated with salt metabolism and homeostatic regulation (ectABC and betABC), nitrogen intake through nitrate and nitrite transporters (nasA, and narGI), and phosphorus uptake (pstABCS) were shared among the Halomonas isolates.
Conclusions
The isolated bacteria demonstrate consistent adaptation to high salt concentrations, and their nitrogen and phosphorus uptake mechanisms are highly optimized. This optimization is expected in an extremophile environment characterized by minimal disturbances or abrupt seasonal variations. The primary significance of this study lies in the dearth of genomic information available for this saline and low-disturbance environment. This makes it important for ecosystem conservation and enabling an exploration of its biotechnological potential. Additionally, the study presents the first two draft genomes of H. janggokensis.
Similar content being viewed by others
Introduction
Isabel Island (or Isabela Island) has a volcanic origin and is mainly composed of basaltic stone [1]. Its geographic location is 21.846621 N, 105.883377 W, and it is considered a continental island, although it is located ~30 km from San Blas port in Nayarit, Mexico [2]. Within the exposed surface, there are numerous scattered craters that originated from ancient explosions [3]. The most remarkable and well-preserved one hosts a small circular (maar) lake called 'Lago Crater' or 'Laguna Fragatas' [4]. This lake has a diameter of 160 m. It’s classified as a meromictic thalassohaline lake [5], with some fluctuations in its water levels depending on the rains and less probably related to the communication through porous rocks with seawater [5]. The lake does not appear to have a direct connection to the ocean, as it does not display fluctuations corresponding to marine tides [6]. As with most thalassohaline lakes, the dominant ions found in Laguna Fragatas are sodium and chloride, similar to those found in the ocean [7]. The exposed rocks lining the walls of the lake have likely undergone alterations as a result of the annual fluctuations in water levels or due to the chemical effects of guano. Isabel Island is home to numerous marine birds, and their excrement, plays a significant role in the lake's ecosystem. The guano serves as the primary source of carbon, nitrogen, and phosphorus for the lake community [8, 9]. Due to the significant presence of seabirds and the ecological importance of the island, the Mexican government declared Isabel Island a National Park (Ecological Reserve) in 1980 [10]. As part of an environmental management plan implemented in 2006, human activities on the island were restricted to ecotourism and responsible fishing [11] . These measures further diminished the already minimal human impact on the lacustrine ecosystem of Laguna Fragatas, ensuring its preservation and protection.
Hypersaline environments have salt concentrations higher than regular seawater (greater than 0.6 M). Halophiles organisms are microorganisms capable of live in hypersaline environments and often require a high salt concentration for their growth [12]. In 1978, Kushner [13] proposed an internal division based on the amount of salt they required for proper development, categorizing them as slight, moderate, and extreme halophiles. These microorganisms can be found in all three domains of life and are distinguished by their requirement of high salinity conditions for growth [14, 15]. Halophiles, halotolerant, and non-halophilic organisms can be closely related in phylogenetic trees. Despite this heterogeneity, some phylogenetically coherent groups include only halophile organisms. Archaea belonging to the class Halobacteria and Bacteria in the order Halanaerobiales, or family Halomonadaceae are examples of taxonomic groups comprising only these organisms. Halomonas is the type genus of the family Halomonadaceae, and H. elongata is the type species of the genus [16]. It is not monophyletic and comprises two separated phylogenetic groups containing many species [17]. It was proposed as a genus in 1980 [18]. The members of this genus have been used as models for studies of halophily. They grow better aerobically, but some species can grow using nitrate, nitrite, or fumarate as an electron acceptor in the presence of glucose [19]. Some of their representatives are highly halophilic bacteria [20], adapted to a wide range of saline concentrations [21]. Despite these common characteristics, the genus has other heterogeneous features shared among its members, and some of its species have promising industrial uses, as the production of betaine, ectoine, polyhydroxyalkanoates, biosurfactants, among others and the bioremediation of industrial wastes [22]. The mining of complete genomes from isolated halophile organisms allows for the identification of previously uncharacterized biosynthetic gene clusters within the genomes of sequenced organisms [23] and facilitates the understanding of the environmental dynamics within those halophilic sites. This process involves not only the computational prediction of biosynthesis-related genes but also functional interrogation, ideally leading to a comprehensive understanding of the related chemistry [24].
The main relevance of this study lies in the lack of genomic information from organisms isolated from this low disturbed hypersaline environment. The analysis utilizes high-quality genomes from Halomonas strains deposited in the NCBI database, with KEGG database serving as a guide (due to its thorough curation, substantial number of entries, and citations) for genome mining of the moderate halophilic strains genomes (Hven4, Hven7, Hven9, Hven10, Hjan13, and Hjan14) isolated from Laguna Fragatas by Aguirre [25] between 2016 and 2020 for this. This work also reports the first two draft genomes of strains from H. janggokensis.
Materials and methods
The genomic material used in this study came from bacteria isolated by Aguirre-Garrido et al. [26] (H. venusta strains Hven4, Hven7, Hven9, H. janggokensis Hjan13, and Hjan14), H. venusta Hven10 was isolated specifically for this study. The genome from H. venusta DSM 4347 T was originally isolated from marine water in Hawaii, and its genome was assembled and annotated by Martinez-Abarca, et al. [27]. Briefly, the strains were cultivated in LBS10 medium and were identified by 16S rRNA gene Sanger sequencing to [26].
Genome sequencing
Whole-genome shotgun sequencing of genomic DNA was done at the Integrated Microbiome Resource (IMR) from Dalhousie University, Canada. The libraries were prepared using the Illumina Nextera Flex Kit for the MiSeq platform (150 + 150 bp PE) [28]. The quality of the obtained sequences was checked using Fastqc 0.11.9 [29]. Reads with qualities lower than Phred 20 and lengths smaller than 280 bp were removed using Trimmomatic 0.38 [30]. Genome assemblies were done with Unicycler [31] enhancing SPAdes [32]. The quality of the assembled genomes was calculated using QUAST [33]. Contigs smaller than 1000 bp were discarded as they could lead to mismatched assemblies, and bigger ones were mapped to the same species reference genome against H. venusta DSM 4743 T, Hjan strains have no reference genome available using Geneious mapper [34]. Contigs were visualized with Mauve to check the order of the genomes due to its capability to leverage synteny to facilitate the analysis of genome alignments [35]. The genomes assembled and the database obtained were graphically represented utilizing BRIG [36].
Functional annotation and pangenome analysis
The assembled genomes from our isolates were compared against a set of complete genomes of Halomonas spp. downloaded from the NCBI database [37] using KEGG (Kyoto Encyclopedia of Genes and Genomes) database as a guide for selecting genomes with known metabolic pathways (Table 1). The annotation of our assembled genomes was carried out using the NCBI’s Prokaryotic Genome Annotation Pipeline (PGAP) [38]. Annotated genomes were used as input to obtain the pangenome with Roary 3.13.0 [39, 40] and Anvio 7.1 [41] by a Diamond alignment [42]. Anvi’o was also used for inferring the metabolic pathways modules compared against KEGG [43,44,45]. Data was organized, visualized, and plotted using R [46], Tidyverse [47], and ggplot2 [48]. TRIBE-MCL algorithm, based on Markov clustering [49] for the assignment of proteins into families based on pre-computed sequence similarity information is the approach used by Roary and Orthofinder-tools package [39, 50], for calculating the relationships between true orthologs genes (OG). Roary makes a classification into core and accessory genomes. The core genes present in the genomes are further divided into hard-core (present in > 99% of genomes) and soft-core (present in 95–99% of genomes) genes. Additionally, there are shell genes (found in 15–95% of genomes) and cloud genes (present in less than 15% of genomes) that comprise the accessory genome [51]. For the phylogenetic analyses, ANI values were calculated using Anvi’o 7.1 [52] via pyANI [53, 54]. OrthoANI was used for comparing the genomic similarity between the coding regions of the genomes [55].
Results and discussion
Genome sequencing of five Halomonas isolates
The six genomes got assembled successfully, getting complete genomes for Hven4, Hven7, Hven10, and Hven9; and draft genomes for Hjan13 and Hjan14. Quality parameters of all the assembled genomes are show in Table 1. N50 values are less than 5 contigs, and the gotten coverages oscillated from 56 × to 94x. The length of the genomes was around 4 Mb (Table 1), coinciding with the length of the Halomonas genomes, which fluctuated from 3.5 to 5 Mb. The genomes are graphically represented in Supplementary Fig. 1 indicating the GC distribution and GC skew. The focus was specially directed towards H. janggokensis due to the lack of previous records regarding the complete genome sequencing of the type strain M24T [56] This study marks the initial stride in offering genomic information concerning H. janggokensis.
Phylogenetic analysis of Halomonas spp. reveals major subdivisions inside the genus
Halomonas genomes obtained and assembled were compared against the highest quality genomes available in databases to assure the reliability of the comparations. This was particularly important to improve the genomic mining necessary for further comparisons. Members of the genus Halomonas exhibit moderate halophilicity. While certain individuals within this genus may employ metabolisms involving nitrate, nitrite, and other electron acceptors, the majority of their species demonstrate a chemoorganotrophic metabolism [15]. Conversely, in other saline lakes, microbial communities are dominated by other Arthrospira [57], Burkholderia [58], and Pseudoalteromonas [59]. Halomonas has also been identified as a significant source of carbohydrate-degrading enzymes from soda lakes in Ethiopia [60]. Sorokin et al. [61] suggest that several members of the genus Halomonas are the most metabolically diverse in soda lakes, in addition to having the capability to fully mineralize Glycine Betaine, which is one of the most widely used osmolytes by halophilic bacteria.
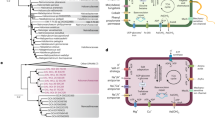
An average nucleotide identity (ANI) tree was constructed showing the phylogenetic relationship of Laguna Fragatas among the Halomonas selected genomes (Fig. 1a). The tree has two main clades with six different phylogroups. Phylogroup I, include H. alkaliphila, H. campaniensis, all the group of H. venusta isolated in this study, and its type strain DSM 4743 T. Phylogroup II includes H. axialensis, H. meridiana, and H. piezotolerans, and H. hydrothermalis well known for sharing the capability of living in high deep oceans dealing with high pressures in salty environments [62,63,64]. Phylogroup III relates H. olivaria, H. titanicae, H. sulfidaeris, and the pair of H. janggokensis isolated from Isabel Island Lake; in this comparison level, those isolates seem to be more related to each other than to H. venusta from phylogroup II. Phylogroup IV showed a closed relationship between H. campisalis, H. sulfidivorans, H. sulfidivorans, H. chromatireducens and H. sulfidoxydans strains reported to have metabolisms specialized in sulfur bioconversion [65]. Finally, H. huangheensis and did not fully integrate into its own phylogroup. In addition, the phylogenetic tree showed distant species, including H. aestuari, H. beimeninsis, and H. elongata.
a Phylogenetic tree of the 24 Halomonas strains studied whose genomes are available in metabolic pathways studies in KEGG. The tree was generated using the average nucleotide identity (ANI) values. It shows the phylogenetic relationships of the isolated strains among the Halomonas genus. b Average nucleotide sequence identity analysis shows them to belong to the same species. Identity percentages above 70% indicates affiliation to the same genus; and 95 to 100% identity to the same species
Within the phylogroup isolated from the crater, H. venusta strains Hven4 and Hven9 showed a closer relationship based on average nucleotide identity, comparing similarity between the coding regions of the genomes. Despite belonging to the same species, strains Hven9 and Hven10 have a slightly divergence. Hjan strains are completely related each other and the discrepancy against Hven strains is notable (Fig. 1b).
The Halomonas pangenome is large and open
This analysis defined the size of the pangenome increases steadily with the addition of each other genome suggesting Halomonas has a large, open pangenome. This was based on the translated amino acid sequence comparison of the 24 analyzed genomes. The pangenome comprises 100,043 genes gathered in 7479 orthogroups (OGs), representing 95% of the total. From those, 1565 OGs are present in the hard-core and 286 in the soft-core, a conservative portion of the pangenome. Meanwhile, the shell genome has 3162 OGs, and the cloud genome has 2466 (Fig. 2a). Hven isolates and the type strain showed a robust core genome having slight changes located mainly in the cloud genome. Meanwhile, despite the lack of information, Hjan13 and Hjan14 showed specific metabolic signatures in the cloud genome and some concordance between the core genomes (Fig. 2b). The identified signatures are depicted in the metabolic pathways involved in the biosynthesis of terpenes and isoprenoids with chain lengths ranging from 5 to 20 carbons, as illustrated in Fig. 7.
The Pangenome of Halomonas genus. A A pie chart representation of the pangenome of 24 Halomonas strains is cited in Table 1. The chart shows the proportion of genes classified according to their presence within the genus. Core genes are found in > 99% of genomes, soft core genes are found in 95–99% of genomes, shell genome are found in 15–95%, and cloud genome are present in less than 15% of genomes. B Gene presence/absence matrix from pangenome analysis of 24 Halomonas strains. Each row shows each isolate's gene profile and how conserved the Halomonas core genome is
Halomonas genomes have substantial and diverse biosynthetic potential
The metabolic functions assigned according to the KEGG modules are shown in Fig. 3. There are differences between species and completeness of the modules showing the calculated metabolic capabilities of each one; those capabilities are closely related to the developed mechanisms for adaptation to the environmental selection pressures present in their natural environments.
Heatmap showing KEGG modules categories and subcategories found in the complete genomes. Each row represents one KEGG Orthologous module. Completeness higher than 0.70 is necessary to assume that the module has the number of genes to be considered functional. Darker squares show more density of genes
Osmotic regulation
Most Halomonas species in this study presented a similar osmotic regulation strategy mediated by producing compatible solutes, with betaine and ectoine being the main osmoprotectants (Fig. 4). These molecules can be synthesized de novo or captured from the environment [66, 67]. Despite the clusters of genes involved in ectoine synthesis, ectABC, and betaine, betABC, were present in most of the studied genomes. The ectoine transportation system TeaABC was also detected in the Halomonas genomes. This transporter allows cells to accumulate suitable solutes when they are available in their medium [68]. Halomonas strains from Isabel Island showed a salt-out strategy related to the presence of the ectABC and betABC gene cassettes. Although the biosynthesis of compatible solutes is energetically more expensive than other strategies [69, 70], it is not a problem in an environment that has a significant amount of nutrients available for intake, allowing the members of the community to deal with the annual changes in salinity mediated by the evaporation of water levels [6].
Heatmap of gene presence (black) and absence (white) of genes associated with the metabolism and homeostatic regulation of salt (ectABC, betABC, and trkAHI), intake of nitrogen as nitrate and nitrite transporters (nasA, and narGI), phosphorus uptake (pstABCS, and phoBRU), transporters (mrpABCDEF) and production of polyhydroxyalkanoates (phaABC)
Strains in this study that did not present ectABC nor betABC operons possessed the trk system that encodes Trk proteins responsible for K + ion intake. The Trk system requires ATP and drives potassium uptake through the transmembrane electrochemical proton gradient [71, 72], a mechanism of adaptation to higher saline levels [73]. Other mechanisms include the presence of the operon mrpABCDEF, related to adaptation to osmotic pressures and pH homeostasis by changing salinity and pH via the efflux of monovalent cations such as K + , Na + , and protons) [74, 75].
Nitrogen metabolism
Halomonas is known because of its variation in the inventory of denitrification genes [65]. Besides this, dissimilatory nitrate reduction is widely spread among the genus (Fig. 5a). A set of genes required for denitrification was found in the H. venusta strains, including membrane-associated nitrate reductase genes narGH, nitrite reduction genes (either nirK or nirS) were not detected, but Isabel Island Halomonas have nirD presence, gene implied in that metabolism. The gene encoding periplasmic nitrate reductase napA and its chaperone napD were identified in some of the genomes. Assimilatory nitrate reduction is less present among the genus, but it could be detected. Nitrate reductase nasA was identified, but the genes encoding assimilatory nitrite reduction to ammonia (NIT-6 and nirA) were not identified in the genome. The inorganic nitrogen compounds acquisition and regulator gene amtB was present in only one of the genomes sets. The presence of this metabolic set can be related to the constant intake of seabird guano [1, 76]. Guano is rich in nitrogen in nitrate form [77]; the rapid action of the native bacteria explains the low levels of nitrate and high ammonia [6]. The presence of complete cassettes implied in dissimilatory nitrate reductions reinforces the importance of this compound for the metabolism of nitrogen in these domaining species (Fig. 5b).
A Heatmap showing KEGG modules implied in the metabolism of nitrogen found in the complete genomes. Each row represents one KEGG Orthologous module. B Closeup to the nitrate metabolism among Halomonas isolated. Completeness higher than 0.70 is necessary to assume that the module has the number of genes to be considered entirely functional. Darker squares show more density of genes
Phosphate metabolism
The intake of phosphorus by many bacteria is mediated by an ABC–transport complex, Pst (phosphate-specific transport), coupled to PhoU (a phosphate-specific transport system accessory protein) (Ma et al., 2016). It is an active transport system with a high affinity to Pi and has been well-characterized in Bacteria [78,79,80]. The Pst system is coded by the pstSCAB-phoU operon [81]. This system was detected in 15 of the genomes preserving the cassette in almost all of them, demonstrating the importance of facilitating and controlling phosphorus intake, as it is a limiting element, the genes implied in the intake of phosphorous are shown in Table 2.
As with nitrogen cycling, the seabird feces in Isabel Island form a eutrophic system with a large amount of available phosphorus in different forms. H. venusta strains have all the operon; meanwhile, H. janggokensis only some of the genes implied in the intake.
The utilization of phosphorus by specific members of the genus has been previously documented. Employing this capability for bioremediation to address highly eutrophicated waters, with a distinct emphasis on the removal of nitrogen and phosphorus, has yielded promising outcomes [82,83,84]. For this removal process, halophilic conditions and an alkaline pH (above 8.3) were necessary, which is consistent with Laguna Fragatas due to its alkaline pH [85].
Differences between H. venusta and H. janggokensis
Other notable differences between H. venusta and H. janggokensis are associated with dTDP-L-rhamnose biosynthesis (Fig. 6), previously identified in a strain of H. beimeninsis [72]. Although this compound has been linked to the motility of pathogens, neither of the two species (H. venusta or H. beimeninsis) has been reported as pathogenic. However, it is worth noting that dTDP-L-rhamnose can serve as a substrate for the synthesis of rhamnolipids, which have biosurfactant capabilities [86]. H. janggokensis strains show complete cassettes related to C10-C20 isoprenoids, these compounds have been synthetized using microorganisms because of higher efficiency and more environmental friendliness than traditional plant extraction and chemical synthesis methods, improving the implemented for isoprenoid production in industry in the past few decades [87].
Hven4 and Hven10 show some interesting biosynthetic capabilities related to aromatics compounds (Fig. 7) like the genes related to the biotransformation of anthranilate into catechol. These have been reported in E. coli expressing P. aeruginosa genes [88].
Conclusions
In many saline environments members of the genus Halomonas are considered minor components of the community [89, 90] the study of Laguna Fragatas, an environment dominated by this family [26] gives a novel approach towards a non-typical hypersaline environment and its community dynamics; highlighting its capacity as a genetic, and biotechnological reservoir. This report compares the relationships between high-quality genomes of Halomonas and genomes of strains isolated from Laguna Fragatas using genomic, phylogenomic, pangenomic, and metabolic approaches, revealing spread similarities in metabolic adaptations and differences related to aromatic compounds metabolisms, isoprenoid biochemistry and phosphorous intake that make the genus highly adaptable to halophilic environments.
This analysis allows us to conclude that several genes are associated with metabolism and homeostatic regulation shared by all the species of Halomonas studied. This fact, particularly in Laguna Fragatas isolates, indicates that the adaptation to high concentrations of salt, nitrogen, and phosphorus uptake are highly optimized, as could be expected from an extremophile environment characterized by no disturbances or abrupt variations in the values of ecosystem parameters. Therefore, it is a result consistent with the presence of stable microbial communities, as has already been reported in other works for archaea, viruses, and bacteria [91, 92].
In addition, this study is the first to analyze complete genomes of isolated bacteria from Isabel Island maar. It provides exploratory results about its metabolic potential, encouraging further preservation of the area and future research related to the production of metabolites of biotechnological interest, such as PHAs production. In this sense, the results of this study are consistent with other works where the relevance of the diversity of enzymes and metabolic pathways with biotechnological interest is found in this type of extreme environment. Finally, this work also reports the first two draft genomes of H. janggokensis strains.
Availability of data and materials
Genome sequences of Halomonas venusta, strains Hven4, Hven7, Hven9, Hven10 have been deposited in GenBank under accession numbers CP090952 (https://www.ncbi.nlm.nih.gov/nuccore/CP090952.1), CP090951 (https://www.ncbi.nlm.nih.gov/nuccore/CP090951.1), CP090950 (https://www.ncbi.nlm.nih.gov/nuccore/CP090950.1), JAUYZU000000000 (https://www.ncbi.nlm.nih.gov/nuccore/JAUYZU000000000.1/) respectively; while genome sequences of Halomonas janggokensis, strains Hjan13 and Hjan14 have accession numbers JAKNQU010000001 to JAKNQU010000010 (https://www.ncbi.nlm.nih.gov/nuccore/JAKNQU000000000.1/), and JAKNQT010000001 to JAKNQT010000005 (https://www.ncbi.nlm.nih.gov/nuccore/NZ_JAKNQT000000000.1), respectively. The raw genomic data were deposited under SRA accession numbers SRR17542732, SRR17542733, SRR17542734, SRR17543027, and SRR17543028.
References
Alcocer J, Lugo A, Del Rosario Sanchez M, Escobar E. Isabela crater-lake: a Mexican insular saline lake. Hydrobiologia. 1998;381:1–7.
Vargas-Marquez F. Parques Nacionales de México Vol. II Zonas Norte y Sur. Parques Nacionales de México. Mexico City: Instituto Nacional de Ecología; 1997. Available from: http://www.paot.org.mx/centro/ine-semarnat/anp/AN07.pdf.
Housh TB, Aranda-Gómez JJ, Luhr JF. Isla Isabel (Nayarit, México): quaternary alkalic basalts with mantle xenoliths erupted in the mouth of the Gulf of California. J Volcanol Geotherm Res. 2010;197(1–4):85–107. https://doi.org/10.1016/j.jvolgeores.2009.06.011.
Alvarez R, Figueroa G. Radiometric Mapping of Hydrothermal Alterations in Isla Isabel, Mexico. In: Kanlı AI, editor. Applied geophysics with case studies on environmental, exploration and engineering geophysics. London: IntechOpen; 2019. p. 1–24.
Hernández-Palacios E, Filonov A, Sánchez-Román O, López-Barba M. Hydrography of crater lake of Isabel Island, Nayarit, México. Limnology. 2022;23(1):137–51. https://doi.org/10.1007/s10201-021-00678-8.
Kienel U, Plessen B, Schettler G, Weise S, Pinkerneil S, Böhnel H, et al. Sensitivity of a hypersaline crater lake to the seasonality of rainfall, evaporation, and guano supply. Fundam Appl Limnol. 2013;183(2):135–52.
Litchfield CD. Saline Lakes. In: Reitner J, Thiel V, editors. Encyclopedia of Geobiology Encyclopedia of Earth Sciences Series. Dordrecht: Springer Netherlands; 2011 . p. 765–9. Available from: https://link.springer.com/referenceworkentry/10.1007/978-1-4020-9212-1_178. Cited 2022 Jan 8.
Kienel U, Plessen B, Schettler G, Weise S, Pinkerneil S, Böhnel H, et al. Seasonality of rainfall, evaporation, and guano supply. 2013.
Romero-Viana L, Kienel U, Sachse D. Lipid biomarker signatures in a hypersaline lake on Isabel Island (Eastern Pacific) as a proxy for past rainfall anomaly (1942-2006AD). Palaeogeography, Palaeoclimatology, Palaeoecology. 2012;350–352:49–61. Available from: https://doi.org/10.1016/j.palaeo.2012.06.011.
Diario Oficial de la Federación. Decreto por el que se declara Parque Nacional a la Isla Isabel, ubicada frente a las costas del Estado de Nayarit, declarándose de interés público la conservación y aprovechamiento de sus valores naturales, para fines recreativos, culturales y de investig. Mexico; 1980. Available from: https://dof.gob.mx/nota_detalle.php?codigo=4862220&fecha=08/12/1980#gsc.tab=0.
Diario Oficial de la Federación. AVISO por el que se informa al público en general, que la Secretaría de Medio Ambiente y Recursos Naturales ha concluido la elaboración del Programa de Manejo del Area Natural Protegida con el carácter de Parque Nacional Isla Isabel, ubicada frente a las. Mexico; 2006. p. 17–56. Available from: https://simec.conanp.gob.mx/pdf_pcym/19_DOF.pdf.
Flannery WL. Current status of knowledge of halophilic bacteria. Bacteriol Rev. 1956;20(2):49–66.
Kushner DJ. Life in high salt and solute concentrations : halophilic bacteria. In: Kushner DJ, editor. Microbial Life in Extreme Environments. London: Academic Press; 1978. p. 317–68. Available from: http://ci.nii.ac.jp/naid/10016643410/en/. Cited 2020 Mar 27.
Amoozegar MA, Safarpour A, Noghabi KA, Bakhtiary T, Ventosa A. Halophiles and their vast potential in biofuel production. Frontiers in Microbiology. Volume 10. Frontiers Media S.A; 2019.
De La Haba RR, Sanchez-Porro C, Marquez MC, Ventosa A. Taxonomy of Halophiles. In: Horikoshi K, editor. Extremophiles Handbook. Springer; 2011. p. 256–84.
Oren A. Microbial life at high salt concentrations: phylogenetic and metabolic diversity. Saline Syst. 2008;4(2). Available from: http://www.salinesystems.org/content/4/1/2.
De La Haba RR, Arahal DR, Márquez MC, Ventosa A. Phylogenetic relationships within the family Halomonadaceae based on comparative 23S and 16S rRNA gene sequence analysis. Int J Syst Evol Microbiol. 2010;60(4):737–48. Available from: https://www.microbiologyresearch.org/content/journal/ijsem/10.1099/ijs.0.013979-0. Cited 2022 Jan 11.
Vreeland RH, Litchfield CD, Martin EL, Elliot E. Halomonas elongata, a new genus and species of extremely salt-tolerant bacteria. Int J Syst Bacteriol. 1980;30(2):485–95.
Mata JA, Martínez-Cánovas J, Quesada E, Béjar V. A detailed phenotypic characterisation of the type strains of halomonas species. Syst Appl Microbiol. 2002;25(3):360–75.
Ventosa A, Nieto JJ, Oren A. Biology of moderately halophilic aerobic bacteria. Microbiol Molecular Biol Rev. 1998;62(2):504 (Available from: /pmc/articles/PMC98923/. Cited 2022 Jan 19).
Arahal DR, Ventosa A. The Family Halomonadaceae. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K, Stackebrandt E, editors. The prokaryotes. New York: Springer; 2006. p. 1689–99.
Kim KK, Lee JS, Stevens DA. Microbiology and epidemiology of Halomonas species. Future Microbiol. 2013;8(12):1559–73.
Albarano L, Esposito R, Ruocco N, Costantini M. Genome mining as new challenge in natural products discovery. Mar Drugs. 2020;18(4):1–17.
Nett M. Genome mining: concept and strategies for natural product discovery Vol. 99. Progress in the chemistry of organic natural products. 2014. p. 199–245.
de la Torre-Hernández ME, Salinas-Virgen LI, Aguirre-Garrido JF, Fernández-González AJ, Martínez-Abarca F, Montiel-Lugo D, et al. Composition, structure, and PGPR traits of the Rhizospheric bacterial communities associated with wild and cultivated echinocactus platyacanthus and neobuxbaumia polylopha. Front Microbiol. 2020;11:11(June).
Aguirre-Garrido JF, Ramírez-Saad HC, Toro N, Martínez-Abarca F. Bacterial diversity in the soda saline crater lake from Isabel Island, Mexico. Microb Ecol. 2016;71(1):68–77.
Martinez-Abarca F, Hernández-Soto LM, Ramírez-Saad HC, Aguirre-Garrido JF. Complete genome sequence of Halomonas venusta type strain DSM 4743, a moderately Halophilic Marine Bacterium. Microbiol Resour Announc. 2021;10(18):11–2.
Comeau AM, Douglas GM, Langille MGI. Microbiome helper: a custom and streamlined workflow for microbiome research. mSystems. 2017;2(1). Available from: https://journals.asm.org/journal/msystems. Cited 2021 Sep 2.
Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010.
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–20. Available from: https://academic.oup.com/bioinformatics/article/30/15/2114/2390096. Cited 2021 Sep 3.
Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13(6):1–22.
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–77.
Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29(8):1072–5.
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647–9.
Darling ACE, Mau B, Blattner FR, Perna NT. Mauve: Multiple Alignment of Conserved Genomic Sequence With Rearrangements. Genome Res. 2004;14(7):1394. Available from: /pmc/articles/PMC442156/ . Cited 2021 Sep 4.
Alikhan N-F, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 2011;12(1):1–10. Available from: https://bmcgenomics.biomedcentral.com/articles/10.1186/1471-2164-12-402. Cited 2021 Sep 1.
Agarwala R, Barrett T, Beck J, Benson DA, Bollin C, Bolton E, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2016;44(Database issue):D7. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4702911/. Cited 2022 May 19.
Tatusova T, Dicuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, et al. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016;44(14):6614–24. Available from: https://www.ncbi.nlm.nih.gov/genome/. Cited 2021 Sep 4.
Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31(22):3691–3. Available from: https://academic.oup.com/bioinformatics/article/31/22/3691/240757. Cited 2022 May 7.
Tange O. GNU Parallel 20220222 (‘Donetsk Luhansk’). 2022. Available from: https://zenodo.org/record/6213471. Cited 2022 Jun 6.
Chen LX, Anantharaman K, Shaiber A, Murat Eren A, Banfield JF. Accurate and complete genomes from metagenomes. Genome Res. 2020;30(3):315–33.
Buchfink B, Reuter K, Drost HG. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat Methods. 2021;18(4):366–8. Available from: https://www.nature.com/articles/s41592-021-01101-x. Cited 2021 Sep 3.
Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. Available from: http://www.genome.ad.jp/kegg/. Cited 2021 Sep 1.
Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019;28(11):1947–51. Available from: https://onlinelibrary.wiley.com/doi/full/10.1002/pro.3715. Cited 2022 May 16.
Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 2021;49(D1):D545–51. Available from: https://academic.oup.com/nar/article/49/D1/D545/5943834. Cited 2022 May 16.
Team RD, Core A, Language, and Environment for Statistical Computing. R Foundation for Statistical Computing. 2018;2. Available from: http://www.r-project.org.
Wickham H, Averick M, Bryan J, Chang W, Mcgowan LD et al. Welcome to the Tidyverse. J Open Source Softw. 2019;4(43):1686. https://doi.org/10.21105/joss.01686. Cited 2022 May 16.
Wickham H. ggplot2: elegant graphics for data analysis. New York: Springer-Verlag; 2016.
Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30(7):1575–84. Available from: https://pubmed.ncbi.nlm.nih.gov/11917018/. Cited 2022 May 20.
Roder T. Orthofinder-tools. 2022. Available from: https://github.com/MrTomRod/orthofinder-tools.
Livingstone PG, Morphew RM, Whitworth DE. Genome sequencing and Pan-genome Analysis of 23 Corallococcus spp. Strains reveal unexpected diversity, with Particular plasticity of predatory gene sets. Front Microbiol. 2018;9: 3187.
Eren AM, Kiefl E, Shaiber A, Veseli I, Miller SE, Schechter MS, et al. Community-led, integrated, reproducible multi-omics with anvi’o. Nature Microbiology. 2021;6(1):3–6. https://doi.org/10.1038/s41564-020-00834-3.
Pritchard L, Glover RH, Humphris S, Elphinstone JG, Toth IK. Genomics and taxonomy in diagnostics for food security: soft-rotting enterobacterial plant pathogens. Anal Methods. 2016;8(1):12–24.
Pritchard L, Harrington B, Cock P, Davey R, Waters N, Sharma A et al. widdowquinn/pyani: v0.2.12. Zenodo; 2022. Available from: https://zenodo.org/record/6927794. Cited 2022 Jul 29.
Lee I, Kim YO, Park SC, Chun J. OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol MicroBiol. 2016;66(2):1100–3.
Kim KK, Jin L, Yang HC, Lee ST. Halomonas gomseomensis sp. nov., Halomonas janggokensis sp. nov., Halomonas salaria sp. nov. and Halomonas denitrificans sp. nov., moderately halophilic bacteria isolated from saline water. Int J Syst Evol Microbiol. 2007;57(4):675–81. Available from: https://www.microbiologyresearch.org/content/journal/ijsem/10.1099/ijs.0.64767-0. Cited 2022 Jan 10.
Leboulanger C, Agogué H, Bernard C, Bouvy M, Carré C, Cellamare M, et al. Microbial Diversity and Cyanobacterial Production in Dziani Dzaha Crater Lake, a Unique Tropical Thalassohaline Environment. PLoS One. 2017;12(1):e0168879. Available from: https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0168879. Cited 2022 May 14.
Wang Y, Bao G. Diversity of prokaryotic microorganisms in alkaline saline soil of the Qarhan Salt Lake area in the Qinghai-Tibet Plateau. Scientific Reports. 2022;12(1):1–14. https://doi.org/10.1038/s41598-022-07311-3.
Naghoni A, Emtiazi G, Amoozegar MA, Cretoiu MS, Stal LJ, Etemadifar Z, et al. Microbial diversity in the hypersaline Lake Meyghan Iran. Scientific Rep. 2017;7(1):1–13. https://doi.org/10.1038/s41598-017-11585-3.
Jeilu O, Gessesse A, Simachew A, Johansson E, Alexandersson E. Prokaryotic and eukaryotic microbial diversity from three soda lakes in the East African Rift Valley determined by amplicon sequencing. Front Microbiol. 2022;13(December):1–14.
Sorokin DY. Microbial utilization of Glycine Betain in hypersaline soda lakes. Microbiology. 2021;90(5):569–77.
Yan F, Fang J, Cao J, Wei Y, Liu R, Wang L, et al. Halomonas piezotolerans sp. Nov., a multiple-stress-tolerant bacterium isolated from a deep-sea sediment sample of the new britain trench. Int J Syst Evol MicroBiol. 2020;70(4):2560–8.
Tsurumaki M, Deno S, Galipon J, Arakawa K. Complete genome sequence of Halophilic Deep-Sea Bacterium Halomonas axialensis strain Althf1. Microbiol Res Announcements. 2019;5(30):9–10.
Takahashi Y, Takahashi H, Galipon J, Arakawaa K. Complete genome sequence of Halomonas meridiana strain Slthf1, isolated from a deep-sea thermal vent. Microbiol Res Announcements. 2020;9(March):1–2.
Wang L, Shao Z. Aerobic denitrification and heterotrophic sulfur oxidation in the genus halomonas revealed by six novel species characterizations and genome-based analysis. Front Microbiol. 2021;12:390.
Gregory GJ, Boyd EF. Stressed out: bacterial response to high salinity using compatible solute biosynthesis and uptake systems, lessons from vibrionaceae. Computational Struct Biotechnol J. 2021;19:1014–27. https://doi.org/10.1016/j.csbj.2021.01.030.
Hou Q, Fang Z, Zhu Q, Dong H. Microbial diversity in Huguangyan Maar Lake of China revealed by high–throughput sequencing. J Oceanol Limnol. 2019;37(4):1245–57. Available from: https://link.springer.com/article/10.1007/s00343-019-8016-1. Cited 2022 Jan 12.
Grammann K, Volke A, Kunte HJ. New type of osmoregulated solute transporter identified in halophilic members of the Bacteria domain: TRAP transporter TeaABC mediates uptake of ectoine and hydroxyectoine in Halomonas elongata DSM 2581T. J Bacteriol. 2002;184(11):3078–85.
Ventosa A, Nieto J, Oren A. Biology of moderately halophilic aerobic bacteria. Microbiol Mol Biol Rev. 1998;62. Available from: http://mmbr.asm.org/. Cited 2020 Feb 10.
Oren A. Halophilic microbial communities and their environments. Current Opin Biotechnol. 2015;33:119–24. https://doi.org/10.1016/j.copbio.2015.02.005.
Kraegeloh A, Amendt B, Kunte HJ. Potassium Transport in a Halophilic Member of the Bacteria Domain: Identification and Characterization of the K Uptake Systems TrkH and TrkI from Halomonas elongata DSM 2581 T. J Bacteriol. 2005;187(3):1036–43. Available from: http://jb.asm.org/. Cited 2020 Mar 22.
Chen YH, Lu CW, Shyu YT, Lin SS. Revealing the Saline Adaptation Strategies of the Halophilic Bacterium Halomonas beimenensis through High-throughput Omics and Transposon Mutagenesis Approaches OPEN. Sci Rep. 2017;7:13037. Available from: www.nature.com/scientificreports/. Cited 2020 Mar 19.
Gunde-Cimerman N, Plemenitaš A, Oren A. Strategies of adaptation of microorganisms of the three domains of life to high salt concentrations. FEMS Microbiol Rev. 2018;009:353–75. Available from: https://academic.oup.com/femsre/article-abstract/42/3/353/4909803. Cited 2020 Mar 13.
Mormile MR, Edwards T, Frank R, Geurin Z, Haendiges J, Hoffmann M et al. Whole-Genome Analysis of Halomonas sp. Soap Lake #7 Reveals It Possesses Putative Mrp Antiporter Operon Groups 1 and 2. Genome Biology and Evolution. 2019;11(6):1706–9. Available from: https://academic.oup.com/gbe/article/11/6/1706/5520441. Cited 2021 Sep 30.
Fang H, Qin XY, Zhang KD, Nie Y, Wu XL. Role of the group 2 Mrp sodium/proton antiporter in rapid response to high alkaline shock in the alkaline- and salt-tolerant Dietzia sp. DQ12-45-1b. Appl Microbiol Biotechnol. 2018;102(8):3765–77.
Calixto-Albarrán I, Osorno JL. The diet of the magnificent frigatebird during chick rearing. Condor. 2000;102(3):569–76.
Zwolicki A, Zmudczyńska-Skarbek KM, Iliszko L, Stempniewicz L. Guano deposition and nutrient enrichment in the vicinity of planktivorous and piscivorous seabird colonies in Spitsbergen. Polar Biology. 2013;36(3):363–72 (Available from: https://link.springer.com/article/10.1007/s00300-012-1265-5. Cited 2022 Jan 11 ).
Furtwängler K, Tarasov V, Wende A, Schwarz C, Oesterhelt D. Regulation of phosphate uptake via pst transporters in Halobacterium salinarum R1. Mol Microbiol. 2010;76(2):378–92.
Saona LA, Soria M, Durán-Toro V, Wörmer L, Milucka J, Castro-Nallar E, et al. Phosphate-arsenic interactions in Halophilic microorganisms of the Microbial Mat from Laguna Tebenquiche: from the Microenvironment to the genomes. Microb Ecol. 2021;81(4):941–53.
Williams TJ, Allen MA, Demaere MZ, Kyrpides NC, Tringe SG, Woyke T, et al. Microbial ecology of an Antarctic hypersaline lake: genomic assessment of ecophysiology among dominant haloarchaea. ISME J. 2014;8(8):1645–58.
Alex A, Antunes A. Pyrosequencing characterization of the microbiota from Atlantic intertidal marine sponges reveals high microbial diversity and the lack of co-occurrence patterns. PLoS ONE. 2015;10(5):1–17.
Li XW, Zeng FJ, Jiang ZZ, Liu WF, Zhu YM, Zhang LH. Optimization of nitrogen and phosphorus removal from saline wastewater using Halomonas sp. H12 strain. IOP Conference Series: Earth Environ Sci. 2018;191(1):1–8.
Kim J, Kang S, Kim HS, Kim S, Lee SS. Pilot plant study on nitrogen and phosphorus removal in marine wastewater by marine sediment with sequencing batch reactor. PLoS ONE. 2020;15(5):1–16. https://doi.org/10.1371/journal.pone.0233042.
Nguyen HTT, Nielsen JL, Nielsen PH. Candidatus Halomonas Phosphatis, a novel polyphosphate-accumulating organism in full-scale enhanced biological phosphorus removal plants. Environ Microbiol. 2012;14(10):2826–37.
Aguirre-Garrido JF, Ramirez-Saad HC, Toro N, Martinez-Abarca F. Bacterial Diversity in the Soda Saline Crater Lake from Isabel Island, Mexico. Environ Microbiol. 2015;71(1):68–77. Available from: http://link.springer.com/10.1007/s00248-015-0676-6.
Aguirre-Ramírez M, Medina G, González-Valdez A, Grosso-Becerra V, Soberón-Chávez G. The Pseudomonas aeruginosa rmlBDAC Operon, encoding dTDP-L-rhamnose biosynthetic enzymes, is regulated by the quorum-sensing transcriptional regulator RhlR and the alternative sigma factor σ S. Microbiology. 2012;158(4):908–16.
Wang Z, Zhang R, Yang Q, Zhang J, Zhao Y, Zheng Y et al. Recent advances in the biosynthesis of isoprenoids in engineered Saccharomyces cerevisiae [Internet]. 1st ed. Vol. 114, Advances in Applied Microbiology. Elsevier Inc.; 2021. 1–35 p. https://doi.org/10.1016/bs.aambs.2020.11.001.
Balderas-Hernández VE, Treviño-Quintanilla LG, Hernández-Chávez G, Martinez A, Bolívar F, Gosset G. Catechol biosynthesis from glucose in Escherichia coli anthranilate-overproducer strains by heterologous expression of anthranilate 1,2-dioxygenase from Pseudomonas aeruginosa PAO1. Microb Cell Fact. 2014;13(1):1–11.
Ghai R, Pašić L, Fernández AB, Martin-Cuadrado AB, Mizuno CM, McMahon KD, et al. New abundant microbial groups in aquatic hypersaline environments. Sci Rep. 2011;1: 135.
Fernández AB, Vera-Gargallo B, Sánchez-Porro C, Ghai R, Papke RT, Rodriguez-Valera F, et al. Comparison of prokaryotic community structure from Mediterranean and Atlantic Saltern concentrator ponds by a metagenomic approach. Front Microbiol. 2014;5(MAY):1–12.
Rodriguez-Brito B, Li LL, Wegley L, Furlan M, Angly F, Breitbart M, et al. Viral and microbial community dynamics in four aquatic environments. ISME J. 2010;4(6):739–51.
Zhang M, Han F, Liu Z, Han Y, Li Y, Zhou W. Ammonium-assimilating microbiome: a halophilic biosystem rationally optimized by carbon to nitrogen ratios with stable nitrogen conversion and microbial structure. Bioresour Technol. 2022;350: 126911.
Acknowledgements
The authors express their gratitude to the staff of Ecomata Dive Center for their assistance in providing transportation to the island, accommodations during their stay, and assistance in collecting samples from the crater lake.
Funding
This work was supported by grant: Universidad Autónoma Metropolitana Unidad Lerma, México arrangement 135-(18.21.4); PID2020-113207GBI00 funded by MCIN/AEI/10.13039/501100011033 and by “ERDF A way of making Europe”. LMHS had a Consejo Nacional de Ciencia y Tecnología (CONACyT) grant.
Author information
Authors and Affiliations
Contributions
LMHS, MLP, HRS, FMA, and JFAG designed the study. LMHS did the bioinformatics analysis under the supervision of FMA and JFAG. LMHS wrote the manuscript following the suggestions of FMA, JFAG, HRS, and MLP.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Supplementary Fig. 1.
Graphical representation of the genomes of Halomonas indicating from innermost ring: distribution of the GC content (black), GC skew (purple/green), and represented with different colors, the homology with the species within the genus. Currently, Halomonas elongata is recognized as the type species among the genus. Because of this, the comparison was made against its genome.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Hernández-Soto, L.M., Martínez-Abarca, F., Ramírez-Saad, H. et al. Genome analysis of haloalkaline isolates from the soda saline crater lake of Isabel Island; comparative genomics and potential metabolic analysis within the genus Halomonas. BMC Genomics 24, 696 (2023). https://doi.org/10.1186/s12864-023-09800-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-023-09800-9