Abstract
Background
Plasmodium falciparum malaria is a leading cause of pediatric morbidity and mortality in holoendemic transmission areas. Severe malarial anemia [SMA, hemoglobin (Hb) < 5.0 g/dL in children] is the most common clinical manifestation of severe malaria in such regions. Although innate immune response genes are known to influence the development of SMA, the role of natural killer (NK) cells in malaria pathogenesis remains largely undefined. As such, we examined the impact of genetic variation in the gene encoding a primary NK cell receptor, natural cytotoxicity-triggering receptor 3 (NCR3), on the occurrence of malaria and SMA episodes over time.
Methods
Susceptibility to malaria, SMA, and all-cause mortality was determined in carriers of NCR3 genetic variants (i.e., rs2736191:C > G and rs11575837:C > T) and their haplotypes. The prospective observational study was conducted over a 36 mos. follow-up period in a cohort of children (n = 1,515, aged 1.9–40 mos.) residing in a holoendemic P. falciparum transmission region, Siaya, Kenya.
Results
Poisson regression modeling, controlling for anemia-promoting covariates, revealed a significantly increased risk of malaria in carriers of the homozygous mutant allele genotype (TT) for rs11575837 after multiple test correction [Incidence rate ratio (IRR) = 1.540, 95% CI = 1.114–2.129, P = 0.009]. Increased risk of SMA was observed for rs2736191 in children who inherited the CG genotype (IRR = 1.269, 95% CI = 1.009–1.597, P = 0.041) and in the additive model (presence of 1 or 2 copies) (IRR = 1.198, 95% CI = 1.030–1.393, P = 0.019), but was not significant after multiple test correction. Modeling of the haplotypes revealed that the CC haplotype had a significant additive effect for protection against SMA (i.e., reduced risk for development of SMA) after multiple test correction (IRR = 0.823, 95% CI = 0.711–0.952, P = 0.009). Although increased susceptibility to SMA was present in carriers of the GC haplotype (IRR = 1.276, 95% CI = 1.030–1.581, P = 0.026) with an additive effect (IRR = 1.182, 95% CI = 1.018–1.372, P = 0.029), the results did not remain significant after multiple test correction. None of the NCR3 genotypes or haplotypes were associated with all-cause mortality.
Conclusions
Variation in NCR3 alters susceptibility to malaria and SMA during the acquisition of naturally-acquired malarial immunity. These results highlight the importance of NK cells in the innate immune response to malaria.
Similar content being viewed by others
Background
Plasmodium falciparum, the most prevalent malaria parasite in sub-Saharan Africa, causes 99% of the estimated malaria cases in the region [1]. The burden of malaria largely impacts children under 5 years of age, accounting for 80% of all malaria-related mortality [1]. Life-threatening complications of malaria include hyper-parasitemia, hypoglycemia, hyper-lactatemia, kidney failure, metabolic acidosis, cerebral malaria, severe malarial anemia [SMA, hemoglobin (Hb) < 5.0 g/dL with peripheral parasitemia], and respiratory distress [2]. In holoendemic P. falciparum transmission regions, such as western Kenya, severe malaria presents primarily as SMA [3, 4]. Despite malaria control efforts in the region, SMA has remained one of the major causes of morbidity and mortality in children aged less than 5 years [4, 5].
The selective pressure of malaria on the human genome is epitomized by the sickle hemoglobin (HbS) allele of hemoglobin beta gene (HBB) for which heterozygous carriers are protected against severe malaria in areas of high malaria prevalence [6, 7]. Our previous studies in western Kenya illustrate that genetic variants, particularly those in innate immune response genes, influence susceptibility to malaria and subsequent development of SMA [8,9,10,11,12,13,14,15]. Previous linkage analysis in different ethnic groups from Africa identified a linkage between the 6p21.3 locus of the major histocompatibility complex (MHC) region and susceptibility to mild malaria [16,17,18]. Centrally located in the MHC region is the natural cytotoxicity-triggering receptor 3 (NCR3). A variant located in the promoter region of NCR3, rs2736191:C > G, has been associated with increased mild malaria episodes in carriers of the mutant allele (C allele, which corresponds to G allele in the current study) in Burkina Faso and Congo [19, 20]. The wildtype allele of rs2736191 was found to enhance NCR3 promoter activity [20]. Investigation of the rs2736191 polymorphism in a Senegalese population revealed no relationship between the SNP and cerebral malaria [21].
Although the role of NCR3 in the pathogenesis of SMA remains elusive, it is known that this protein is constitutively expressed on the surface of natural killer (NK) cells and plays an important role in NK cell activation, degranulation, and cytotoxicity [22, 23]. Moreover, NCR3 recognizes parasitized red blood cells (pRBCs), and directly binds to the Duffy Binding Like (DBL1-α) domain of P. falciparum erythrocyte membrane protein-1 (PfEMP-1) without the requirement of accessory molecules, such as MHC class I [24]. Binding of pRBCs to NCR3 on NK cells also activates cell cytotoxicity and promotes the release of cytokines and chemokines known to influence the pathogenesis of SMA [25,26,27,28,29].
Both rs2736191 and another NCR3 variant rs11575837:C>T (located in the non-coding region of exon 1) have been shown to be inversely associated with risk of primary Sjögren’s syndrome (pSS), and the minor T allele of rs11575837 is associated with reduced NCR3 gene transcription [30, 31]. Similar to severe malarial anemia, the pathogenesis of pSS involves immune dysregulation.
To extend previous studies on the potential role of NK cells in the pathogenesis of malaria, we examined the relationship between NCR3 variants (rs2736191 and rs11575837) and longitudinal clinical outcomes (i.e., malaria and SMA episodes) in a cohort of Kenyan children residing in a holoendemic P. falciparum area. Here, we report the impact of the two NCR3 variants on susceptibility to pediatric malaria and SMA throughout a 36-mos. follow-up period during the development of naturally-acquired malarial immunity.
Results
Clinical, demographic, and laboratory characteristics of the study participants at enrollment
The enrollment characteristics of the cohort, stratified according to presence of malaria and malarial anemia status [aparasitemic (n = 289), non-SMA (n = 962) and SMA (n = 264)], are presented in Table 1. The sex ratio was comparable among the three groups (P = 0.752), while age differed across the groups (P = 0.003) with SMA patients being the youngest. Children with SMA exhibited the lowest hematocrit levels as well as RBC counts (P < 0.001 and P < 0.001, respectively). White blood cell counts progressively increased across the groups (P < 0.001) and were highest in children with SMA. The non-SMA group presented with a higher parasite density than the SMA group (P = 0.043).
HIV-1 status and bacteremia were assessed in the cohort since we have shown previously that they enhance the risk of SMA [32, 33]. The distribution of HIV-1 was comparable across the groups (P = 0.116), whereas bacteremia significantly differed (P = 0.036) and was highest in the aparasitemic group. Examination of genetic factors known to influence the development of severe malaria [34,35,36] revealed that the distribution of α3.7-thalassemia variants and sickle cell trait (HbAS) differed across the groups (P = 0.014 and P < 0.001, respectively). Children with SMA had the highest carriage of heterozygous and homozygous α3.7-thalassemia variants, the lowest inheritance of HbAS, and the highest proportion of HbSS. The distribution of glucose-6-phosphate dehydrogenase (G6PD) deficiency was comparable across the groups (P = 0.387).
Characteristics of the NCR3 variants in the study population
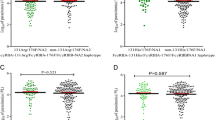
NCR3 is located on chromosome 6p21.33 (GRCh38.p12). The two SNPs selected for investigation, shown in Fig. 1A, have been shown to functionally impact gene expression and clinical outcomes in mild malaria (i.e., rs2736191) and primary pSS (i.e., rs11575837), respectively [30, 31]. Genotyping of the two SNPs in the population revealed a MAF for rs2736191 at 0.30 and rs11575837 at 0.03 (Fig. 1B). Transcription factor binding site (TFBS) analysis, using TFBIND [37], showed that the conversion of C to G in rs2736191 caused ablation of the binding site for AP-2 alpha (i.e., TFAP2A or AP2) and creation of binding sites for transcription factor 3 (i.e., TCF3 or E47) and zinc finger and BTB domain containing 6 (i.e., ZBTB6 or ZID, Fig. 1B). TFBS analysis of rs11575837 revealed that conversion of C to T results in loss of the TFAP2A binding site and the gain of a hepatocyte nuclear factor 4 (HNF4A) binding site (Fig. 1B). LD analysis between the selected SNPs yielded D′ as 0.882, LOD as 6.61, and r2 as 0.011 (Fig. 1C).
Chromosome location of NCR3 and linkage disequilibrium of SNPs under investigation. A Human natural cytotoxicity-triggering receptor 3 gene (NCR3) is located on chromosome 6p21.33 (GRCh38.p12). The SNPs under investigation were rs2736191:C > G and rs11575837:C > T, respectively. rs2736191 is located in the proximal promoter region, while rs11575837 is a 5’ UTR variant and located in exon 1. B NCR3 minor allele frequencies (MAF) for the Luhya (LWK) and Luo populations. Transcription factor binding analyses of the NCR3 variants. Transcription Factor AP-2 alpha, TFAP2A; Transcription Factor 3, TCF3; Zinc finger and BTB domain containing 6, ZBTB6; and Hepatocyte nuclear factor 4, HNF4A. C Linkage disequilibrium between the selected NCR3 SNPs (D’: 0.882, LOD: 6.61, r2: 0.011)
The distribution of genotypes and haplotypes for rs2736191 and rs11575837 is shown in Table 2. The observed frequencies of the rs2736191 genotypes in the overall population were 0.503 for CC, 0.386 for CG, and 0.111 for GG, displaying significant departure from HWE (χ2 = 11.58, P < 0.001). The observed frequencies of the rs11575837 genotypes in the overall population were 0.941 for CC, 0.054 for CT and 0.005 for TT, displaying significant departure from HWE as well (χ2 = 28.57, P < 0.001). The distribution of the genotypes and the four haplotypes were comparable across the three clinical groups.
Longitudinal risk of malaria and SMA episodes for the NCR3 genotypes and haplotypes
The impact of the genotypes and haplotypes on malaria and SMA episodes across the 36-mos. follow-up period was determined by fitting a Poisson regression model selecting for covariates [i.e., age, sex, co-infections (HIV-1 and bacteremia), G6PD deficiency, sickle-cell trait, and alpha-thalassemia] known to influence susceptibility to malaria and SMA. The results of the regression modeling and occurrence of events is shown in Table 3. There were 7,228 malaria episodes, for which 406 SMA events occurred during the cohort observational period. Significant effects on susceptibility to malaria were observed for rs11575837 in which carriage of homozygous recessive genotype (TT) increased the risk of malaria (IRR = 1.540, 95% CI = 1.114–2.129, P = 0.009, significant after Holm-Bonferroni correction). Neither rs2736191 nor any of the haplotypes derived from the two SNPs significantly impacted the longitudinal susceptibility to malaria.
A similar Poisson regression selecting for the same covariates revealed an increased risk of SMA over 36-mos. in carriers of rs2736191 genotypes CG (IRR = 1.269, 95% CI = 1.009–1.597, P = 0.041), and in the additive model (IRR = 1.198, 95% CI = 1.030–1.393, P = 0.019). There was a trend of increased risk of SMA in carriers of genotype GG without significance (IRR = 1.378, 95% CI = 0.991–1.915, P = 0.057). However, none of the results remained significant after correction for multiple comparisons. Carriage of CT or TT genotypes for rs11575837 showed a non-significant increased risk of developing SMA [(IRR = 1.294, 95% CI = 0.828–2.020, P = 0.258) and (IRR = 1.911, 95% CI = 0.609–6.000, P = 0.267), respectively]. Analysis of the impact of the haplotypes on susceptibility to SMA revealed an additive protective effect for individuals who inherited the CC haplotype (IRR = 0.823, 95% CI = 0.711–0.952, P = 0.009, significant after Holm-Bonferroni adjustment). Conversely, there was an enhanced risk of SMA in individuals who inherited the GC haplotype (IRR = 1.276, 95% CI = 1.030–1.581, P = 0.026), and in the additive model (IRR = 1.182, 95% CI = 1.018–1.372, P = 0.029), but neither retained significance after Holm-Bonferroni adjustment. There was no association between an altered risk profile of SMA for either the CT or GT haplotypes.
A Cox proportional hazard modeling was fitted to examine the relationship between the NCR3 genotypes/haplotypes and all-cause mortality. No significant relationships with all-cause mortality were observed for either rs2736191, rs11575837, or their haplotypes (Additional file 1: Table S1).
Discussion
Nearly every child under 5 years of age in the holoendemic P. falciparum transmission area where the study was conducted experience repeated episodes of malaria, yet only a subset of individuals develop SMA, typically within the first year of life. Young children (< 12 mos.) are especially susceptible to severe malaria as maternal immunity begins to wane and adaptive immunity starts to develop with repeated episodes of malaria [38]. Since childhood immunity to malaria gradually develops across successive episodes [39], the impact of genetic variants of malaria and SMA is best captured in longitudinal studies during the development of naturally-acquired malarial immunity [40]. To the best of our knowledge, we report the first study on the longitudinal risk of clinical malaria and SMA in carriers of rs2736191 and rs11575837, and their haplotypes.
The SNPs targeted for investigation were selected based on previous reports showing that the variants impart functional changes in gene expression and influence mild malaria and pSS [19, 20, 30, 31]. Notably, the substitution of C by G in rs2736191 creates TFBSs for TCF3 or ZBTB6 and ablates a binding site for TFAP2A (AP2). Previous studies show that TFAP2A (AP2) regulates the transcription of IFN-γ Receptor 1 (IFNGR1), and that elevated IFN-γ production is protective against infection with malaria [41,42,43]. In addition, TCF3 was shown to promote the development of γδ T cells, differentiation of memory CD8 T cells, and increase γδ T cells and CD8 + T cell responses in children with SMA [44,45,46,47]. Although not reported in malaria, human ZBTB6 was one of the most differentially expressed genes in the whole blood of patients with Crohn’s disease [48]. Conversion of C to T in rs11575837 leads to the loss of a TFAP2A (AP2) binding site and the gain of a HNF4A binding site. Although HNF4A was shown to promote erythropoiesis during embryonic development in a murine model, it remains to be determined if children who are carriers of the T-allele have altered binding to HNF4A and subsequent alterations in erythropoiesis [49].
The MAFs of the variants under investigation were higher in the study participants than for the LWK ethnic group included in the 1000 Genome Project [31]. Additionally, the overall distribution of rs2736191 and rs11575837 genotypes in our study population displayed significant departure from HWE, indicative of an influence of evolutionary forces on the human genome in the study population. There is mounting evidence that natural selection influences the frequencies of disease-associated genetic variants in different populations [50]. For example, malaria exerts a strong evolutionary force on risk-associated alleles [7, 40] and may, at least in part, explain findings presented here on the allelic distribution.
LD analysis revealed that the two SNPs are co-inherited (D′ = 0.882). Since D′ values are known to fluctuate upwards for less common alleles (i.e., rs11575837), we also determined the D′ confidence intervals and r2 [51]. D′ captures the recombination events (inheritance) between an allele of one SNP and that of another, while r2 is a statistical measure of correlation between the SNPs, therefore, both should be considered when deciphering the degree of association between SNPs [52]. Thus, both SNPs were investigated since the D′ was high and the r2 value was low (r2 = 0.01), suggesting that the two SNPs may convey different information.
Although previous studies found an association between carriage of the minor allele for rs2736191 and increased mild malaria [19, 20], there was no relationship between the different genotypes and malaria episodes in our study population. In the former investigation in Congolese children [20] the relationship between the mutant allele and mild malaria was only significant in children greater than 5 years of age, but not in younger children [20]. Moreover, in the parent-sib study conducted in Burkina Faso, the mean age of the sibs was 12.1 ± 6.2 years [19]. Collectively, these studies suggest an age-specific relationship between carriage of the minor allele and susceptibility to malaria that may not have been detected in our study since the population was considerably younger (primarily < 5 years). Although this hypothesis remains to be tested, there was a significant relationship between carriage of the G-allele and increased susceptibility to SMA in carriers of GG and in the additive model. However, this relationship was not significant after testing for multiple comparisons.
Although the previous family-based association study in Burkina Faso did not include rs11575837 in the analyses due to a low MAF (< 0.01) in the study population, we investigated this SNP based on its association with functional properties and susceptibility to pSS [30, 31]. The MAF in the current study for rs11575837 was 0.03 with TT carriers having significantly increased susceptibility to acute malaria across the follow-up period. The minor allele T has been associated with lowered NCR3 expression [30]. Since NCR3 can activate NK cells to clear malaria parasites by directly recognizing pRBCs [23, 24, 27], decreased NCR3 expression in TT carriers could reduce antiparasitic effects and result in increased susceptibility to malaria. Thus, even though there was a low MAF for rs11575837 in the study cohort, carriage of both T-alleles did impact on susceptibility to malaria, consistent with results found for other diseases in European populations in which the MAF was also low (0.02) [30, 31].
While there was also an increased risk for SMA (RR = 1.9) in children who inherited TT for rs11575837, the results were not significant, likely due to the low number of events for rare variant. Although requiring further evidence, it is possible that the rarity of the TT genotype maintained in the study population could be explained by increased susceptibility to malaria, SMA, and subsequent mortality. There was a progressive increase in the risk of mortality associated with carriage of T-alleles but the low carriage rate in the context of a low mortality rate cannot confirm this hypothesis. In the study region, childhood mortality from malaria has historically been very high [3]. As such, failure to reach reproductive age has had a strong impact on selection.
Investigations on the impact of coinheritance of the two SNPs did not elucidate any significant findings for altered susceptibility to longitudinal malaria episodes. However, the additive model revealed that a progressive carriage rate of both wild type alleles (CC) significantly reduced the risk of SMA. This finding is consistent with selective effects for the variants in the population, particularly for the rarer rs11575837 mutant allele. Additional modeling efforts revealed that inheritance of the CG genotype increased the risk of SMA in carriers versus non-carriers and in the additive model, but the results were not significant after correction for multiple comparisons. Nonetheless, in the context of the significant risk found for carriage of G-alleles for rs2736191 in the genotypic model, it appears that carriage of the G-allele in the GC haplotype certainly influences the longitudinal development of SMA.
Secondary analyses were performed to determine if the SNPs (or their haplotypes) predict all-cause mortality. Inheritance of either of the two SNPs individually, or in combination, were not significantly associated with all-cause mortality. However, based on the reduced number of carriers for several of the variants in the context of an overall low mortality rate in the cohort, the influence of the selected variants on childhood mortality could not be determined with a high level of statistical confidence. Additionally, there are limitations of the current study: the covariates in the statistical model did not include other potential anemia-promoting factors such as nutrition status, and the observed deviation from the HWE may introduce a bias in a genetic association study [53].
Conclusions
We provide the first evidence that carriage of the TT genotype for rs11575837 is linked to increased longitudinal malaria episodes. Additional findings include the protective effect of CC haplotype carriage against SMA in the additive model, and conversely, the additive effect of GC haplotype carriage on enhanced susceptibility to SMA. Collectively, these novel findings support further studies to define the molecular mechanisms by which NK cells and their pathways influence the pathogenesis of SMA. Such findings could facilitate the development of improved strategies for the control and clinical management of severe malaria.
Methods
Study site and study participants
The prospective observational study over 36 mos. was conducted in children presenting at Siaya County Referral Hospital (SCRH) to determine the relationship between NCR3 variants and susceptibility to malaria and SMA. SCRH is in western Kenya, a region of holoendemic P. falciparum malaria transmission [54, 55]. Severe malaria in western Kenya primarily manifests as SMA [56, 57]. Study participants (n = 1,515, aged 1.9–40 mos.), who either reported for their first documented hospital visit for febrile episodes or routine childhood vaccinations, were recruited at SCRH. Written informed consent in the language of choice (i.e., English, Swahili or Dholuo) was obtained from the parent or legal guardian of all children participating in the study. Questionnaires were used to collect demographic and clinical information. Based on P. falciparum parasite density and Hb levels in peripheral blood, study participants were grouped into three categories upon enrollment: aparasitemic (n = 289), non-SMA (Hb ≥ 5.0 g/dL, n = 962), and SMA (Hb < 5.0 g/dL, n = 264). Exclusion criteria included: children with cerebral malaria (a rare occurrence in this holoendemic area); clinical evidence of acute respiratory infection; and prior hospitalization. Patients were treated according to the Ministry of Health-Kenya guidelines.
Longitudinal follow-ups
Following enrollment, parents/guardians were requested to bring their child to hospital every three mos. throughout the 36-mo. longitudinal follow-up period and during any acute febrile episodes. A complete physical exam and clinical laboratory tests were performed at each quarterly and acute visit. All-cause mortality data was collected throughout the follow-up period. The geographic information system coordinates of each child’s address were recorded upon enrollment. For the participants who did not report for a scheduled follow-up visit, our study field team went to the residence to determine the health status of the child, which included a verbal autopsy in cases of mortality.
Laboratory investigations
Heel or finger-prick blood samples (< 100 μL) were obtained and used to determine key variables such as parasitemia and Hb concentrations according to previous published methods [54]. Complete blood counts (CBC) were assessed using the Beckman Coulter ACT diff2™ (Beckman-Coulter Corporation, Miami, FL, USA). To account for the common causes of severe anemia in the region, anemia-promoting conditions including HIV-1, bacteremia, HbAS status, α3.7-thalassemia, and G6PD deficiency were determined. Pre- and post-HIV test counseling was provided to the parents/guardians of all participants. HIV-1 exposure was determined serologically (i.e., Unigold™ and Determine™) and HIV-1 infection was determined by pro-viral DNA PCR testing according to our previous methods [32]. Bacteremia was determined according to our published methods [33]. The presence of the HbAS trait was determined by cellulose acetate electrophoresis as per manufacturer’s instructions (Helena Bio-Sciences, Oxford, United Kingdom), α3.7-thalassemia deletion was determined using a PCR-based method [58], and G6PD deficiency was determined by a fluorescent spot test using the manufacturer’s methods (Trinity Biotech Plc., Bray, Ireland).
Genotyping of rs2736191 and rs11575837 variants
Genomic DNA was extracted from buccal swabs using the MasterAmp™ Buccal swab DNA Extraction kit (Epicentre Biotechnologies, Madison, WI, USA) and amplified using GenomiPhi® system (GE Healthcare, 174 NJ, USA) before genotyping. rs2736191 and rs11575837 were genotyped using our established PCR conditions [15] and the TaqMan® 5’ allelic discrimination Assay-By-Design high-throughput method according to the manufacturer’s instructions [Assay ID: C_16286876_10 for rs2736191 and C_27834902_10 for rs11575837; Thermofisher Scientific, Carlsbad, CA, USA). StepOne™ Software (Version 2.3) was used for allelic discrimination (Thermofisher Scientific, Carlsbad, CA, USA).
Data analysis
Demographic, clinical and laboratory characteristics of participants at enrolment were analyzed using SPSS® v23.0 (IBM SPSS Inc., Chicago, IL, USA). Data across the study groups was compared using Pearson’s Chi-square (χ2) test and the Kruskal-Wallis test. Differences in parasitological variables between SMA and non-SMA were computed using Mann–Whitney U test. Haplotypes composed of rs2736191 and rs11575837 were constructed using HPlus software program v2.5 (Fred Hutchinson Cancer Research Center, Seattle, WA, USA). Proportions of alleles, genotypes, and haplotypes of the NCR3 variants were compared across the study groups using χ2 tests of homogeneity. Hardy–Weinberg Equilibrium (HWE) was evaluated using a χ2 goodness-of-fit test for the genotype frequencies of both SNPs. Since there was a low minor allele frequency (MAF) for rs11575837, the exact test for HWE was used [59]. Linkage disequilibrium (LD) was calculated using Multiallelic Interallelic Disequilibrium Analysis (MIDAS) software version 1.0 [60].
The association between NCR3 genotypes/haplotypes and longitudinal clinical outcomes were analyzed in R (version 3.6.1) by fitting a Poisson rate regression using a forward-backward model selected based on the Akaike Information Criterion (AIC). The following covariates were subject to model selection: age, sex, co-infections (HIV-1 and bacteremia), G6PD deficiency, sickle-cell trait, and alpha-thalassemia, since these covariates can influence malarial anemia [32, 33, 61, 62]. The Poisson regression, accounted for the varying length of the follow-up observational period by treating the logarithm of that length as an offset to the logarithm of the expected number of events (Poisson rate regression). In the “additive model” the Poisson rate regression was carried out by coding the NCR3 genotypes/haplotypes metrically (indicating 0, 1, or 2 copies of the mutant allele or haplotype, respectively) rather than categorically. Survival analyses were performed by a non-parametric Cox proportional hazard model to determine the relative risk of all-cause mortality associated with NCR3 genotypes/haplotypes over a 36-mos. follow-up period. For each model, Holm-Bonferroni correction was applied to control for familywise error rates in multiple comparisons. The statistical significance for all analyses was set at P ≤ 0.05.
Availability of data and materials
The data that support the results of this research are available at clinVAR (https://www.ncbi.nlm.nih.gov/clinvar/). The public data can be found at: accession numbers SCV002762723 for rs11575837, and SCV002762724 for rs2736191.
Abbreviations
- SMA:
-
Severe malarial anemia
- NCR3:
-
Natural cytotoxicity-triggering receptor 3;
- HbS:
-
Sickle hemoglobin
- HBB:
-
Hemoglobin beta
- MHC:
-
Major histocompatibility complex
- NK cell:
-
Natural killer cell
- pRBCs:
-
Parasitized red blood cells
- DBL1-α:
-
Duffy Binding Like
- PfEMP-1:
-
P. falciparum Erythrocyte membrane protein-1
- pSS:
-
Primary Sjögren’s syndrome
- SCRH:
-
Siaya county referral hospital
- CBC:
-
Complete blood counts
- HbAS:
-
Sickle-cell trait
- G6PD:
-
Glucose-6-phosphate dehydrogenase deficiency
- HWE:
-
Hardy–Weinberg equilibrium
- MAF:
-
Minor allele frequency
- LD:
-
Linkage disequilibrium
- MIDAS:
-
Multiallelic interallelic disequilibrium analysis
- AIC:
-
Akaike information criterion
- TFBS:
-
Transcription factor binding site
- ZBTB6:
-
Zinc finger and BTB domain containing 6
- HNF4A:
-
Hepatocyte nuclear factor 4
References
WHO. World Malaria Report 2021. 2021.
Marsh K, Forster D, Waruiru C, Mwangi I, Winstanley M, Marsh V, et al. Indicators of life-threatening malaria in African children. N Engl J Med. 1995;332(21):1399–404.
Akech S, Chepkirui M, Ogero M, Agweyu A, Irimu G, English M, et al. The clinical profile of severe pediatric malaria in an area targeted for routine RTS, S/AS01 malaria vaccination in Western Kenya. Clin Infect Dis. 2019;71(2):372–80.
Okoyo C, Githinji E, Muia RW, Masaku J, Mwai J, Nyandieka L, et al. Assessment of malaria infection among pregnant women and children below five years of age attending rural health facilities of Kenya: a cross-sectional survey in two counties of Kenya. PLoS One. 2021;16(9):e0257276.
Paton RS, Kamau A, Akech S, Agweyu A, Ogero M, Mwandawiro C, et al. Malaria infection and severe disease risks in Africa. Science (New York, NY). 2021;373(6557):926–31.
Flint J, Harding RM, Boyce AJ, Clegg JB. The population genetics of the haemoglobinopathies. Baillieres Clin Haematol. 1998;11(1):1–51.
Tishkoff SA, Williams SM. Genetic analysis of African populations: human evolution and complex disease. Nat Rev Genet. 2002;3(8):611–21.
Ouma C, Davenport GC, Awandare GA, Keller CC, Were T, Otieno MF, et al. Polymorphic variability in the interleukin (IL)-1beta promoter conditions susceptibility to severe malarial anemia and functional changes in IL-1beta production. J Infect Dis. 2008;198(8):1219–26.
Awandare GA, Martinson JJ, Were T, Ouma C, Davenport GC, Ong’echa JM, et al. MIF (macrophage migration inhibitory factor) promoter polymorphisms and susceptibility to severe malarial anemia. J Infect Dis. 2009;200(4):629–37.
Ong’echa JM, Raballah EO, Kempaiah PM, Anyona SB, Were T, Davenport GC, et al. Polymorphic variability in the 3’ untranslated region (UTR) of IL12B is associated with susceptibility to severe anaemia in Kenyan children with acute Plasmodium falciparum malaria. BMC Genet. 2011;12:69.
Okeyo WA, Munde EO, Okumu W, Raballah E, Anyona SB, Vulule JM, et al. Interleukin (IL)-13 promoter polymorphisms (-7402 T/G and -4729G/A) condition susceptibility to pediatric severe malarial anemia but not circulating IL-13 levels. BMC Immunol. 2013;14:15.
Munde EO, Okeyo WA, Raballah E, Anyona SB, Were T, Ong’echa JM, et al. Association between Fcγ receptor IIA, IIIA and IIIB genetic polymorphisms and susceptibility to severe malaria anemia in children in western Kenya. BMC Infect Dis. 2017;17(1):289.
Achieng’ AO, Hengartner NW, Raballah E, Cheng Q, Anyona SB, Lauve N, et al. Integrated OMICS platforms identify LAIR1 genetic variants as novel predictors of cross-sectional and longitudinal susceptibility to severe malaria and all-cause mortality in Kenyan children. EBioMedicine. 2019;45:290–302.
Kisia LE, Kempaiah P, Anyona SB, Munde EO, Achieng AO, Ong’echa JM, et al. Genetic variation in interleukin-7 is associated with a reduced erythropoietic response in Kenyan children infected with Plasmodium falciparum. BMC Med Genet. 2019;20(1):140.
Anyona SB, Hengartner NW, Raballah E, Ong’echa JM, Lauve N, Cheng Q, et al. Cyclooxygenase-2 haplotypes influence the longitudinal risk of malaria and severe malarial anemia in Kenyan children from a holoendemic transmission region. J Hum Genet. 2020;65(2):99–113.
Jepson A, Sisay-Joof F, Banya W, Hassan-King M, Frodsham A, Bennett S, et al. Genetic linkage of mild malaria to the major histocompatibility complex in Gambian children: study of affected sibling pairs. BMJ (Clinical research ed). 1997;315(7100):96–7.
Flori L, Sawadogo S, Esnault C, Delahaye NF, Fumoux F, Rihet P. Linkage of mild malaria to the major histocompatibility complex in families living in Burkina Faso. Hum Mol Genet. 2003;12(4):375–8.
Brisebarre A, Kumulungui B, Sawadogo S, Atkinson A, Garnier S, Fumoux F, et al. A genome scan for Plasmodium falciparum malaria identifies quantitative trait loci on chromosomes 5q31, 6p21.3, 17p12, and 19p13. Malar J. 2014;13:198.
Delahaye NF, Barbier M, Fumoux F, Rihet P. Association analyses of NCR3 polymorphisms with P. falciparum mild malaria. Microbes Infect. 2007;9(2):160–6.
Baaklini S, Afridi S, Nguyen TN, Koukouikila-Koussounda F, Ndounga M, Imbert J, et al. Beyond genome-wide scan: association of a cis-regulatory NCR3 variant with mild malaria in a population living in the Republic of Congo. PLoS One. 2017;12(11):e0187818.
Thiam A, Baaklini S, Mbengue B, Nisar S, Diarra M, Marquet S, et al. NCR3 polymorphism, haematological parameters, and severe malaria in Senegalese patients. PeerJ. 2018;6:e6048.
Moretta A, Bottino C, Vitale M, Pende D, Cantoni C, Mingari MC, et al. Activating receptors and coreceptors involved in human natural killer cell-mediated cytolysis. Annu Rev Immunol. 2001;19:197–223.
Wang H, Zheng X, Wei H, Tian Z, Sun R. Important role for NKp30 in synapse formation and activation of NK cells. Immunol Invest. 2012;41(4):367–81.
Mavoungou E, Held J, Mewono L, Kremsner PG. A Duffy binding-like domain is involved in the NKp30-mediated recognition of Plasmodium falciparum-parasitized erythrocytes by natural killer cells. J Infect Dis. 2007;195(10):1521–31.
Artavanis-Tsakonas K, Eleme K, McQueen KL, Cheng NW, Parham P, Davis DM, et al. Activation of a subset of human NK cells upon contact with Plasmodium falciparum-infected erythrocytes. J Immunol (Baltimore, Md: 1950). 2003;171(10):5396–405.
Korbel DS, Finney OC, Riley EM. Natural killer cells and innate immunity to protozoan pathogens. Int J Parasitol. 2004;34(13–14):1517–28.
Baratin M, Roetynck S, Pouvelle B, Lemmers C, Viebig NK, Johansson S, et al. Dissection of the role of PfEMP1 and ICAM-1 in the sensing of Plasmodium-falciparum-infected erythrocytes by natural killer cells. PLoS One. 2007;2(2):e228.
Fauriat C, Long EO, Ljunggren HG, Bryceson YT. Regulation of human NK-cell cytokine and chemokine production by target cell recognition. Blood. 2010;115(11):2167–76.
Perkins DJ, Were T, Davenport GC, Kempaiah P, Hittner JB, Ong’echa JM. Severe malarial anemia: innate immunity and pathogenesis. Int J Biol Sci. 2011;7(9):1427–42.
Rusakiewicz S, Nocturne G, Lazure T, Semeraro M, Flament C, Caillat-Zucman S, et al. NCR3/NKp30 contributes to pathogenesis in primary Sjogren’s syndrome. Sci Transl Med. 2013;5(195):195ra96.
Auton A, Abecasis GR, Altshuler DM, Durbin RM, Abecasis GR, Bentley DR, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74.
Otieno RO, Ouma C, Ong’echa JM, Keller CC, Were T, Waindi EN, et al. Increased severe anemia in HIV-1-exposed and HIV-1-positive infants and children during acute malaria. AIDS. 2006;20(2):275–80.
Were T, Davenport GC, Hittner JB, Ouma C, Vulule JM, Ong’echa JM, et al. Bacteremia in Kenyan children presenting with malaria. J Clin Microbiol. 2011;49(2):671–6.
Richer J, Chudley A. The hemoglobinopathies and malaria. Clin Genet. 2005;68(4):332–6.
Taylor SM, Parobek CM, Fairhurst RM. Haemoglobinopathies and the clinical epidemiology of malaria: a systematic review and meta-analysis. Lancet Infect Dis. 2012;12(6):457–68.
Weatherall DJ. The evolving spectrum of the epidemiology of thalassemia. Hematol Oncol Clin North Am. 2018;32(2):165–75.
Tsunoda T, Takagi T. Estimating transcription factor bindability on DNA. Bioinformatics. 1999;15(7–8):622–30.
CDC. Malaria’s impact worldwide 2021. https://www.cdc.gov/malaria/malaria_worldwide/impact.html. Accessed 18 Jul 2022.
Griffin JT, Hollingsworth TD, Reyburn H, Drakeley CJ, Riley EM, Ghani AC. Gradual acquisition of immunity to severe malaria with increasing exposure. Proc Biol Sci. 1801;2015(282):20142657.
Kwiatkowski DP. How malaria has affected the human genome and what human genetics can teach us about malaria. Am J Hum Genet. 2005;77(2):171–92.
Ahmed CM, Johnson HM. IFN-gamma and its receptor subunit IFNGR1 are recruited to the IFN-gamma-activated sequence element at the promoter site of IFN-gamma-activated genes: evidence of transactivational activity in IFNGR1. J Immunol (Baltimore, Md: 1950). 2006;177(1):315–21.
Chen C, Guo L, Shi M, Hu M, Hu M, Yu M, et al. Modulation of IFN-γ receptor 1 expression by AP-2α influences IFN-γ sensitivity of cancer cells. Am J Pathol. 2012;180(2):661–71.
King T, Lamb T. Interferon-γ: the Jekyll and Hyde of malaria. PLoS Pathog. 2015;11(10):e1005118.
Hviid L, Kurtzhals JA, Dodoo D, Rodrigues O, Rønn A, Commey JO, et al. The gamma/delta T-cell response to Plasmodium falciparum malaria in a population in which malaria is endemic. Infect Immun. 1996;64(10):4359–62.
Safeukui I, Gomez ND, Adelani AA, Burte F, Afolabi NK, Akondy R, et al. Malaria induces anemia through CD8+ T cell-dependent parasite clearance and erythrocyte removal in the spleen. mBio. 2015;6(1):e02493-14.
Schauder DM, Shen J, Chen Y, Kasmani MY, Kudek MR, Burns R, et al. E2A-regulated epigenetic landscape promotes memory CD8 T cell differentiation. Proc Natl Acad Sci U S A. 2021;118(16):e2013452118.
Roels J, Van Hulle J, Lavaert M, Kuchmiy A, Strubbe S, Putteman T, et al. Transcriptional dynamics and epigenetic regulation of E and ID protein encoding genes during human T cell development. Front Immunol. 2022;13:960918.
Mamoor S. ZBTB6 is hyper-methylated and differentially expressed in the blood of patients with Crohn’s disease. 2020.
Makita T, Hernandez-Hoyos G, Chen TH, Wu H, Rothenberg EV, Sucov HM. A developmental transition in definitive erythropoiesis: erythropoietin expression is sequentially regulated by retinoic acid receptors and HNF4. Genes Dev. 2001;15(7):889–901.
Gorlov IP, Gorlova OY, Amos CI. Allelic spectra of risk SNPs are different for environment/lifestyle dependent versus independent diseases. PLoS Genet. 2015;11(7):e1005371.
Mueller JC. Linkage disequilibrium for different scales and applications. Brief Bioinform. 2004;5(4):355–64.
Bush WS, Moore JH. Chapter 11: genome-wide association studies. PLoS Comput Biol. 2012;8(12):e1002822.
Zintzaras E. Impact of Hardy-Weinberg equilibrium deviation on allele-based risk effect of genetic association studies and meta-analysis. Eur J Epidemiol. 2010;25(8):553–60.
Ong’echa JM, Keller CC, Were T, Ouma C, Otieno RO, Landis-Lewis Z, et al. Parasitemia, anemia, and malarial anemia in infants and young children in a rural holoendemic Plasmodium falciparum transmission area. Am J Trop Med Hyg. 2006;74(3):376–85.
Minakawa N, Dida GO, Sonye GO, Futami K, Njenga SM. Malaria vectors in Lake Victoria and adjacent habitats in western Kenya. PLoS One. 2012;7(3):e32725.
Bloland PB, Boriga DA, Ruebush TK, McCormick JB, Roberts JM, Oloo AJ, et al. Longitudinal cohort study of the epidemiology of malaria infections in an area of intense malaria transmission II. Descriptive epidemiology of malaria infection and disease among children. Am J Trop Med Hyg. 1999;60(4):641–8.
Obonyo CO, Vulule J, Akhwale WS, Grobbee DE. In-hospital morbidity and mortality due to severe malarial anemia in western Kenya. Am J Trop Med Hyg. 2007;77(6 Suppl):23–8.
Dodé C, Krishnamoorthy R, Lamb J, Rochette J. Rapid analysis of -α3.7 thalassaemia and αααanti 3.7 triplication by enzymatic amplification analysis. Br J Haematol. 1993;83(1):105–11.
Engels WR. Exact tests for Hardy-Weinberg proportions. Genetics. 2009;183(4):1431–41.
Gaunt TR, Rodriguez S, Zapata C, Day INM. MIDAS: software for analysis and visualisation of interallelic disequilibrium between multiallelic markers. BMC Bioinformatics. 2006;7(1):227.
Aidoo M, Terlouw D, Kolczak M, McElroy P, Kuile F, Kariuki S, et al. Protective effects of the sickle cell gene against malaria morbidity and mortality. Lancet. 2002;359(9314):1311–2.
Preuss J, Jortzik E, Becker K. Glucose-6-phosphate metabolism in Plasmodium falciparum. IUBMB Life. 2012;64(7):603–11.
Acknowledgements
We are grateful to the Siaya County Referral Hospital for clinical support and UNM-Maseno Global Health Program Laboratories. We also extend our appreciation to the parents/guardians of the study participants and children who took part in the study.
Funding
This work was supported by grants from the National Institute of Health [R01-TW008306 (CO), R01 AI30473 (DJP), and D43 TW05884 (DJP)].
Author information
Authors and Affiliations
Contributions
DJP conceived the project and acquired the funding. DJP and CO supervised the work. COO, QC, DJP, and CO designed the study. COO performed the experiments. COO, QC, EOM, ER, SBA, BHM, CGL, PO, KAS, DJP, and CO analyzed the data. COO, QC, DJP, and CO drafted the manuscript. All authors reviewed, revised, and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The study was approved by the Institutional Review Board of the University of New Mexico, USA (16-284), and the Ethics Review Committee of Maseno University, Kenya (MUERC; MSU/DRPI/MUERC/00510/18). Parent or legal guardian of every pediatric study participant gave written informed consent during enrollment. All methods were carried out in accordance with relevant guidelines and regulations.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
Survival analysis for all-cause mortality.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Onyango, C.O., Cheng, Q., Munde, E.O. et al. Human NCR3 gene variants rs2736191 and rs11575837 alter longitudinal risk for development of pediatric malaria episodes and severe malarial anemia. BMC Genomics 24, 542 (2023). https://doi.org/10.1186/s12864-023-09565-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-023-09565-1