Abstract
Background
Along with global warming, resulting in crop production, exacerbating the global food crisis. Therefore, it is urgent to study the mechanism of plant heat resistance. However, crop resistance genes were lost due to long-term artificial domestication. By analyzing the potential heat tolerance genes and molecular mechanisms in other wild materials, more genetic resources can be provided for improving the heat tolerance of crops. Elephant grass (Pennisetum purpureum Schum.) has strong adaptability to heat stress and contains abundant heat-resistant gene resources.
Results
Through sequence structure analysis, a total of 36 RWP-RK members were identified in elephant grass. Functional analysis revealed their close association with heat stress. Four randomly selected RKDs (RKD1.1, RKD4.3, RKD6.6, and RKD8.1) were analyzed for expression, and the results showed upregulation under high temperature conditions, suggesting their active role in response to heat stress. The members of RWP-RK gene family (36 genes) in elephant grass were 2.4 times higher than that of related tropical crops, rice (15 genes) and sorghum (15 genes). The 36 RWPs of elephant grass contain 15 NLPs and 21 RKDs, and 73% of RWPs are related to WGD. Among them, combined with the DAP-seq results, it was found that RWP-RK gene family expansion could improve the heat adaptability of elephant grass by enhancing nitrogen use efficiency and peroxidase gene expression.
Conclusions
RWP-RK gene family expansion in elephant grass is closely related to thermal adaptation evolution and speciation. The RKD subgroup showed a higher responsiveness than the NLP subgroup when exposed to high temperature stress. The promoter region of the RKD subgroup contains a significant number of MeJA and ABA responsive elements, which may contribute to their positive response to heat stress. These results provided a scientific basis for analyzing the heat adaptation mechanism of elephant grass and improving the heat tolerance of other crops.
Similar content being viewed by others
Background
Since 1980s, the global climate has imposed serious impacts on the environment, especially high temperatures have been affecting crop yield and reducing arable land worldwide [1]. High temperature is one of the major factors affecting plant reproductive development and is also a driving force for plant evolution. Research has shown that heat stress can impact plant chromosome pairing, leading to natural polyploidy [2]. African regions “tropical continent” contains rich varieties of genetic resources to study long-term adaptive evolution in plants under heat stress [3]. Elephant grass (Pennisetum purpureum Schum., AABB, 2n = 4x = 28), from Africa commonly known as Napier grass which is cultivated worldwide as energy and forage grass due to its high biological yield and strong heat stress tolerance ability [4, 5].
The RWP-RK gene family consists of two subfamilies: NIN-Like Protein (NLP) and RWP domain-containing (RKD) are specific to plants and function as transcription factors [6]. It has been discovered in recent years that NLP plays a crucial role in answering nitrogen stress and regulating plant nitrogen metabolism [7]. This gene family is widely found in Arabidopsis thaliala [8], Oryza sativa [9], Brachypodium distachyum [10], Triticum aestivum [6] and Zea mays L. [11]. In addition, NLP showed responses against different types of abiotic stresses in rice and Arabidopsis thaliana, among which significant increase in AtNLP4 and AtNLP9 expression pattern was found under heat stress [12].
It has been reported that RKD involves in gametophyte growth, while compared with NLP studies in plants, its functions are relatively less studied in plant abiotic stress research [13, 14]. Arabidopsis thaliana has five RKD genes, AtRKD4 is preferentially expressed in early embryos (several stages of development, such as seeds), and the deletion of this gene leads to abnormal development of fertilized egg cells [15]. In wheat, the expression of TaRKD1 and TaRKD2 in egg cells was detected by single-cell RT-PCR [16]. Specific expression of CitRWP, a homolog of Arabidopsis thaliana RKD, was observed in the ovules of Citrus sinensis, with higher expression in polyembryonal varieties compared to monoembryos, indicating that the gene exerts a critical effect on the development of nucellus embryo and apomixis in citrus [17]. In order to reveal the relationship between heat adaptation evolution and the expansion of the RWP-RK gene family in elephant grass, we conducted a systematic analysis of the RWP-RK gene family. By analyzing the evolution of the RWP-RK gene family and its response mechanisms under abiotic stress, we have provided new insights for studying plant heat tolerance.
Results
RWPs in elephant grass
In the elephant grass genome, 36 genes were identified to contain the RWP conserved domain after removing redundant sequences. Among these genes, 15 belong to the NLPs and 21 belong to the RKDs. After performing a phylogenetic analysis to avoid any confusion and overlapping gene names, the genes were renamed as CpNLPs and CpRKDs and ranked based on the subfamily to which they belonged, as presented in Table 1. The amino acid content of CpNLP protein ranges from 719 (CpNLP6.2) to 1047(CpNLP3.4), with relative molecular weights of 75.79 and 114.4 kDa. The CpRKD protein contains amino acids ranging from 194 (CpRKD6.4) to 865 (CpRKD6.6) with relative molecular weights of 21.81 kDa and 96.7kDA, respectively. The CpNLP contains more amino acids and has a larger molecular weight because it has one more Phox and Bem1 (PB1) domains than CpRKD. Interestingly, 93% of the CpNLP protein theoretical isoelectric point (pI) was less than 7, while 51% of the CpRKD protein theoretical pI was greater than 7. 94% of the RWPs through BUSCA (http://busca.biocomp.unibo.it, [18]) for subcellular localization prediction are expressed in the cell nucleus, except for CpRKD6.6 and CpRKD1.1 expressed in the cell membrane and chloroplasts.
Expansion of the RWPs in Pennisetum
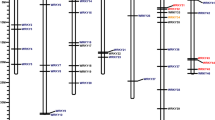
The full-length protein sequences of elephant grass (Pennisetum purpureum Schum.), sorghum (Sorghum bicolor), rice (Oryza sativa), and pearl millet (Pennisetum. glaucum) were aligned to reveal the phylogenetic relationship of RWPs in elephant grass. In addition, to more clearly compare the evolutionary differences between RWPs in elephant grass and other species, we carried out phylogenetic analyses on the NLP and RKD subfamilies of RWP-RK gene family (Fig. 1). The 31 NLPs were classified into three major clades consisting of 12, 8, and 12 proteins, which is consistent with the previous evolutionary analysis (Fig. 1A). Compared with sorghum, the NLPs of Pennisetum (pearl millet and elephant grass) were more widely distributed, especially forming specific clades in Clade I. In addition, NLPs of subgenome A in elephant grass had higher homology with pearl millet and were distributed evenly in all branches.
Different from the clustering results of NLPs and RKDs of elephant grass could not clearly distinguish gene categories (Fig. 1B). Here, we found that CpRKD8.1 in elephant grass was significantly different from other CpRKDs and formed a separate branch. Comparing CpRKD8.1 with all RWPs in elephant grass, it was found that CPRKD8.1 was more similar to NLPs in elephant grass, especially CpNLP1.1 and CpNLP1.2 (Fig. 1C). This may be related to the evolution of RWPs in elephant grass.
WGD promoted RWPs gene sequence recombination and membership increase
Due to the high similarity between the sequences of CpRKD8.1 and NLPs in elephant grass, to further analyze the evolutionary relationship between RKDs and NLPs in elephant grass, phylogenetic analysis was conducted on the RWP conserved domain sequences of 36 RWPs genes in elephant grass (Fig. S1). In elephant grass, RKDs and NLPs had independent branches, and the clustering results of RKDs showed an evolutionary trend from CpRKD1 to CpRKD6, while NLPs formed two independent evolutionary directions, CpNLP3 and CpNLP1. This may indicate a functional divergence between RKDs and NLPs under environmental selection. By analyzing the distribution of RWP gene family on elephant grass chromosomes, it was found that CpRKD8.1 and CpNLP6.1 were located on chromosome 6B and closely related (Fig. S2B). Therefore, the production of CpRKD8.1 may be related to the self-replication of CpNLP6.1 caused by WGD.
Because the differentiation time of sorghum was earlier than elephant grass and pearl millet, we analyzed the WGD influence of RWPs gene evolution in elephant grass by collinearity analysis between elephant grass and sorghum. MCScanX analysis identified 26 RWPs associated with WGD, including 15 RKDs and 11 NLPs (Fig. S2A). Interestingly, the distribution of RWPs related to WGD in elephant grass B and A subgenomes was also 15 and 11 (Fig. S2B), See Table S1 for details. In addition, a total of 29 gene pairs were found by collinearity analysis between elephant grass and sorghum, and all gene pairs of Ka/ KS < 1, suggesting that RWPs in elephant grass are highly conserved and subject to strong purification selection.
WGD promoted the increase of RKDs and NLPs sequence diversity
Although RWPs are strongly selected for purification in elephant grass, phylogenetic analysis of NLPs and RKDs showed that CpNLPs are more conserved than CpRKDs. Therefore, to further analyze the functional differences between NLPs and RKDs in elephant grass, we analyzed the sequence structure of NLPs and RKDs respectively.
The motifs of NLPs and RKDs genes in the elephant were visualized using the Simple MEME Wrapper tool in TBTools [19]. Twelve conserved motifs were identified from CpNLPs, including motif 9 and motif 1, which were identified as the N-terminal and C-terminal of RWP conserved motif by NCBI analysis (Fig. 2B). In addition, Motif7 and Motif4 were identified as the N-and C-terminus of the PB1 conserved area from 15 CpNLPs. Compared with CpNLPs, the amino acid sequences of RKDs showed greater diversity, with only 3 conserved motifs identified among 21 RKDs (Fig. 2A). Unlike CpNLPs, the RWP conserved domain (Motif1) of RKDs was well conserved in all genes, although RKDs showed large differences in amino acid sequences. In general, NLPs and RKDs in elephant grass changed due to the WGD sequence, which may be related to the selection of living environment and the adaptive evolution of genes.
ABA and MeJA affect RWPs expression in elephant grasses under abiotic stress
CpNLPs were expressed (0.02–32.61 TPM) in all tissues (stem tip, leaf, stem, root, and flower), mainly in leaves and stem, and the highest expression levels in leaves and stems were CpNLP3.1 and CpNLP2.2, respectively (Fig. 3B). Compared with CpNLPs, the expression of CpRKDs is more specific, and some genes are not detected in various tissues, which may be activated in response to specific environmental signals or signals at developmental stages (Fig. 3A). In addition, we found that CpNLPs and CpRKDs of elephant grass A subgenome had expression advantages, and the expression of NLPs and RKDs in elephant grass A subgenome was higher than that of genes belonging to B subgenome in all Clade (Table S2). According to plantcare cis-acting factors prediction, RWPs in elephant grass responded to a variety of phytohormone signals (IAA, GA, MeJA, BHA and ABA), MeJA and ABA had the most binding sites. In addition, by comparing the differences in hormone signal binding sites between CpRKDs and CpNLPs, it was found that CpRKDs contained more MeJA and ABA signal binding sites. Among all the cis-acting elements in response to hormones, the type of element that responded to ABA was the largest (Table S3). The expression trend of RWPs in elephant grass was detected by external application of ABA (100 mM) and qRT-PCR, and it was found that external application of ABA could affect its expression trend (Fig. 4).
Effects of ABA on RWPs expression in elephant grass. A Expression trends of NLPs in Elephant grass at 6, 12 and 48 h after ABA (100 mM) treatment; B Expression trends of RKDs in Elephant grass at 6, 12 and 48 h after ABA (100 mM) treatment; C Distribution of abscisic acid-responsive elements in promoters (upstream < 2100) CpNLP2.2, CpNLP3.1 and CpNLP5.2 in elephant grass; D Distribution of abscisic acid elements in CpRKD1.1, CpRKD4.3 and CpRKD8.1 promoters (upstream < 2100) in elephant grass. (*: p < 0.05; ** p < 0.01)
For further analyzing the expression differences of RKDs and NLPs in elephant grass under stress, RT-PCR was adopted for detecting the gene expression of RKDs and NLPs in elephant grass under salt (NaCl 100 mM), drought (PEG 20%) and high temperature(40 °C/38 °C) stress, respectively. CpNLP5.2 in elephant grass can rapidly respond to a variety of abiotic stress signals, and its expression significantly increased after 2 h of high temperature, drought, and salt stress, indicating that CpNLP5.2 exerts a core effect on the adaptation of elephant grasses to stress. Meanwhile, four randomly selected RKDs were differentially expressed under high-temperature stress, explaining that they have special effects under heat stress(Fig. 5). In addition, we found that the expressions of CpAPX, CpSOD, and CpPOD in elephant grass were also significantly increased under the three kinds of stresses (Fig. S3). Among them, CpRKD1.1, CpRKD4.3, and CpRKD6.6 were correlated with CpPOD expression, which means that RKD might be related to peroxide synthesis (Fig. S4).
Functional of NLP and RKD of RWP gene family
Gene expression is closely related to function. In order to further reveal the influence of the RWP gene family expansion on grass thermal adaptability, the DAP-seq target gene information of AtNLP7 and AtRKD2 was provided by Plant Cistrome Database for subsequent RWPs functional analysis.
Based on the functional annotations of AtNLP7 target genes, not only genes taking part in the control of N metabolisms, including ferredoxin-nitrite reductase (nirA), NR reductase (NR) and NRT /nitrite (NRT) but also significantly enriched carbon metabolic pathways (Value = 0.002571), associated genes such as phosphoenolpyruvate carboxylase (PPC), malate dehydrogenase (MDH1) and triosephosphate isomerase (TPI) suggested that NLPs could regulate the balance between nitrogen uptake and C fixation in plants(Fig. 6, Table S5). Although some studies have shown that AtNLP7 mutants may be involved in physiological and metabolic processes related to drought stress [20], functional annotation analysis of AtNLP7 target genes showed that NLPs were more important for stable plant growth. Regulating nitrogen-nitrogen related balance-related genes nitrogen-mediated tiller growth response 5 (NGR5) and growth-regulating factor 4 (GRF4) successfully improved nitrogen application efficiency and yield in rice [21]. Therefore, it is of great significance to work on the biological function and regulatory mechanism of NLPs for crop improvement.
Functional annotation analysis of RKDs and NLPs downstream genes. Blue areas represent 790 target genes bound by AtRKD2 (FRIP > 0.05); The purple area shows the 910 target genes bound by AtNLP7(FRIP > 0.05); The intersection of blue and purple represents the 39 target genes shared by AtNLP7 and AtRKD2 (FRIP > 0.05)
Compared with NLPs, the RKDs have been rarely studied in plants, and only a few studies have shown that RKDs may be related to gamete development in the plants [22]. In the functional annotation enrichment analysis results of AtRKD2 target genes, no genes related to nitrogen metabolism were found, but the enrichment of more genes was realized in the biosynthesis of secondary metabolites, including phenylpropanoid, pantothenate and CoA biosynthesis, etc. (Fig. 6, Table S6). These secondary metabolites can assist plants to cope with abiotic stressful condition and are also often used as physiological indicators of plant response to stresses, such as peroxidase and sinapate 1-glucosyltransferase (BRT1). In addition, the expression of a related peroxidase gene in elephant grass under heat, drought, and salt stress was found highly upregulated in the present study, suggesting that RKDs played a positive role in elephant grass resisting abiotic stresses. Therefore, RKDs may play an important role to enhance the resistance of elephant grass against abiotic stresses besides participating in the formation and development of gametes.
Discussion
High temperature is a typical climate characteristic in tropical and subtropical regions, especially in Africa, which enables native plants to constantly evolve in the process of adapting to the variable environments and also provides more genetic resources for the study of plant heat tolerance [3]. Whole-genome duplication (WGD) is widely present in plant genomes and is an important event that drives plant evolution [23]. WGD, also known as polyploidization, rapidly reorganizes plant genomes by losing many genes, increasing structural variations, and playing a crucial role in plant evolution and diversity [24]. WGD not only promotes the formation of elephant grass species but also provides rich genetic resources to help elephant grass adapt to a heated environment. 73% of RWPs production was associated with WGD, which means that WGD is the source driving force of RWP gene family expansion in elephant grass.
The functional adaptive evolution of the NBS_LRR gene family in plants enables them to adapt to complex natural environments [25]. The expansion of the RWP gene family in elephant grass is closely related to Africa's harsh ecological environment. The expression of CpNLP5.2 increased significantly after 2 h of heat, drought, and salt stress, and the four randomly selected RKDs also increased expression under heat stress, suggesting that the RWP gene family exerts a core effect on the adaptation of elephant grass to heat stress. Recent studies on the function of the RWP gene of its related species Pearl millet showed that the increase of the RWP gene family was closely related to the adaptation of elephant grass to the natural environment of high temperature and drought in Africa. MeJA and ABA are two major plant hormones in response to abiotic stress and exert an important role in plant answer to abiotic stress. In this study, the promoter regions of NLP and RKD are rich in the response elements of these two hormones, In vitro addition of ABA hormone showed that ABA could rapidly induce RWP gene expression. However, the expressions of NLP and RKD were significantly different under the three different stress treatments. For example, NLP was more active to salt stress signals, while RKD was more active to heat stress. This difference in the expression of RWP gene family may result from the functional differentiation of NLP and RKD.
NLP is a core transcription factor that mainly regulates the nitrate content in plant cytoplasm [26]. The root-to-shoot nitrate allocation response (SINAR) is a key mechanism for plants to adapt to stressful environments by increasing the distribution of nitrate in roots under stress conditions [27]. When high levels of nitrate are detected, NLP is transported to the nucleus and activates the expression of genes taking part in nitrate transport and metabolism, including NRT1.1, NRT2.1, and NIA1 [28]. NRT1.5 [29] and NRT1.8 [30] can enhance the tolerance of Arabidopsis to various abiotic stresses and affect the accumulation of malondialdehyde and proline. AtNLP7 can bind to key nitrate pathway genes and regulate nitrate assimilation and metabolism by activating or inhibiting downstream transcriptional regulatory factor TCP20 [31]. AtNLP6 promotes root meristem growth under nutrient stress conditions by controlling the expression of nitrate-responsive genes [32]. Additionally, AtNLP7 can also regulate nitrate assimilation and metabolism through a similar mechanism, helping plants adapt to stressful environments [31]. Moreover, the discovery of the nitrate-CPK-NLP signaling pathway highlights the importance of NLP phosphorylation in plant vegetative growth [33].
RKD, another subfamily of the RWP gene family, was observed in this research to be mainly involved in the synthesis of Phenylpropanin secondary metabolites. These secondary metabolites are mainly polyphenolic compounds, which can remove harmful free radicals in plants and improve plant tolerance under heat, drought, and salt stress [34]. Additionally, studies have demonstrated that RKD is closely associated with gamete formation and development [35]. AtRKD1 and AtRKD2 transcripts are only detectable during the later phases of female gametophyte growth, while AtRKD3, AtRKD4, and AtRKD5 transcripts are detectable during both early and late phases, according to Tedeschi et al. [36]. AtRKD1 and AtRKD2 are highly expressed in the egg apparatus and egg cells. However, overexpressing AtRKD1 and AtRKD2 can result in cell proliferation, differentiation and regeneration defects, and a change in gene expression towards an egg cell-like transcriptome [16]. During their reproductive period, plants are particularly vulnerable to high temperatures. High temperatures can inhibit spikelet development, reduce seed set, impair rice grain filling, decrease grain weight, and affect grain quality. Therefore, RKDs may exert a key effect on plant response to heat stress, particularly during the reproductive stage.
The expansion of RWP gene family improves the heat tolerance of elephant grass in the two fields below: the expansion of NLP subfamily improves the transport and absorption of nitrate nitrogen and reduces the influence of heat stress on its development; RKD expansion promotes the synthesis of phenylpropanoid secondary metabolites under heat stress and reduces stress damage. Although the first RWP protein was identified in chlamydomonas as early as 1997, the functional mechanism of the RWP gene family in abiotic stress has been rarely studied [37]. By analyzing the evolution and function of the significantly expanded RWP gene family in elephant grass under heat stress, it was further confirmed that the conserved gene family had rich genetic diversity and functional specificity in non-model plants. Hence, analyzing the specific heat tolerance mechanism of non-model plants is crucial to understand the evolution of plant heat adaptation and reducing the impact of global warming on crop safety.
Conclusions
The hot climate in Africa has driven the thermal adaptation evolution in elephant grass, while whole genome duplication (WGD) has facilitated the expansion of the RWP-RK gene family. Under high temperature stress, the RKD subgroup demonstrates a more rapid response compared to the NLP subgroup. This could be attributed to the significant presence of MeJA and ABA responsive elements in the promoter region of the RKD subgroup, which contribute to their active response to high temperature stress. These research findings provide a scientific basis for analyzing the heat adaptation mechanism in elephant grass and improving the heat tolerance of other crops. Further research into the heat adaptation characteristics of elephant grass will help us better address the challenges posed by climate change and provide valuable guidance for crop breeding and improvement.
Materials and methods
Plant materials and treatments
The elephant grass (Pennisetum purpureum Schum. ‘LSPR’) accessions were collected from Chongzhou, Sichuan, China. The accessions were grown in a nutrient solution under glasshouse conditions with a 14-h light and 10-h dark cycle at 28 °C and 25 °C, respectively. At the three-leaf stage (4 weeks of age), potted seedlings with similar phenotypes were selected for abiotic stress experiments. For heat treatment, elephant grass leaves were collected at 0, 6, 12, and 48 h after exposure to a temperature of 40 °C with a 14-h light and 10-h dark cycle. Drought and NaCl treatments were applied using 20% polyethylene glycol (PEG) and 100 mM NaCl solution instead of nutrient solution, respectively. Leaves were collected at 0, 2, 4, and 6 h after treatment. Each treatment was collected in triplicate per time point.
RNA separation and quantitative real-time polymerase chain reaction analysis
The Plant Total RNA Isolation Kit (FOREGENE, Chengdu, China) was adopted to collect elephant grass leaves and extract total RNA, followed by reverse transcription with the HiScript III 1st Strand cDNA Synthesis Kit. Gene expression was normalized with elongation factor-1-alpha (EF1α) as the reference gene. qRT-PCR was made with the CFX96 qRT-PCR System and ChamQ Universal SYBR qPCR Master Mix, with the reaction program following the standard protocol of the kit. Through comparing the relative expression level of target genes, the calculation of control treatments was made with the 2 − ΔΔCt approach. Primer Premier 5.0 software was adopted to design quantitative real-time polymerase chain reaction (qRT-PCR) primers, and Table S4 lists the qRT-PCR primer sequences.
Identification of RWP genes
For identifying members of the RWP gene family in each species, the potential Markov model of RWP was offered by PFAM (http://pfam.janelia.org/, [38]). Candidate genes were then verified using NCBI CD-search (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi, [39]) to confirm their conservative structure domain (E-value < 0.01).
Multiple-sequence alignment and phylogenetic analysis
For comparing full-length protein sequences across each species, a sequence alignment was created using the MUSCLE tool (http://www.ebi.ac.uk/Tools/msa/muscle/) and saved in ClustalW format. Subsequently, a phylogenetic analysis was performed with 1000 bootstrap replications with the MEGA 6.0 program [40]. The Poisson model method and the same alignment file were adopted to construct an unrooted Neighbor-joining and Minimum-Evolution tree.
Comparison of chromosomal distributions, exon/intron structures, and protein domains in elephant grass
For the analysis of the genomic distribution of RWP genes in elephant grass, we utilized MapInspect software (https://mapinspect1.software.informer.com/, [41]). Additionally, we employed the Gene Structure Display Server 2.0 program (http://gsds.cbi.pku.edu.cn/) to examine intron/exon structures. To predict RWP-RK domains, we used the MEME (Multiple Expectation Maximization for Motif Elicitation) online tool (http://meme-suite.org/tools/meme) with the parameters motif width 6–200 residues and a maximum number of motifs = 20 [42]. Subsequently, we downloaded the conserved motif logos from MEME, and we generated motif images with TBtools_master (version 1.098769; https://github.com/CJ-Chen/TBtools).
Genomic collinearity analysis
Tandem duplications were detected in elephant grass and sorghum genomes using BLAST and MCScanX. Multilocus genes were classified as tandem duplicates if they were in nearby areas or separated by uniform intergenic areas, with coverage and similarity levels above 90% and 95%, respectively.
Analyze of the promoter cis-regulatory elements
To analyze potential cis-acting regulatory factors in the putative promoter areas of elephant grass RWPs, the 2-kb sequences upstream of each gene were acquired through BLAST searches of the elephant grass genome with whole gene IDs. The PlantCARE database (http://bioinformatics.psb. ugent.be/webtools/plantcare/html/, [43]) was adopted to analyze the obtained sequences for cis-elements identification. A custom script in R program version 4.1.0 (https://www.r-project.org/) was adopted to visualize the identified cis-elements.
Biophysical properties of RWP genes in elephant grass
To determine the physical and chemical properties of the RWP proteins in the elephant grass genome, their amino acid number, molecular weight, and theoretical pI were calculated with the ExPASy-ProtParam tool [44]. Furthermore, the subcellular localization of these proteins was forecast with ProComp 9.0 [45].
Gene expression patterns analyzed using published transcriptome data
Raw transcriptome data were processed with reference to the analysis by Di Bella et al., [46] and the log10 transformed Transcripts Per Kilobase Million fragments (TPM) mapped values of the 36 RWP genes were calculated. Heatmaps were generated using the local Multiple Arrays Viewer program (https://sourceforge.net/projects/mev-tm4/files/mev-tm4/).
Statistical analysis
The statistical analysis of data was made with one-way analysis of variance (ANOVA) and significance was determined with Dunnett’s test with a p-value threshold of 0.05. Differential expression was evaluated with a two-fold cut-off value to consider the biological significance of the observed changes [47]. To investigate the relationship between CpPOD and CpRKD (CpRKD1.1, CpRKD4.3, and CpRKD6.6), the calculation of Pearson correlation coefficients was made. All statistical analyses were made with R version 3.6.1.
KEGG annotation analysis of AtNLP7 and AtRKD2 target genes
Firstly, potential regulators of target genes were identified from the Plant Cistrome Database [48] based on a FRIP score of > 0.05, with AtNLP7 and AtRKD2 identified as potential transcription factors. Subsequently, functional enrichment analysis was made on the identified target genes with KOBAS (KEGG Orthology Based Annotation System, http://kobas.cbi.pku.edu.cn/, [49]). The resulting p-values were corrected for FDR, with a significance threshold of p ≤ 0.05, to determine significantly enriched pathways related to peak-related genes.
Availability of data and materials
The following sources provided the genome and protein sequence for rice, elephant grass, pearl millet, and sorghum: https://ftp.ebi.ac.uk/ensemblgenomes/pub/release-56/plants/fasta/oryza_sativa/dna ([50] rice), http://milletdb.novogene.com/home/ ([51], elephant grass, CIAT6263), https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA749489 ([52], pearl millet, PRJNA749489), and https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJEB24962 ([53], sorghum, PRJEB24962). To investigate the expression patterns of RWPs in different vegetative tissues of elephant grass, we utilized high-throughput transcriptome sequencing data from PRJNA649119 (https://www.ncbi.nlm.nih.gov/bioproject/649119, [54]). The potential regulators of AtNLP7 and AtRKD2 target genes raw data download from http://neomorph.salk.edu/dap_web/pages/browse_table_aj.php. The data used to support the findings of this study are included within the main text and supplementary files of this article.
References
Pörtner HO, et al. Climate change 2022: Impacts, adaptation and vulnerability, IPCC Sixth Assessment Report. 2022.
Waadt R, Seller CA, Hsu PK, Takahashi Y, Munemasa S, Schroeder JI. Plant hormone regulation of abiotic stress responses. Nat Rev Mol Cell Biol. 2022;23(10):680–94.
Couvreur TLP, Dauby G, Blach-Overgaard A, et al. Tectonics, climate and the diversification of the tropical African terrestrial flora and fauna. Biol Rev. 2021;96(1):16–51.
Sun M, Huang D, Zhang A, et al. Transcriptome analysis of heat stress and drought stress in pearl millet based on Pacbio full-length transcriptome sequencing. BMC Plant Biol. 2020;20(1):323.
Anderson WF, Dien BS, Brandon SK, Peterson JD. Assessment of bermudagrass and bunch grasses as feedstock for conversion to ethanol. Biotechnology for Fuels and Chemicals. 2008;145(1–3):13–21.
Kumar A, Batra R, Gahlaut V, et al. Genome-wide identification and characterization of gene family for RWP-RK transcription factors in wheat (Triticum aestivum L.). PLoS One. 2018;13(12):e0208409.
Wang YY, Cheng YH, Chen KE, et al. Nitrate transport, signaling, and use efficiency[J]. Annu Rev Plant Biol. 2018;69:85–122.
Konishi M, Yanagisawa S. Arabidopsis NIN-like transcription factors have a central role in nitrate signalling. Nat Commun. 2013;4(1):1–9.
Jagadhesan B, Sathee L, Meena HS, et al. Genome wide analysis of NLP transcription factors reveals their role in nitrogen stress tolerance of rice. Sci Rep. 2020;10(1):1–16.
Liu M, Chang W, Fan Y, et al. Genome-Wide Identification and Characterization of nodule-inception-like protein (NLP) Family Genes in Brassica napus. Int J Molecular Sci. 2018;19(8):2270.
Ge M, et al. Genome-wide analysis of maize NLP transcription factor family revealed the roles in nitrogen response. Plant Growth Regul. 2018;84(1):95–105.
Chardin C, Girin T, Roudier F, Meyer C, Krapp A. The plant RWP-RK transcription factors: key regulators of nitrogen responses and of gametophyte development. J Experimental Botany. 2014;65(19):5577–87.
Liu KH, Liu M, Lin Z, et al. NIN-like protein 7 transcription factor is a plant nitrate sensor. Science. 2022;377(6613):1419–25.
Liu X, Blomme J, Bogaert KA, et al. Transcriptional dynamics of gametogenesis in the green seaweed Ulva mutabilis identifies an RWP-RK transcription factor linked to reproduction. BMC Plant Biol. 2022;22(1):1–12.
Waki T, Hiki T, Watanabe R, Hashimoto T, Nakajima K. The Arabidopsis RWP-RK protein RKD4 triggers gene expression and pattern formation in early embryogenesis. Curr Biol. 2011;21(15):1277–81.
Koszegi D, Johnston AJ, Rutten T, et al. Members of the RKD transcription factor family induce an egg cell-like gene expression program. Plant J. 2011;67(2):280–91.
Wang N, Song X, Ye J, et al. Structural variation and parallel evolution of apomixis in citrus during domestication and diversification[J]. Natl Sci Rev. 2022;9(10):nwac114.
Savojardo C, Martelli PL, Fariselli P, Profiti G, Casadio R. BUSCA: an integrative web server to predict subcellular localization of proteins. Nucleicd Acids Res. 2018;46(W1):W459–66.
Chen C, Chen H, Zhang Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020;13(8):1194–202.
Castaings L, Camargo A, Pocholle D, et al. The nodule inception-like protein 7 modulates nitrate sensing and metabolism in Arabidopsis. Plant J. 2009;57(3):426–35.
Wu K, Wang S, Song W, et al. Enhanced sustainable green revolution yield via nitrogen-responsive chromatin modulation in rice. Science. 2020;367(6478):eaaz2046.
Jeong S, Palmer T M, Lukowitz W. The RWP-RK factor GROUNDED promotes embryonic polarity by facilitating YODA MAP kinase signaling[J]. Curr Biol. 2011;21(15):1268–76.
Clark JW, Donoghue PC. Whole-genome duplication and plant macroevolution. Trends Plant Sci. 2018;23(10):933–45.
Del Pozo JC, Ramirez-Parra E. Whole genome duplications in plants: an overview from Arabidopsis. J Exp Bot. 2015;66(22):6991–7003.
Zhang T, Qiao Q, Novikova PY, et al. Genome of Crucihimalaya himalaica, a close relative of Arabidopsis, shows ecological adaptation to high altitude[J]. Proc Natl Acad Sci. 2019;116(14):7137–46.
Mu X, Luo J. Evolutionary analyses of NIN-like proteins in plants and their roles in nitrate signaling. Cellular Molecular Life Sciences. 2019;76(19):3753–64.
Zhang GB, Yi HY, Gong JM. The Arabidopsis ethylene/jasmonic acid-NRT signaling module coordinates nitrate reallocation and the trade-off between growth and environmental adaptation. The Plant Cell. 2014;26(10):3984–98.
Krouk G. Nitrate signalling: Calcium bridges the nitrate gap. Nature Plants. 2017;3:17095.
Lin SH, Kuo HF, Canivenc G, et al. Mutation of the Arabidopsis NRT1.5 nitrate transporter causes defective root-to-shoot nitrate transport. The Plant Cell. 2008;20(9):2514–28.
Li JY, Fu YL, Pike SM, et al. The Arabidopsis nitrate transporter NRT1.8 functions in nitrate removal from the xylem sap and mediates cadmium tolerance. The Plant Cell. 2010;22(5):1633–46.
Jian W, Zhang D, Zhu F, et al. Nitrate reductase-dependent nitric oxide production is required for regulation alternative oxidase pathway involved in the resistance to Cucumber mosaic virus infection in Arabidopsis[J]. Plant Growth Regul. 2015;77:99–107.
Yan D, Easwaran V, Chau V, et al. NIN-like protein 8 is a master regulator of nitrate-promoted seed germination in Arabidopsis[J]. Nat Commun. 2016;7(1):13179.
Liu K, Niu Y, Konishi M, et al. Discovery of nitrate–CPK–NLP signalling in central nutrient–growth networks[J]. Nature. 2017;545(7654):311–6.
Sharma A, Shahzad B, Rehman A, Bhardwaj R, Landi M, Zheng B. Response of phenylpropanoid pathway and the role of polyphenols in plants under abiotic stress. Molecules. 2019;24(13):2452.
Sakuraba Y, Zhuo M, Yanagisawa S. RWP-RK domain-containing transcription factors in the Viridiplantae: biology and phylogenetic relationships[J]. J Exp Bot. 2022;73(13):4323–37.
Tedeschi F, Rizzo P, Rutten T, et al. RWP-RK domain-containing transcription factors control cell differentiation during female gametophyte development in Arabidopsis[J]. New Phytol. 2017;213(4):1909–24.
Ferris PJ, Goodenough UW. Mating type in Chlamydomonas is specified by mid, the minus-dominance gene. Genetics. 1997;146(3):859–69.
Finn RD, Coggill P, Eberhardt RY, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44(D1):D279–85.
Lu S, Wang J, Chitsaz F, et al. CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 2020;48(D1):D265–8.
Kumar S, Stecher G, Peterson D, Tamura K. MEGA-CC: computing core of molecular evolutionary genetics analysis program for automated and iterative data analysis. Bioinformatics. 2012;28(20):2685–6.
Wang S, Chen J, Zhang W, et al. Sequence-based ultra-dense genetic and physical maps reveal structural variations of allopolyploid cotton genomes. Genome Biol. 2015;16(1):108.
Bailey TL, Elkan C. The value of prior knowledge in discovering motifs with MEME. International Conference on Intelligent Systems for Molecular Biology. 1995;3:21–9.
Lescot M, Déhais P, Thijs G, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30(1):325–7.
Gasteiger E. et al. Protein Identification and Analysis Tools on the ExPASy Server. In: Walker, J.M. (eds) The Proteomics Protocols Handbook. Springer Protocols Handbooks. Humana Press. 2005;571–607
Briesemeister S, Rahnenführer J, Kohlbacher O. YLoc–an interpretable web server for predicting subcellular localization. Nucleic Acids Res. 2010;38(Web Server issue):W497–502.
Di Bella JM, Bao Y, Gloor GB, Burton JP, Reid G. High throughput sequencing methods and analysis for microbiome research. J Microbiol Methods. 2013;95(3):401–14.
Le DT, Nishiyama R, Watanabe Y, et al. Genome-wide survey and expression analysis of the plant-specific NAC transcription factor family in soybean during development and dehydration stress. DNA Res. 2011;18(4):263–76.
O’Malley RC, Huang SC, Song L, et al. Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape. Cell. 2016;165(5):1280–92.
Bu D, Luo H, Huo P, et al. KOBAS-i: intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 2021;49(W1):W317–25.
Zerbino DR, Achuthan P, Akanni W, et al. Ensembl 2018. Nucleic Acids Res. 2018;46(D1):D754–61.
Sun M, Yan H, Zhang A, et al. Milletdb: a multi-omics database to accelerate the research of functional genomics and molecular breeding of millets. Plant Biotechnol J. 2023. https://doi.org/10.1111/pbi.14136.
Yan H, Sun M, Zhang Z, et al. Pangenomic analysis identifies structural variation associated with heat tolerance in pearl millet. Nature Genetic. 2023;55(3):507–18.
Smith O, Nicholson WV, Kistler L, et al. A domestication history of dynamic adaptation and genomic deterioration in Sorghum. Nature Plants. 2019;5(4):369–79.
Yan Q, Wu F, Xu P, et al. The elephant grass (Cenchrus purpureus) genome provides insights into anthocyanidin accumulation and fast growth. Mol Ecol Resour. 2021;21(2):526–42.
Acknowledgements
This work was supported by Modern Agroindustry Technology Research System (CARS-34) and the Sichuan Province Breeding Research grant (2016NYZ0036), Modern Agricultural Industry System Sichuan Forage Innovation Team (SCCXTD-2020-16), Chongqing Research Institution Performance Incentive and Guidance Project(22533), Chongqing financial special fund project(23515C), Sichuan Province's Science Fund for Distinguished Young Scholars under Grant (2021JDJQ001). We thank the Sichuan Key Laboratory of Grass Germplasm Resources Creation and Utilization in Southwest China for providing the experimental platform.
Funding
This work was supported by Chongqing Research Institution Performance Incentive and Guidance Project(22533) and Chongqing financial special fund project(23515C), Modern Agroindustry Technology Research System (CARS-34), the Sichuan Province Breeding Research grant (2016NYZ0036), Modern Agricultural Industry System Sichuan Forage Innovation Team (SCCXTD-2020–16), Sichuan Province's Science Fund for Distinguished Young Scholars under Grant (2021JDJQ001), Chongqing Natural Science Foundation(2022NSCO-MSX5331).
Author information
Authors and Affiliations
Contributions
Yarong Jin, Dejun Huang and Huang Linkai designed this project. Yarong Jin and Jinchan Luo participated in experimental processing, data collection and curation. Huang Linkai supervised the project. Yarong Jin wrote the original draft of article. Yuchen Yang, Jiyuan Jia, Min Sun, Xiaoshan Wang and Imran Khan participated in revising the manuscript. Huang Linkai and Dejun Huang contributed to review and edited the manuscript. All authors carefully read and agreed to publish this manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Elephant grass (Pennisetum purpureum Schum. ‘LSPR’) is preserved in the Herbarium of Sichuan Agricultural University (Chengdu, Sichuan Province). Its voucher specimen number is Pp-20170612–001, identified by Prof. Huang Linkai. All samples were adopted for the total experiment. No specific permits are required for sample collection in this study. We comply with the IUCN Policy Statement on Research In wolving Species at Risk of Extinction and the Convention on the Trade in Endangered Species of Wild Fauna and Flora Each member of the team declared that has read the relevant institutional, national, and international guidelines and legislation before the commencement of the experiment and had complied with each of the legislative requirements during the experiment.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1:
Figure S1. Phylogenetic analysis of RKD and NLP subfamilies of Elephant grass RWP gene family.(the distance scale is 0.9). Figure S2. Mechanism analysis of RWP gene family expansion in elephant grass.(A: Collinearity analysis of RWP gene in elephant grass and sorghum genome; B: Chromosome mapping of RWP gene in elephant grass). Figure S3. Expression trend of CpAPX、CpSOD and CpPOD in elephant grass under abiotic stress(*:p<0.05; **: p<0.01). A:salt(NaCl 100mM; B:drought(PEG 20%); C: heat (40°C/38°C, 12h light / 12h dark). (*:p<0.05 ; ** p<0.01). Figure S4. CpRKD1.1, CpRKD4.3, and CpRKD6.6 were correlated with CpPOD expression.
Additional file 2:
Table S1. Ka/Ks analysis of homologous genes between Elephant grass and sorghum.
Additional file 3:
Table S2. Expression pattern analysis of RWPs in 5 tissues of Elephant grass.
Additional file 4:
Table S3. Analysis of RWPs promoter hormone-responsive elements in elephant grass.
Additional file 5:
Table S4. The primer of RWPs in elephant grass for qRT-PCR.
Additional file 6:
Table S5. KEGG functional annotation enrichment analysis of 910 target genes bound by AtNLP7.
Additional file 7:
Table S6. KEGG functional annotation enrichment analysis of 790 target genes bound by AtRKD2.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Jin, Y., Luo, J., Yang, Y. et al. The evolution and expansion of RWP-RK gene family improve the heat adaptability of elephant grass (Pennisetum purpureum Schum.). BMC Genomics 24, 510 (2023). https://doi.org/10.1186/s12864-023-09550-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-023-09550-8