Abstract
Background
The early death and health problems of calves caused substantial economic losses in the dairy industry. As the immune system of neonates has not been fully developed, the absorption of maternal immunoglobulin (Ig) from colostrum is essential in protecting newborn calves against common disease organisms in their early life. The overwhelming majority of Ig in bovine whey is transported from the serum. Therefore, Ig concentration in the colostrum and serum of dairy cows are critical traits when estimating the potential disease resistance of its offspring.
Results
Colostrum, blood, and hair follicle samples were collected from 588 Chinese Holstein cows within 24 h after calving. The concentration of total IgG, IgG1, IgG2, IgA and IgM in both colostrum and serum were detected via ELISA methods. With GCTA software, genome-wide association studies (GWASs) were performed with 91,620 SNPs genotyped by GeneSeek 150 K (140,668 SNPs) chips. As a result, 1, 5, 1 and 29 significant SNPs were detected associated with the concentrations of colostrum IgG1, IgG2, IgA IgM, and serum IgG2 at the genome-wide level (P < 3.08E–6); 11, 2, 13, 2, 12, 8, 2, 27, 1 and 4 SNPs were found significantly associated with total IgG, IgG1, IgG2, IgA and IgM in colostrum and serum at the suggestive level (P < 6.15E–5). Such SNPs located in or proximate to (±1 Mb) 423 genes, which were functionally implicated in biological processes and pathways, such as immune response, B cell activation, inflammatory response and NF-kappaB signaling pathways. By combining the biological functions and the known QTL data for immune traits in bovine, 14 promising candidate functional genes were identified for immunoglobulin concentrations in colostrum and serum in dairy cattle, they were FGFR4, FGFR2, NCF1, IKBKG, SORBS3, IGHV1S18, KIT, PTGS2, BAX, GRB2, TAOK1, ICAM1, TGFB1 and RAC3.
Conclusions
In this study, we identified 14 candidate genes related to concentrations of immunoglobulins in colostrum and serum in dairy cattle by performing GWASs. Our findings provide a groundwork for unraveling the key genes and causal mutations affecting immunoglobulin concentrations in colostrum and important information for genetic improvement of such traits in dairy cattle.
Similar content being viewed by others
Background
The early survival rate and health of calves are important factors affecting the production efficiency of the dairy industry. It was reported that approximately 31% of preweaning mortality events occurring in the first 3 weeks of life were attributed to the low serum IgG concentration of calf (less than 10 mg/mL when sampled between 24 and 48 h of age) [1, 2]. Indeed, the immune system of neonates has not been fully developed depending almost entirely on the transport of maternal immunoglobulin (Ig) from colostrum after birth. Hence, the absorption of colostrum Ig during the first 24 h after birth is essential for the health and survival of the neonatal calf.
Immunoglobulins are the major protein components of colostrum, comprising 70–80% of the total protein content, while in mature milk, immunoglobulins constitute only 1–2% [3, 4]. There are three major immunoglobulins in bovine serum and milk: IgG, IgM and IgA, with IgG consisting of two subclasses (IgG1 and IgG2). IgG1 accounts for over 75% (46.4 mg/ml), and IgM (6.8 mg/ml), IgA (5.4 mg/ml) and IgG2 (2.9 mg/ml) are followed successively [3]. Immunoglobulins are produced by B1-cells and possess a multitude of functions such as activate complement-mediated bacteriolytic reactions, augment the recognition and phagocytosis of bacteria by leucocytes (opsonization), prevent the adhesion of microbes to surfaces, inhibit bacterial metabolism, agglutinate bacteria, and neutralize toxins and viruses [5]. Bovine colostrum immunoglobulins are notably transported from the serum and accumulated in the mammary gland during the prepartum dry period [6, 7]. Hence, delineation of the genetic architecture underlying the concentrations of immunoglobulins in cows’ colostrum and serum is important for identifying ways to improve the survival rate of neonatal calves in dairy cattle.
Concentrations of immunoglobulins are typical quantitative characteristics controlled by multiple QTLs and polygenes [8]. Heritability estimates for IgG concentrations in blood ranged from 0.27 to 0.64 in humans [9,10,11,12]. Similarly, heritability estimates of immunoglobulins in serum and milk in dairy cattle ranged from 0.08 to 0.45 [13,14,15,16]. Heritability for IgM was higher than IgG ranging from 0.18 to 0.45 and from 0.08 to 0.23, respectively. Estimates of heritability in serum were generally higher than in milk (0.15–0.25) [13].
Genome-wide association studies (GWASs) have been performed for immunoglobulins in serum or mature milk. The first GWAS based on 2247 individuals from four European cohorts (CROATIA-Vis, CROATIA-Korcula, Orkney Complex Disease Study and Northern Swedish Population Health Study) identified 9 genome-wide significant loci associate with IgG glycosylation and 4 out of them contained genes encoding glycosyltransferases [17]. Another GWAS for IgG glycosylation patterns in humans indicated that RUNX family transcription factor 3 (RUNX3) was associated with decreased galactosylation and involved in both IgA class switching and B-cell maturation as well as T-cell differentiation and apoptosis [18]. In pigs, 2 genome-wide and 4 chromosome-wide significant SNPs were detected for IgG blocking percentage to CSF virus in serum by performing GWAS [19]. Especially, a GWAS for blood natural antibodies in Canadian Holstein cows identified 23 SNPs that were significantly associated with IgG concentration at genome-wide level [20]. Another GWAS for milk natural antibodies in Dutch Holstein-Friesian cattle identified some significant SNPs for IgG1 and IgM with candidate genes on Bos taurus autosome (BTA) 3, 17, 18, and 21 that related to immunoglobulin structure and early B cell development [21]. However, there are few studies on gene identification for immunoglobulin concentrations in colostrum in dairy cattle so far. In addition, in bovine colostrum, the immunoglobulins were found mainly derived from serum [6, 7]. Hence, investigation on the concentrations of immunoglobulins in both colostrum and serum can better disentangle the genetic architecture underlying colostrum immunoglobulin traits. Here, we conducted genome-wide association studies for the concentrations of immunoglobulin components in colostrum and serum in a Chinese Holstein population to identify the functional genes that contributed to the phenotypic variation of colostrum immunoglobulins and provide molecular information for genetically improving such traits to increase calves’ disease-resistance.
Results
Statistics of phenotypes
In this study, we measured the concentrations of immunoglobulins in both colostrum and serum for 588 Chinese Holstein cows. As a result, a total of 10 traits were recorded, including concentrations of total IgG, IgG1, IgG2, IgA and IgM. Means and the corresponding standard deviations for the original and corrected phenotypic values were shown in Table 1. The estimated heritability of total IgG, IgG1, IgG2, IgA and IgM concentrations in colostrum and serum ranged from was at 0.094 to 0.48 and from 0.087 to 0.295, respectively (Table 2).
Genome-wide association study
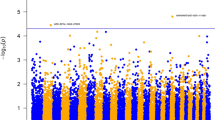
After LD analysis, a total of 16,257 effectively independent tests number were suggested. Thus, the threshold P-value for genome-wide significant association was set at 3.08E–6 (0.05/16,257) and that for suggestive significant association was 6.15E–5 (1/16,257) [22]. Based on the QQ plots (Figs. 1 and 2) and the estimated inflation factor (λ) of 0.98–1.03 for all traits, no population stratification was observed.
Manhattan and Q-Q plots of the observed P-values for the concentrations of immunoglobulins in the colostrum. A and B Indicated IgG concentrations. C and D Indicated IgG1 concentrations. E and F Indicated IgG2 concentrations. G and H Indicated IgA concentrations. I and J Indicated IgM concentrations. The Manhattan plots presented −log10 (P-values) for genome-wide SNPs (y-axis) plotted against their respective positions on each chromosome (x-axis), the horizontal red and red dashed lines in the Manhattan plots indicated the genome-wide (3.08E–6) and suggestive significance (6.15E–5) thresholds, respectively. The Q-Q plots showed the observed −log10-transformed P-values (y-axis) and the expected −log10-transformed P-values (x-axis)
With GCTA 1.90.2, we performed the GWASs for the 10 traits. In colostrum (Fig. 1 and Table 3), significant associations between 11 SNPs and total IgG were found at the suggestive level (P < 6.15E–5). The significant SNPs were located on BTA 2 (1 SNP), 4 (3 SNPs), 6 (5 SNPs), 18 (1 SNP), 22 (1 SNP). For IgG1, one genome-wide significant SNP (P < 3.08E-6) on BTA 13 and two suggestive significant SNPs (P < 6.15E–5) on BTA 11 were detected. For IgG2, five genome-wide significant SNPs (P < 3.08E-6) and 13 suggestive significant SNPs (P < 6.15E–5) were observed, locating on BTA 20 (1 SNP) and 21 (17 SNPs). There was one genome-wide significant SNP (P < 3.08E-6) and two suggestive significant SNPs (P < 6.15E–5) detected associated with IgA, locating on BTA 5 (1 SNP) and 11 (2 SNPs). Twelve SNPs were significantly associated with IgM at the suggestive level (P < 6.15E–5), distributing on BTA 1 (2 SNPs), 10 (4 SNPs), 15 (5 SNPs) and 17 (1 SNP).
In serum (Fig. 2 and Table 4), eight SNPs were found significantly associated with total IgG at the suggestive level (P < 6.15E–5), locating on BTA 3 (3 SNPs), 6 (3 SNPs), 22 (1 SNP) and 30 (1 SNP). Two SNPs located on BTA 9 and 19 had significant associations with total IgG1 at the suggestive level (P < 6.15E–5). For IgG2, 29 genome-wide significant SNPs (P < 3.08E-6) and 27 suggestive significant SNPs (P < 6.15E–5) were detected, locating on 7 (1 SNP), 12 (3 SNPs), BTA 20 (1 SNP) and 21 (51 SNPs). Additionally, one and four SNPs were significantly associated with the concentration of IgA and IgM at the suggestive level (P < 6.15E–5), respectively. Significant SNPs were located on BTA 15 (1 SNP), BTA6 (2 SNPs), 7 (1 SNP) and 17 (1 SNP).
Candidate genes and function analysis
After comparing to the reference genes (UMD 3.1), a total of 423 genes that contained or were adjacent to (± 1 Mb) the significant SNPs were mapped, including 392 protein-coding genes, 30 non-coding RNAs and 1 pseudogene (Additional file 1: Table S1).
To further investigate the biological functions of these candidate genes, we performed GO and KEGG analysis and observed that 73 genes were enriched in immune-related biological processes and pathways such as adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains, immune response, B cell activation, inflammatory response, and NF-kappaB signaling pathways (Additional file 2: Table S2). Simultaneously, we compared the physical positions of the 423 genes with the peak of the known QTLs that have been shown associated with immune capacity in dairy cattle (Cattle QTLdb), including IgG level, FMDV peptide-induced cell proliferation, ConA-induced cell proliferation and Clinical mastitis. Consequently, 226 genes were found located within the QTL regions with a distance to the peak positions of less than 1.0 cM.
Integrating the results of GO/KEGG and QTL data, 14 overlapping genes were considered as promising candidates for the concentrations of immunoglobulins in colostrum and serum (Table 5). They were fibroblast growth factor receptor 4 (FGFR4), fibroblast growth factor receptor 2 (FGFR2), neutrophil cytosolic factor 1 (NCF1), inhibitor of nuclear factor kappa B kinase regulatory subunit gamma (IKBKG), sorbin and SH3 domain containing 3 (SORBS3), immunoglobulin heavy variable 4–59 (IGHV1S18), KIT proto-oncogene, receptor tyrosine kinase (KIT), prostaglandin-endoperoxide synthase 2 (PTGS2), BCL2 associated X, apoptosis regulator (BAX), growth factor receptor bound protein 2 (GRB2), Thousand and one kinase 1 (TAOK1), intercellular adhesion molecule 1 (ICAM1), transforming growth factor beta 1 (TGFB1), and Rac family small GTPase 3 (RAC3).
Discussion
In this study, we identified the chromosome regions related with immunoglobulin concentrations in colostrum and serum in dairy cattle by performing GWASs with high density SNP genotypes. Consequently, we detected 19, 5, 74, 4 and 16 significant SNPs associated with the total IgG, IgG1, IgG2, IgA and IgM, respectively. To our knowledge, this is the first investigation on the genetic architecture of colostrum immunoglobulins in dairy cattle.
In general, a genomic inflation factor λ of < 1.05 suggests no population stratification [23]. In this study, the calculated λ values ranged from 0.99 to 1.03 for concentrations of Ig concentration in colostrum and serum, suggesting population stratification was well controlled.
In the present study, the significant SNPs associated with the concentration of IgG2 in colostrum and serum were almost entirely distributed on BTA21 from 63.3 to 71.5 Mb. Similarly, two previous GWASs in Canadian and Dutch Holstein populations observed that the significant SNPs for IgG in serum and IgG1 in mature milk were mainly located in BTA21 from 55.5 to 70.6 Mb and 66.0 to 71.6 Mb, which contained the region identified in our study [20, 21]. The previous studies revealed that the main locus of bovine immunoglobulin heavy chain variable genes was located on approximately 71.5 Mb of BTA21 [24, 25], indicating this region may be related to the formation of immunoglobulin. Concurrently, the significant SNPs for the total IgG and IgG1 concentrations in colostrum and serum distributed on multiple chromosomes, including BTA2, 3, 4, 6, 9, 11, 18, 19, 22 and 30, which is inconsistent with the previous two GWASs for IgG in serum and IgG1 in mature milk. Such inconsistency was most likely due to the huge difference of Ig formation mechanism and concentration between colostrum and mature milk. The majority of bovine colostrum Ig was transported from serum and accumulate in the mammary gland during the prepartum dry period, under the influence of prolactin and ceases abruptly at parturition, resulting in 200 times difference between Ig concentration in colostrum and mature milk [26]. Furthermore, 2 significant SNPs associated with IgM concentrations in colostrum and serum were detected on 73.3 and 74.2 Mb of BTA17, very close to 2 significant SNPs on BTA17 (72.5 to 73.6 Mb) identified for IgM in mature milk in a previous GWAS in Dutch Holstein populations [20]. The remaining significant SNPs were first reported in this study.
Combing the biological functions of the 423 functional genes that contained or were closed to the significant SNPs with less than 1 Mb and the known QTL data for immune traits in bovine,14 promising genes were identified for Ig. Of these, 2, 2, 3 and 1 candidate genes were selected for the total IgG, IgG1, IgG2 and IgM concentration in colostrum, respectively. FGFR2 and FGFR4 belong to the fibroblast growth factor receptor family which has been shown to mediate pro-inflammatory signaling in the liver and airway epithelium in chronic obstructive pulmonary disease [27]. NCF1 encodes a cytosolic subunit of neutrophil NADPH oxidase, an enzyme responsible for reactive oxygen species (ROS) production, which is pivotal in both host defense and the control of inflammation [28, 29]. IKBKG encodes the regulatory subunit of the inhibitor of kappaB kinase (IKK) complex, which activates NF-kappaB resulting activation of genes involved in inflammation and immunity [30, 31]. SORBS3 encodes an SH3 domain-containing adaptor protein that regulates cell adhesion and signal transduction. The deficiency of adaptor protein could suppress vascular inflammation and inactivate Akt–nuclear factor κB signaling [32]. IGHV1S18 is an immunoglobulin heavy chain variable region that encodes Ig heavy chain and is directly related to the formation of immunoglobulins. KIT encodes a receptor tyrosine kinase that is associated with the earliest neutrophil developmental stages [33]. PTGS2 could activate the NF-κB signaling pathway which plays a key role in regulating the immune response to infection [34].
Simultaneously, 2, 1, 1 and 2 candidate genes were opted for the concentration of IgG, IgG1, IgG2 and IgM in serum, respectively. Of these, BAX belongs to the BCL2 protein family which could regulate B cell homeostatic proliferation and apoptotic process [35]. GRB2 encodes growth factor receptor-bound protein 2, which could regulate B-cell maturation, B-cell memory responses and inhibits B-cell Ca2+ signaling [36]. Thousand and one kinase 1 (TAOK1) could as a negative regulator of IL-17 to mediate signal transduction and inflammation, controlling colitis of inflammatory bowel disease [37]. ICAM1 encodes a cell surface glycoprotein which is typically expressed on endothelial cells and cells of the immune system. Upregulation of ICAM1 in a mechanism involving NF-қB could inhibit the Epstein-Barr virus infection [38]. The expression level of TGFB1 was associated with melanoma immune response [39]. The protein encodes by RAC3 is a member of the p160 family of nuclear receptor coactivators that plays an important role in NF-kappaB activation [40].
Generally, all these genes played vital roles in the inflammation, neutrophil activation, resistance to viruses, NF-kappaB, B cell homeostasis and immune-related process, which indicated the potentially important roles of Ig in colostrum and serum in resistance to infectious diseases.
In the present study we identified 8 and 6 first-time candidate genes for immunoglobulins in dairy cattle colostrum and serum, respectively. From the breeding perspective, our findings provide important molecular information for the genetic improvement program on health and disease-resistance traits in dairy cattle. On the other hand, as the absence of biological validation and the measurements of serum Ig in the offspring, further in-depth investigations are needed to better understand the genetic mechanisms on how these genes regulated and impacted the formation of immunoglobulins in colostrum before applying them on the breeding of dairy cattle.
Conclusion
In this study, we conducted genome-wide association studies for the concentrations of immunoglobulins in colostrum and serum in Chinese Holstein. A total of 36 genome-wide and 82 suggestive significant SNPs were detected for the total IgG, IgG1, IgG2, IgA and IgM traits, in which the main quantitative trait loci for immunoglobulins were on BTA6 and 21. Combining the identified significant SNPs, functional enrichment, and the known QTL data, we identified 14 promising candidate genes for the concentration of IgG and IgM in colostrum and serum, including FGFR4, FGFR2, NCF1, IKBKG, SORBS3, IGHV1S18, KIT, PTGS2, BAX, GRB2, TAOK1, ICAM1, TGFB1 and RAC3. Our findings provided new insights into the genetic architectures underlying immunoglobulins concentrations in colostrum and important molecular information for the genetic improvement program on these traits in dairy cattle.
Methods
Animals and phenotypes
The animals used in this study consist of 588 Chinese Holstein cows daughters of 44 sires from 10 dairy farms in the Beijing Dairy Cattle Center and the Beijing Sunlon Livestock Development Company Limited. The pedigree contained 1839 animals and was provided by the Beijing Dairy Cattle Center. The average number of daughters per sire was 13.4. Cows ranged from parity 1 to 4 (mean = 2.52). The blood serum and colostrum samples were taken from each cow during the first milking within 24 h after calving for measurement of immunoglobulins. Hair follicle samples were collected from each animal for SNP chip genotyping as well. The whole procedure for collection of the samples (blood, hair and colostrum) was implemented in strict accordance with the protocol approved by the Animal Welfare Committee of China Agricultural University (Permit number: DK996). The animals used in this study were all released to their own population for normal production after sample collection.
The concentrations of immunoglobulins of each colostrum and serum sample were measured, including total IgG (Bovine IgG ELISA Quantitation Set E10–118, Bethyl Laboratories, Montgomery, TX, USA), IgG1 (Bovine IgG1 ELISA Quantitation Set, E10–116), IgG2 (Bovine IgG2 ELISA Quantitation Set, E10–117), IgA (Bovine IgA ELISA Quantitation Set, E10–131) and IgM (Bovine IgM ELISA Quantitation Set, E10–101). For further statistical analysis, the phenotypic values for the concentrations of total IgG, IgG1 and IgG2 in colostrum or serum were square root transformed to fit a normal distribution, simultaneously phenotypes for IgA and IgM concentrations were log-transformed.
Genotyping and quality control
Genomic DNA was extracted from the hair follicle samples with the QIAamp® DNA Mini Kit (QIAGEN, Valencia, CA, USA) for genotyping. A total of 588 individuals were genotyped with the GeneSeek GGP_HDv3 chip (150 K, including 140,668 SNP markers: GeneSeek, Lincoln, NE, USA).
Quality control was conducted on PLINK 1.90 software and the filtering processes were as follows: firstly, samples with all SNPs genotyping rate < 95% were deleted; then, SNPs with call rates < 90%, minor allele frequencies (MAF) < 0.1 and Hardy–Weinberg equilibrium (HWE) p-values < 10–6 were discarded [41, 42]. Thus, 563 individuals with 91,620 SNPs were kept for further analysis (Additional files 3 and 4).
Statistical analysis
Mixed Model based single locus Regression Analyses (MMRA)
We performed single-SNP association analysis for the individual phenotype in GCTA 1.90.2 with the following mixed linear model:
Where y is a vector of transformed phenotypes (the concentration of IgG, IgG1, IgG2, IgA and IgM in colostrum and serum) of all cows; μ is the overall mean; f is the vector of fixed effects, including herd (classes: 1 to 10), parity (classes: 1 = parity 1, 2 = parity 2, 3 = parity 3 and 4 = parity 4) and season of calving (classes: 1 = March to May, 2 = June to August, 3 = September to November and 4 = December to February), X is an incidence matrix relating elements of f to y; c is the vector of the SNP genotype indicators which take values 0, 1 or 2 corresponding to the three genotypes 11, 12 and 22 (assuming 2 is the allele with a minor frequency), b is the regression coefficient of y on c; g is the vector of residual polygenic effects with g ~ N (0, Gσg2) (where G is the genomic relationship matrix and σg2 is the additive variance), Z is the incidence matrix of g; e is the vector of residual errors with e ~ N (0, Iσe2) (where I is the indentity matrix and σe2 is the residual variance). The heritability estimation were carried out by GCTA 1.90.2 software.
The existence of linkage disequilibrium (LD) of SNPs in every chromosome may lead to over-correction when using Bonferroni adjustments [41, 43]. Hence, we used an effectively independent test number to define the thresholds for genome-wide/suggestive significant associations based on the assessed number of independent markers and linkage disequilibrium blocks for markers on every chromosome [22].
Population stratification can result in spurious association findings in a GWAS [42]. Thus, we calculated the genomic inflation factor (λ) and depicted quantile-quantile (Q-Q) plot to assess stratification in our study population using qqman packages in R 3.6.0.
Identification of candidate genes
To further identify the candidate genes associated with the concentrations of immunoglobulins, we selected the functional genes that contained or were adjacent to the significant SNPs with less than 1 Mb based on the bovine gene set in RefSeq database (Bos_taurus_UMD_3.1; http://hgdownload.cse.ucsc.edu/goldenPath/bosTau6/database/). Additionally, to figure out the biological functions of these genes, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment were implemented with DAVID Bioinformatics Resources (https://david.ncifcrf.gov). In addition, we also compared the physical position of these functional genes with the reported quantitative traits loci (QTLs) for immune capacity traits in the Cattle QTL database (https://www.animalgenome.org/cgi-bin/QTLdb/BT/index).
Availability of data and materials
All supporting data can be found within the additional files.
Abbreviations
- Ig:
-
Immunoglobulin
- GWAS:
-
Genome-wide association study
- SNP:
-
Single nucleotide polymorphisms
- RUNX3:
-
RUNX family transcription factor 3
- BTA:
-
Bos taurus autosome
- MMRA:
-
Mixed model based single locus regression analyses
- GRM:
-
Genetic relationship matrix
- REML:
-
Restricted maximum likelihood
- LD:
-
Linkage disequilibrium
- QQ:
-
Quantile-quantile
- GO:
-
Gene Ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- QTL:
-
Quantitative trait locus
- FGFR4 :
-
Fibroblast growth factor receptor 4
- FGFR2 :
-
Fibroblast growth factor receptor 2
- NCF1 :
-
Neutrophil cytosolic factor 1
- IKBKG :
-
Inhibitor of nuclear factor kappa B kinase regulatory subunit gamma
- SORBS3 :
-
Sorbin and SH3 domain containing 3
- IGHV1S18 :
-
Immunoglobulin heavy variable 4–59
- KIT :
-
KIT proto-oncogene receptor tyrosine kinase
- IGHV1S18 :
-
Receptor tyrosine kinase
- PTGS2 :
-
Prostaglandin-endoperoxide synthase 2
- BAX :
-
BCL2 associated X, apoptosis regulator
- GRB2 :
-
Growth factor receptor bound protein 2
- TAOK1 :
-
Thousand and one kinase 1
- ICAM1 :
-
Intercellular adhesion molecule 1
- TGFB1 :
-
Transforming growth factor beta 1
- RAC3 :
-
Rac family small GTPase 3
- IKK:
-
Inhibitor of kappaB kinase
- ROS:
-
Reactive oxygen species
References
Weaver DM, Tyler JW, VanMetre DC, Hostetler DE, Barrington GM. Passive transfer of colostral immunoglobulins in calves. J Vet Intern Med. 2000;14(6):569–77.
Wells SJ, Dargatz DA, Ott SL. Factors associated with mortality to 21 days of life in dairy heifers in the United States. Prev Vet Med. 1996;29(1):9–19.
Porter P. Immunoglobulins in bovine mammary secretions. Quantitative changes in early lactation and absorption by the neonatal calf. Immunology. 1972;23(2):225–38.
Leach JL, Sedmak DD, Osborne JM, Rahill B, Lairmore MD, Anderson CL. Isolation from human placenta of the IgG transporter, FcRn, and localization to the syncytiotrophoblast - implications for maternal-fetal antibody transport. J Immunol. 1996;157(8):3317–22.
Korhonen H, Marnila P, Gill HS. Milk immunoglobulins and complement factors. Br J Nutr. 2000;84(Suppl 1):S75–80.
Newby TJ, Bourne J. The nature of the local immune system of the bovine mammary gland. J Immunol. 1977;118(2):461–5.
Sasaki M, Davis CL, Larson BL. Production and turnover of IgG1 and IgG2 immunoglobulins in the bovine around parturition. J Dairy Sci. 1976;59(12):2046–55.
Falconer DS, Gauld IK, Roberts RC, Williams DA. The control of body size in mouse chimaeras. Genet Res. 1981;38(1):25–46.
Grundbacher FJ. Heritability estimates and genetic and environmental correlations for the human immunoglobulins G, M, and a. Am J Hum Genet. 1974;26(1):1–12.
Kacprazak-Bergman I. Sexual dimorphism of heritability of immunoglobulin levels. Ann Hum Biol. 1994;21(6):563–9.
Outschoorn IM, Talor MV, Hoffman WH, Rowley MJ, Mackay IR, Rose NR, et al. Autoantibody heritability in thyroiditis: IgG subclass contributions. Autoimmunity. 2011;44(3):195–200.
Outschoorn IM, Talor MV, Burek CL, Hoffman WH, Rose NR. Heritability analysis of IgG4 antibodies in autoimmune thyroid disease. Autoimmunity. 2014;47(5):320–6.
de Klerk B, Ducro BJ, Heuven HC, den Uyl I, van Arendonk JA, Parmentier HK, et al. Phenotypic and genetic relationships of bovine natural antibodies binding keyhole limpet hemocyanin in plasma and milk. J Dairy Sci. 2015;98(4):2746–52.
Ploegaert TC, Wijga S, Tijhaar E, van der Poel JJ, Lam TJ, Savelkoul HF, et al. Genetic variation of natural antibodies in milk of Dutch Holstein-Friesian cows. J Dairy Sci. 2010;93(11):5467–73.
Thompson-Crispi KA, Miglior F, Mallard BA. Genetic parameters for natural antibodies and associations with specific antibody and mastitis in Canadian Holsteins. J Dairy Sci. 2013;96(6):3965–72.
Wijga S, Bovenhuis H, Bastiaansen JWM, van Arendonk JAM, Ploegaert TCW, Tijhaar E, et al. Genetic parameters for natural antibody isotype titers in milk of Dutch Holstein-Friesians. Anim Genet. 2013;44(5):485–92.
Lauc G, Huffman JE, Pucic M, Zgaga L, Adamczyk B, Muzinic A, et al. Loci associated with N-glycosylation of human immunoglobulin G show Pleiotropy with autoimmune diseases and Haematological cancers. PLoS Genet. 2013;9(1):e1003225.
Wahl A, van den Akker E, Klaric L, Stambuk J, Benedetti E, Plomp R, et al. Genome-Wide Association Study on Immunoglobulin G Glycosylation Patterns. Front Immunol. 2018;9:277.
Lu X, Liu J, Fu W, Zhou J, Luo Y, Ding X, et al. Genome-wide association study for cytokines and immunoglobulin G in swine. PLoS One. 2013;8(10):e74846.
de Klerk B, Emam M, Thompson-Crispi KA, Sargolzaei M, van der Poel JJ, Mallard BA. A genome-wide association study for natural antibodies measured in blood of Canadian Holstein cows. BMC Genomics. 2018;19(1):694.
Cordero-Solorzano J, Parmentier HK, Arts JAJ, van der Poel J, de Koning DJ, Bovenhuis H. Genome-wide association study identifies loci influencing natural antibody titers in milk of Dutch Holstein-Friesian cattle. J Dairy Sci. 2019;102(12):11092–103.
Nicodemus KK, Liu WL, Chase GA, Tsai YY, Fallin MD. Comparison of type I error for multiple test corrections in large single-nucleotide polymorphism studies using principal components versus haplotype blocking algorithms. BMC Genet. 2005;6:S78.
Price AL, Zaitlen NA, Reich D, Patterson N. New approaches to population stratification in genome-wide association studies. Nat Rev Genet. 2010;11(7):459–63.
Niku M, Liljavirta J, Durkin K, Schroderus E, Iivanainen A. The bovine genomic DNA sequence data reveal three IGHV subgroups, only one of which is functionally expressed. Dev Comp Immunol. 2012;37(3–4):457–61.
Walther S, Czerny CP, Diesterbeck US. Exceptionally long CDR3H are not isotype restricted in bovine immunoglobulins. PLoS One. 2013;8(5):e64234.
Foley JA, Otterby DE. Availability, storage, treatment, composition, and feeding value of surplus colostrum - review. J Dairy Sci. 1978;61(8):1033–60.
Easter M, Garth J, Harris ES, Shei RJ, Helton ES, Wei Y, et al. Fibroblast growth factor receptor 4 deficiency mediates airway inflammation in the adult healthy lung? Front Med (Lausanne). 2020;7:317.
Kennedy AD, DeLeo FR. Neutrophil apoptosis and the resolution of infection. Immunol Res. 2009;43(1–3):25–61.
Geering B, Simon HU. Peculiarities of cell death mechanisms in neutrophils. Cell Death Differ. 2011;18(9):1457–69.
Medunjanin S, Putzier M, Nothen T, Weinert S, Kahne T, Luani B, et al. DNA-PK: gatekeeper for IKKgamma/NEMO nucleocytoplasmic shuttling in genotoxic stress-induced NF-kappaB activation. Cell Mol Life Sci. 2020;77(20):4133–42.
Wang Y, Cui L, Yang G, Zhan J, Guo L, Chen Y, et al. Hepatitis B e antigen inhibits NF-kappaB activity by interrupting K63-linked Ubiquitination of NEMO. J Virol. 2019;93(2):e00667–18.
Guan H, Cheng WL, Guo J, Chao ML, Zhang Y, Gong J, et al. Vinexin beta ablation inhibits atherosclerosis in Apolipoprotein E-deficient mice by inactivating the Akt-nuclear factor kappaB inflammatory Axis. J Am Heart Assoc. 2017;6(2):e004585.
Dinh HQ, Eggert T, Meyer MA, Zhu YP, Olingy CE, Llewellyn R, et al. Coexpression of CD71 and CD117 identifies an early Unipotent neutrophil progenitor population in human bone marrow. Immunity. 2020;53(2):319–334 e316.
Tan C, Liu L, Liu X, Qi L, Wang W, Zhao G, et al. Activation of PTGS2/NF-kappaB signaling pathway enhances radiation resistance of glioma. Cancer Med. 2019;8(3):1175–85.
Brunner C, Marinkovic D, Klein J, Samardzic T, Nitschke L, Wirth T. B cell-specific transgenic expression of Bcl2 rescues early B lymphopoiesis but not B cell responses in BOB.1/OBF.1-deficient mice. J Exp Med. 2003;197(9):1205–11.
Ackermann JA, Radtke D, Maurberger A, Winkler TH, Nitschke L. Grb2 regulates B-cell maturation, B-cell memory responses and inhibits B-cell Ca2+ signalling. EMBO J. 2011;30(8):1621–33.
Zhang Z, Tang Z, Ma X, Sun K, Fan L, Fang J, et al. TAOK1 negatively regulates IL-17-mediated signaling and inflammation. Cell Mol Immunol. 2018;15(8):794–802.
Guo Q, Gao J, Cheng L, Yang X, Li F, Jiang G. The Epstein-Barr virus-encoded G protein-coupled receptor BILF1 upregulates ICAM-1 through a mechanism involving the NF-B pathway. Biosci Biotechnol Biochem. 2020;84(9):1810–9.
Fang S, Xu T, Xiong M, Zhou X, Wang Y, Haydu LE, et al. Role of immune response, inflammation, and tumor immune response-related cytokines/chemokines in melanoma progression. J Invest Dermatol. 2019;139(11):2352–2358 e2353.
Coste A, Antal MC, Chan S, Kastner P, Mark M, O'Malley BW, et al. Absence of the steroid receptor coactivator-3 induces B-cell lymphoma. EMBO J. 2006;25(11):2453–64.
International HapMap C. A haplotype map of the human genome. Nature. 2005;437(7063):1299–320.
Haldar T, Ghosh S. Effect of population stratification on false positive rates of population-based association analyses of quantitative traits. Ann Hum Genet. 2012;76:237–45.
Johnson RC, Nelson GW, Troyer JL, Lautenberger JA, Kessing BD, Winkler CA, et al. Accounting for multiple comparisons in a genome-wide association study (GWAS). BMC Genomics. 2010;11:724.
Acknowledgements
We appreciate the Beijing Dairy Cattle Center and the Beijing Sunlon Livestock Development Company Limited for providing the colostrum, serum and hair samples, and pedigree data for the Chinese Holstein cows used in this study.
Funding
This work was financially supported by the National Natural Science Foundation of China (31530070, 31872330), the National key Research and Development Project of China (2021YFF1000700), Beijing Science and Technology Program (20200105, D171100002417001), and the Program for Changjiang Scholar and Innovation Research Team in University (IRT_15R62). Each of the funding body took part in the design of the study and collection, analysis, and interpretation of data, and the writing of the manuscript.
Author information
Authors and Affiliations
Contributions
SL performed statistical and bioinformatics analysis, and was a major contributor to manuscript preparation. CK performed sample collection and measurement of immunoglobulins concentrations and participated in manuscript preparation, LL, YG and LX performed sample collection and data analysis. BH, YZ and SZ participated in result interpretation. DS conceived and designed the experiments and wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The whole study has been submitted to and approved by the Institutional Animal Care and Use Committee (IACUC) at China Agricultural University (Beijing, China; permit number: DK996). Sample collection specifically for this study followed the standard procedures with the full agreement of the Beijing Dairy Cattle Center and the Beijing Sunlon Livestock Development Company Limited who owned the animals.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
The features of genes contained or were near to (within 1 Mb) the identified significant SNPs for immunoglobulins.
Additional file 2: Table S2.
Functional enrichment results of GO and Pathway analysis on the 1083 genes.
Additional file 3.
MAP file of SNP data.
Additional file 4.
PED file of SNP data.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Lin, S., Ke, C., Liu, L. et al. Genome-wide association studies for immunoglobulin concentrations in colostrum and serum in Chinese Holstein. BMC Genomics 23, 41 (2022). https://doi.org/10.1186/s12864-021-08250-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-021-08250-5