Abstract
Background
Genetic improvement of fillet quality attributes is a priority of the aquaculture industry. Muscle composition impacts quality attributes such as flavor, appearance, texture, and juiciness. Fat and moisture make up about ~ 80% of the tissue weight. The genetic architecture underlying the fat and moisture content of the muscle is still to be fully explored in fish. A 50 K gene transcribed SNP chip was used for genotyping 789 fish with available phenotypic data for fat and moisture content. Genotyped fish were obtained from two consecutive generations produced in the National Center for Cool and Cold Water Aquaculture (NCCCWA) growth-selective breeding program. Estimates of SNP effects from weighted single-step GBLUP (WssGBLUP) were used to perform genome-wide association (GWA) analysis to identify quantitative trait loci (QTL) associated with the studied traits.
Results
Using genomic sliding windows of 50 adjacent SNPs, 137 and 178 SNPs were identified as associated with fat and moisture content, respectively. Chromosomes 19 and 29 harbored the highest number of SNPs explaining at least 2% of the genetic variation in fat and moisture content. A total of 61 common SNPs on chromosomes 19 and 29 affected the aforementioned traits; this association suggests common mechanisms underlying intramuscular fat and moisture content. Additionally, based on single-marker GWA analyses, 8 and 24 SNPs were identified in association with fat and moisture content, respectively.
Conclusion
SNP-harboring genes were primarily involved in lipid metabolism, cytoskeleton remodeling, and protein turnover. This work provides putative SNP markers that could be prioritized and used for genomic selection in breeding programs.
Similar content being viewed by others
Background
Fish are excellent source of protein with lower content of total fat, saturated fat, and cholesterol and higher omega-3 fatty acids compared to other animals. These characteristics make fish fillets an ideal source of nutrition according to a consensus dietary studies and recommendation [1]. Thus, fillet quality traits have economic importance to the aquaculture industry [2], and consumer attitude towards fish is influenced by fillet quality attributes [3]. For profitable aquaculture production, there is a need for fish fillets with optimum nutritional values and consistent organoleptic qualities. Rainbow trout fish fillet contains ~ 4–18% by weight fat. Rainbow trout cultured at Clear Springs Foods Inc. (Buhl, ID, USA), the largest producer in the U.S., contains fat content of 12–13% [4]. Variations in fat content can result in positive and negative impacts on fillet quality [4, 5]. Both the quantity and quality of intramuscular lipid impact fillet juiciness, flavor, color, texture, and shelf-life [5,6,7,8]. Selection on fat content can enhance fillet color and texture [9], feed conversion ratio (FCR), and protein-retention efficiency [10]. However, accumulating excessive lipids in the muscle makes fillet processing difficult and reduces fillet firmness [11,12,13]. In addition, high levels of polyunsaturated fatty acids make the fillet more prone to lipid oxidation, which contributes to the development of rancid flavor and changes in color and nutritional value [14]. Therefore, management of fat content in fish could be used to minimize the variation in eating quality and yield a product of predictable quality [6]. The aquaculture industry usually controls the fat content of fillets by adjusting lipid content in the diet [15,16,17]. However, there are limitations in using the dietary lipids approach without deteriorating fillet quality due to lipid oxidation and diminishing profitability due to increased feed cost and accumulation of fat in the viscera instead of the muscle. Also, a widely adopted culturing triploid in rainbow trout can prevents loss of fillet quality associated with fat mobilization and protein catabolism during sexual maturation [9, 18].
Fish fillet is a highly perishable food, at least partially, due to high moisture content (60–70%), which results in off-flavors and faster flesh spoilage because water facilitates enzymatic activity and bacterial growth [19, 20]. Low-temperature storage is used to control water activity. However, slow enzymatic reactions can still support microbial growth at low temperatures [21]. Previous studies showed a high correlation between fat and moisture content [22]. In mammals, the intramuscular fat content exhibits a significant negative correlation with moisture content [23, 24]. In fish, this correlation depends greatly on the energetic demands associated with various physiological conditions [18, 25, 26]. An antagonistic biological relationship between traits may hinder their simultaneous improvement, which could lead to unwanted changes in fillet quality [27, 28]. Therefore, an optimal balance among important economic traits needs to be established to enhance product quality and industry returns [2]. Knowledge of the heritability and genetic architecture of each trait provides information necessary in developing appropriate multi-trait selection programs.
Selective breeding can be used to enhance phenotypic traits of interest. A two-way program of selection on muscle fat content was initiated in rainbow trout to produce lean and fat lines where the fat percentage increased by ~ 15 to 31% in the fat line depending on the diet [9, 29]. These lines were used as a model to study the effect of muscle fat content on fillet quality [9]. Separately, five generations of family-based selection on body weight of rainbow trout were performed at the USDA NCCCWA [30]. In the third-generation (year class (YC) 2010), fish fillet fat content showed a moderate correlation with whole body weight (coefficient of determination R2 value of 0.50) [31]. Therefore, selection for bodyweight yielded heavier fish with more fat in the muscle. Similarly, gilthead seabream exhibited a 0.1% increase in muscular fat content concomitant with a 0.08% decline in moisture content per increment of ten grams in weight [32]. Muscle fat and moisture content showed moderate heritability in fish, including rainbow trout, implying the existence of genetic variance in a rainbow trout population selected for an enhanced rate of growth [33], thus making genetic responses to selection possible. However, in salmonids, the genetic architecture of fat and moisture content has not been fully explored in a genome-wide scale [34]. Understanding the genetic basis of the phenotypic traits in question and development of fish strains of improved genetic gain will enhance the efficiency of breeding programs, aquaculture industry profitability, and consumer satisfaction.
Genome-wide association (GWA) studies can identify large-effect variants responsible for phenotypic variations, which can be prioritized in genomic selection. A few GWA studies have been conducted on aquaculture species to identify quantitative genomic loci (QTL) responsible for the genetic variability in body weight [35], fillet quality [35, 36], and disease resistance [37]. In fish, a few GWA studies were performed on Atlantic salmon [11, 34] and common carp [38] to identify QTL associated with muscle fat content. In Atlantic salmon, few significant SNPs associated with muscle fat content were identified using a ~ 5 K and 57 K SNP panels [11, 34]. In common carp, a high-density, 250 K SNP array revealed eight SNPs related to muscle fat content; however, none of the SNPs surpassed the genome-wide significance level [38]. The two studies did not identify QTL explaining a large proportion of the genetic variance in fat content in fish. To the best of our knowledge, no GWA studies have been performed in rainbow trout to identify SNP markers associated with genetic variance for fat and moisture content.
A 50 K transcribed SNP-chip, suitable for GWA analyses, has been recently developed in our laboratory. The array has been used to identify large-effect QTL responsible for genetic variance in fillet yield, firmness, protein content, and body weight gain using the same fish population used in this study [36, 39, 40]. The current study aimed to identify QTL associated with the additive genetic variance in fillet fat and moisture content for the same rainbow trout population.
Results and discussion
Muscle fat and moisture contents are interrelated attributes that affect the organoleptic quality and nutritional value of muscle foods [8, 38, 41, 42]. In fish, high-fat content may influence fillet processing and reduce the firmness leading to fillet downgrading [11]; moreover, it significantly impacts texture, juiciness, and flavor [5,6,7]. In mammals, increased marbling scores are positively related to beef tenderness, accounting for ~ 9% of the shear force variation [43]. The inability to retain moisture during postmortem storage, in both fish and mammals, is associated with a high drip loss and, in turn, reduces the industry profitability by influencing processing yield and palatability [44, 45]. In the pork industry, drip loss results in up to 10% product losses affecting profitability at wholesale and retail levels [44]. Similarly, 1.5 to 5% of drip losses were reported in salmon [46, 47]. Muscle quality traits in rainbow trout are complex and controlled by many genes (i.e., polygenic in nature) [31, 39]. Increased knowledge of the genetic basis of muscle quality traits will facilitate to advance the commercial breeding in salmonids. GWA studies are powerful tools to identify genetic variants associated with complex traits [36, 39, 40]. However, no GWA studies were previously conducted to dissect the genetic architecture of fillet fat and moisture contents in rainbow trout. The SNP-based heritability for fat and moisture content was 0.39 and 0.51, respectively, suggesting existence of adequate genetic variability in the NCCCWA fish population to allow genetic improvement through selective breeding. A higher rate of genetic gain is obtained when genomic information is used [48].
In this study, we used genomic windows of 50 SNPs of a 50 K SNP chip to perform GWA analyses, in addition to the single-marker analysis approach, to identify genomic regions associated with the traits. Given that the 50 K, SNP chip contains SNPs of potential association with intramuscular fat content, all fish used to build the SNP-chip were excluded from the GWA analysis in the present study.
The fish population had an average muscle fat content of 9.2 ± 1.91 (%) and moisture content of 69.93 ± 1.75 (%). Variations in fat and moisture content are shown in Fig. 1. Previous studies reported a significant correlation between changes in fat and moisture content in fish [26, 32]. Consistently, our data showed a significant negative correlation between fat and moisture content (R = − 0.88; p-value = 6.3E-262). Further, fat content was regressed on moisture content using phenotypic data from separate year classes to make sure the size and age differences between fish from YC 2010 and YC 2012 are not influencing the correlation between the traits. Interestingly, the negative correlation between fat and moisture content (R = − 0.88) was maintained in fish from the two consecutive generations.
QTL affecting muscle fat and moisture content using WssGBLUP
All 35,322 SNPs (70.6%) that passed QC were used in the WssGBLUP analysis. A complete list of proportions of additive genetic variance for fat content explained by all genomic windows is provided in Table S1. Of them, a total of 137 genomic sliding windows explaining at least 2% (arbitrary value) of the additive genetic variance for fat content are listed in Table S2. Most of the SNP sliding windows (n = 124; ~ 91%) were located within 62 protein-coding genes. Genomic loci affecting the additive variance for fat content were clustered in 5 chromosomes (1, 4, 5, 19, and 29) (Fig. 2).
Manhattan plot showing association between 50 SNP-genomic sliding windows and muscle fat content. Chromosome 19 showed the highest peaks with genomic loci explaining up to 5.51% of the additive genetic variance. The basal blue line represents 2% of the genetic variance explained by the sliding windows
Chromosome 19 harbored the highest number (n = 50) and the most significant peaks affecting fat content (up to 5.51%) (Table S2, Fig. 2). Many of the SNPs were located within the CDS of the SNP-harboring genes (n = 58) as well as their 3’UTR (n = 55). In order to understand the biological significance of the QTL associated with fat content, we annotated the SNP-harboring genes and searched their functions in the literature (described below).
Similarly, a complete list of the proportions of additive genetic variance for moisture content explained by all windows identified in this study is provided in Table S3. A total of 178 genomic sliding windows revealing at least 2% of the additive genetic variance for moisture content are listed in Table S4. Most of the SNP sliding windows (n = 165; ~ 93%) were located within 86 genes coding for proteins. Genomic loci affecting the additive variance for moisture content were clustered on 5 chromosomes (5, 14, 19, 25, and 29) (Fig. 3). Chromosome 29 harbored the highest number (n = 48), whereas the most significant peaks affecting moisture content (up to 4.46%) were identified on chromosome 19 (Table S4, Fig. 3). Many of the SNPs were located within CDS of the SNP-harboring genes (n = 68) as well as their 3’UTR (n = 72).
Manhattan plot showing association between 50 SNP-genomic sliding windows and muscle moisture content. Chromosome 19 showed the highest peaks with genomic loci explaining up to 4.46% of the additive genetic variance. The basal blue line represents 2% of the genetic variance explained by the sliding windows
Common genes affecting muscle fat and moisture content
As shown above, a negative linear relationship has been established between fat and moisture content in this selectively bred rainbow trout population (YC 2010 and YC 2012), suggesting a common mechanism underlying the genetic variation in the two traits. This negative correlation was consistent with other studies in fish and mammals [23, 24, 26, 32]. In rainbow trout, the correlation between fat and moisture content depends on the physiological status of the fish. For instance, gravid fish approaching spawning and maintained on a high plane of nutrition showed reduced intramuscular fat with a concurrent increase in moisture, shear force, and protein content [26]. On the other hand, fat content was not affected during spawning, while moisture content increased [25]. This was explained by a selective mobilization of either fat or protein during sexual maturation. Depleted macromolecules were replenished by water [25, 26]. In addition to sexual maturation, season, feeding, starvation, temperature, salinity, and selection for WBW were found to affect the fat/moisture balance and impact the product quality. For instance, increased fat content due to fast growth of salmon in the summer was accompanied by a high drip loss [49]. The drip loss negatively impacted the sensory attributes and developed unpleasant odors [49]. Exposure of juvenile salmon to a high salinity stimulated lipid depletion that was partially explained by increased depot lipase activity [50]. Channel catfish fed supplemental diets deposited fat concomitant with loss of moisture. Starved channel catfish at 8.9 °C mobilized muscular fat to supply energy for metabolic process, whereas both fat and protein were mobilized at 21.1 °C; in either case, moisture content increased [51]. Selection for WBW in rainbow trout [30] and gilthead seabream [32] led to high muscular fat content associated with a decline in moisture content (in particular, the more loosely bound water).
The current WssGBLUP identified common SNPs affecting the additive genetic variance for fat and moisture content on chromosomes 19 and 29 (Tables S2 & S4). The majority of the common SNPs (n = 47) were located on chromosome 19. Thirty-two SNPs, out of 47, involved in lipid metabolism were identified in 16 protein-coding genes on chromosome 19 (Table 1). Briefly, cathepsin B had a single 3’UTR SNP. Cathepsin B regulates very-low-density lipoprotein (VLDL) secretion and free fatty acid uptake in response to oleic acid exposure in mice [52]. Thioredoxin-related transmembrane protein 1-like (TMX1) had three SNPs. Loss of TMX increases lipid peroxidation in TMX(−/−) mice, which, in turn, enhances oxidative stress [53]. Guanine nucleotide-binding protein GI/GS/GO gamma-2 subunit (GNG2) had a single 3’UTR SNP. GNG2 expression is positively correlated with adipocyte size [54]. SNPs in genes encoding beta-taxilin and Alpha-L-fucosidase 2 (FUCA2) were covering windows explaining the highest proportion of the additive genetic variation for fat and moisture content. Adipose tissue of obesity susceptible and resistant rats differentially expressed beta-taxilin under a high-fat diet [55]. FUCA2 is a glycolipid processing enzyme [56]. Two SNPs in F-box only protein 30 (FBXO30) and the microtubule-binding protein ensconsin were ranked next to beta-taxilin and FUCA2. An SNP in FBXO30 was located in a genomic region, explaining 4.95% of the additive genetic variance for polyunsaturated fatty acids in cattle [57]. Knockdown of microtubule-binding or -associated proteins led to changes in fat accumulation during adipogenesis [58]. Dihydropyrimidinase-related protein 5-like (CRMP5) had a single synonymous SNP. CRMP5 has GO terms belong to lipid metabolic processes [59]. Five SNPs were identified in a gene encoding trifunctional enzyme subunit alpha, mitochondrial (HADHA). This gene is involved in fatty acid beta-oxidation [59]. mRNA decay activator protein ZFP36L1 had three SNPs in the 3’UTR. Knockdown of mammalian ZFP36L1 led to the downregulation of ERK activation and inhibition of adipogenesis [60]. A single 3’UTR SNP was identified in ELM2 and SANT domain-containing protein 1 (ELMSAN1). Epigenome-wide association analysis showed DNA methylation changes in ELMSAN1 were associated with body mass index (a key measure of adiposity) [61]. Prostaglandin reductase 2 (PTGR2) had two nonsynonymous SNPs. This enzyme catalyzes reduction of the conjugated α,β-unsaturated double bond of 15-keto-PGE2 in an NADPH-dependent manner, which is a critical step in inhibition of PPARγ-mediated adipocyte differentiation [62]. Spectrin beta chain, erythrocytic (SPTB) gene had two SNPs. The SPTB interacts with phospholipids in natural [63] and model membrane systems [64] and has a role in controlling the fluidity of the inner lipid leaflet of the cell membrane (reviewed in [65]).

Chromosome 29 had 14 SNPs in genomic windows explaining at least 2% of the additive genetic variance for intramuscular fat and moisture content (Tables S2 & S4). Of them, seven SNPs were involved in lipid metabolism (Table 1). A gene encoding short-chain specific acyl-CoA dehydrogenase, mitochondrial (ACADS) had a single SNP. This enzyme has a role in fatty acid beta-oxidation [66]. An intronic SNP was identified in a gene coding for arrestin domain-containing protein. The latter has GO terms belonging to fat pad and skin development and regulates the body mass [67]. Myocyte enhancer factor 2c (MEF2C) had the highest number of SNPs (n = 5) on chromosome 29. MEF2C is a transcription factor involved in skeletal muscle differentiation; however, it has been reported as a constituent of a mechanism that programs gene expression involved in development of brown adipocytes [68]. MEF2A and MEF2D isoforms exhibited in vivo differential expression in mammalian striated muscle and white adipose tissue of insulin-deficient diabetic mice [69]. To our knowledge, the role of MEF2C in white adipose tissue remains uncertain.
In addition, twelve SNPs in genes involved in transmembrane transport and cytoskeleton remodeling were identified in common QTL affecting additive variance for fat and moisture content (Table 2). The majority of these SNPs were identified on chromosome 19 (n = 11). Three synonymous SNPs were identified in a gene encoding intersectin-2 (ITSN2). This protein is necessary for the clathrin-mediated endocytosis and actin cytoskeleton remodeling [70]. Six SNPs were identified in 3 genes involved in vesicle-mediated transport (i.e., exocytosis); dnaJ homolog subfamily C member 5B, visinin-like protein 1, and syntaxin-binding protein 5. The actin cytoskeleton remodeling controls each step of exocytosis [71]. Three SNPs were identified in microtubule-associated protein RP/EB family member 3 (MAPRE3) and centrin-3. MAPRE3 and centrin-3 control the dynamics of the microtubule cytoskeleton [59, 72].

Overall, the analysis revealed that most of the loci in association with both fat and moisture content are involved in lipid metabolic process. Therefore, our results suggest that variation in moisture content is likely to be driven by changes in fat content in an antagonistic fashion. This notion was previously suggested as depletion of macromolecules under catalytic muscle conditions is likely forming voids for water accumulation [28].
Unique genes affecting the additive genetic variance for moisture
The actin cytoskeleton interacts with the cell membrane to control water transport [73]. Expression of genes involved in the cytoskeletal organization has previously shown a positive correlation with the drip loss [74]. In the current study, thirty-five variants in genes (n = 14) involved in cytoskeleton remodeling were identified, affecting the additive variance for moisture content in rainbow trout (Table 3). Briefly, bone morphogenetic protein receptor type-2 (BMPR2) had a single synonymous SNP. BMPR2 is known to interact with the cytoskeleton, and BMPR2 mutant mice exhibited cytoskeletal defects [75]. A gene encoding muscle associated receptor tyrosine kinase (MUSK) had a single SNP. Activation of MUSK in myotubes regulates the reorganization of the actin cytoskeleton [76]. Two SNPs were identified in THAP domain containing 1 (THAP1), which has a role in regulation of the mitotic cell cycle [77]. The gene encoding asparaginyl-tRNA synthetase (NARS) had 3 SNPs in windows explaining the highest additive variance (up to 3.46%; Table 3). Mutations in NARS leads to cell cycle arrest in the S phase [78]. The actin cytoskeleton undergoes dramatic changes during the cell cycle [79]. Ten SNPs were identified in three genes coding for cyclin-I (CCNI), cyclin-G1 (CCNG1), and cyclin-G2 (CCNG2). Cyclins function as regulators of the cell cycle and actin cytoskeleton dynamics (reviewed in [80]). The serine/threonine-protein, phosphatase 2A (PP2A), had a 3’UTR SNP. This phosphatase is associated with microtubule stabilization, where it binds and dephosphorylates the microtubule-associated proteins [81]. Annexin A6 (ANXA6) had two synonymous SNPs. ANXA6 contributes to membrane and cytoskeleton organization in a Ca2+-dependent manner [82]. Tubulin beta-4B chain (TUBB4B) had four synonymous SNPs within 1Kb of chromosome 25. TUBB4B is a critical component of microtubules [59]. Five SNPs, clustered in ~2Kb, were identified in a gene coding for mid1-interacting protein 1 (MID1IP1). This protein enhances fatty acid biosynthesis [83] and stabilizes microtubule organization [59]. Two SNPs were identified in a gene encoding tubulin-specific chaperone A (TBCE). TBCE is a tubulin-folding protein required for proper microtubule cytoskeleton organization [84]. Additionally, mutations in TBCE drive muscle atrophy [84]. Proteinase-activated receptor 1 (PAR1) and PAR2 had four SNPs. PAR-mediated, RhoA activation is vital for cytoskeletal reorganization [85].

A strong positive correlation between moisture and protein content has been established in different species. A simultaneous decline in protein and moisture content was previously reported in mammals [24]. Moisture content in rainbow trout exhibited a bidirectional relationship with protein content depending on physiological/metabolic status. For example, a negative correlation between moisture and protein content were previously reported under muscle catabolic conditions associated with full sexual maturation (R2 = 0.994, p < 0.01) [25]; whereas, a positive correlation was reported in female trout, on a high plane of nutrition, that were approaching spawning [26]. This was explained by selective mobilization of either protein during spawning or fat before spawning; in either case, the depleted macromolecule was replaced by water. It is noteworthy that protein content variation of the current study was not statistically significant between the 4 high-ranked families versus 4 low-ranked families (data not shown). The current WssGBLUP analysis indicated that thirteen SNPs in genes involved in protein degradation were involved in the additive genetic variance of moisture content (Table 4). Briefly, E3 ubiquitin-protein ligase RNF170 is an E3 ubiquitin-protein ligase that plays an essential role in the ubiquitination and degradation of inositol 1,4,5-trisphosphate receptor type 1 (ITPR1) [59, 86]. The latter controls the calcium release from the endoplasmic reticulum [87], which affects the muscle protein content in rainbow trout [39] and has a profound effect on the regulation of cytoskeleton [88]. Cystatin-1, which possesses a peptidase inhibitor activity, had a single 5’UTR SNP. Thioredoxin-like 1 (TXNL1) had two synonymous SNPs. The knockdown of TXNL1 moderately stabilizes the ubiquitin-protein conjugates suggesting a connection between protein reduction and proteolysis [89]. Pre-mRNA-processing factor 19 (PRPF19) and ubiquitin-conjugating enzyme E2 D2 (UBE2D2) had four SNPs. These ligases catalyze polyubiquitin chain assembly and play a role in proteasomal protein degradation [90, 91]. Nuclear factor NF-kappa-B p105 subunit (NFKB1) had two 3’UTR SNPs. NFKB1 is involved in the negative regulation of cellular protein metabolic process [92] and apoptotic process [93].

In addition to the ubiquitin-protein ligases, SNPs in three genes involved in lysosomal/phagosomal pathways were identified. Ras-related protein rab7 (RAB7A) harbored a 3’UTR SNP. RAB7A is a major regulator of endo-lysosomal maturation/ trafficking and protein targeting to lysosome inducing autophagosome formation [94]. Thus, RAB7A positively regulates the protein catabolic process [95]. V-type proton ATPase subunit B (ATP6V1B2) had a 3’UTR SNP. V-ATPase is responsible for acidifying the intracellular compartments, including lysosomes [96]. The gene encoding the β chain of the adaptor protein-3 (AP-3) complex had a single synonymous SNP. Deletion in AP3B1 perturbs assembly of AP-3 complex and, in turn, trafficking of transmembrane lysosomal proteins [97].
In this study, most of the common genomic loci affecting the highest proportion of the additive variance were involved in lipid metabolism, suggesting a common mechanism underlying intramuscular fat and moisture content and, partially, explaining the strong negative correlation between the fat and moisture content in this selectively bred rainbow trout population. Unique loci affecting moisture content were primarily involved in cytoskeleton regulations and protein turnover. Inhibition of protease activity, such as calpains, reduced degradation of proteins responsible for cell membrane-cytoskeleton attachments and postmortem drip channel formation in muscle [98]. The presence of calcium enhances proteolysis, by μ-Calpain, of myofibrillar and other cytoskeletal proteins during postmortem storage [45, 99]. Further investigation is warranted to determine QTL that could be prioritized in breeding programs to achieve optimal moisture content with low enzymatic activity and drip loss, and optimal fat content that can meet consumer preferences.
Overall, the WssGBLUP analysis has enriched the current understanding of the genetic architecture of the fat and moisture content in rainbow trout. Common SNP windows explained a high proportion of the additive genetic variance associated with both fat and moisture content, suggesting common regulatory mechanisms. Knowledge of the heritability of fat and moisture content and their correlations with other traits is needed for establishment of sustained multi-trait selection programs. The aquaculture industry is interested in implementing genomic selection in the breeding programs; however, applying high-density SNP chips is cost-prohibitive for small-sized hatcheries and companies. SNPs with a large-effect on genetic variances of fat and moisture content, identified in this study, could be prioritized to reduce SNP panel density needed to evaluate the predictive abilities for both traits. In another study, we found that prioritizing SNPs based on the proportion of variance explained for muscle yield and firmness allowed to reduce the SNP panel density down to ~ 800 SNPs. Reduced SNP panels outperformed the traditional PBLUP model in predicting the future fish performance, and maintained predictive abilities comparable to the 50 K SNP panel (data will be published elsewhere).
Single marker GWA analyses
To identify single SNP markers associated with variation in fat and moisture content, we analyzed SNPs that passed QC filtration (n = 29,451) using a generalized score test; this test incorporates multiple covariates in the analysis and accounts for family structure using a kinship matrix [100]. In this study, 8 and 24 significant SNPs, surpassing the genome-wide significance level, had a potential impact on the fat and moisture content (Bonferroni-corrected p < 1.69E-06; Figs. 4 and 5 and Tables S5 & S6), respectively. Whereas, 29 and 46 SNPs surpassing the suggestive significance level (Bonferroni-corrected p < 1E-05; Figs. 4 and 5 and Tables S5 & S6) were detected in association with fat and moisture content, respectively. Suggestive significant SNPs were not considered for the downstream analysis.
Manhattan plot showing single SNP markers significantly associated with variation in muscle fat content using family-based association analysis. Many of the significant SNPs are located on chromosome 5. Blue and red horizontal lines represent suggestive (1e-05) and significant (1.69e-06) threshold p-values, respectively
Single SNP markers showing significant associations with variation in moisture content using family-based association analysis. Most of the significant SNPs are located on chromosome 17. Blue and red horizontal lines represent suggestive (1e-05) and significance (1.69e-06) threshold p-values, respectively
SNPs associated with the fat content were mainly located on chromosome 5 (n = 7), and SNP-harboring genes have roles in lipid metabolism (Table 5). The list includes 78 kDa glucose-regulated protein (GRP78), spindle and kinetochore associated complex subunit 1 (SKA1), apelin receptor B (APLNR-B), desmoplakin, podocan, and calcium-binding mitochondrial carrier protein SCaMC-1 (SLC25A24). Briefly, two missense mutations were identified in genes coding for GRP78 and SKA1. GRP78 is essential for adipocyte differentiation and a balanced secretion of adipokines. Deletion of GRP78 causes lipoatrophy in mice observed as a dramatic reduction in gonadal and subcutaneous adipose tissue [101]. SKA1 was downregulated in adipose tissues between samples from obese and healthy control children and has been suggested as a candidate biomarker for childhood obesity [102]. Three synonymous mutations were identified in genes encoding APLNR, desmoplakin, and podocan. APLNR knockout mice demonstrated excess fatty acid accumulation in skeletal muscle [103]. Abnormalities in desmoplakin have been associated with changes in lipid metabolism [104]. Podocan belongs to the small leucine-rich proteoglycans (SLRPs) that bind to low-density lipoprotein receptor-related protein (LRP-1) [105]. A 3’UTR SNP was identified in a gene coding for SLC25A24. Mice fed a high-fat diet exhibited increased expression level of SLC25A24; whereas, adipocyte differentiation was suppressed in Slc25a24- knockout [106].

Puromycin-sensitive aminopeptidase (NPEPPS), on chromosome 17, had a nonsynonymous SNP explaining the highest variability in fat content (R2 = 3.2%) (Table 5). NPEPPS impacts different physiological processes, including protein turnover and cell cycle regulation. NPEPPS was upregulated in mitten crabs fed with a linseed oil rich in linoleic acid [107]. However, the effect NPEPPS on lipid metabolism in fish needs further investigations.
SNPs associated with moisture content (n = 24) were associated with protein turnover, calcium metabolism, and cytoskeleton integrity (Table S6). Most of these SNPs (n = 11; ~ 46%) were located on chromosome 17. An SNP in a gene coding for acylphosphatase 2 ranked at the top of the list (R2 = 7.4%) (Table 5); however, its physiological role is not clear. Eight SNPs associated with moisture content were identified in five genes engaged in protein metabolism (Table 5). These genes are NPEPPS, eukaryotic initiation factor 4A-III (EIF4A3), eukaryotic translation initiation factor 4B (EIF4B), ribosome binding protein 1 (RRPB1), and F-box only protein 46 (FBXO46). Briefly, the aminopeptidase, NPEPPS, was associated with variation in fat and moisture content, suggesting a correlation between moisture and fat content. Five SNPs were identified in two genes encoding EIF4A3 and EIF4B, suggesting a role for the protein translation machinery in determining variation in moisture content. RRPB1 is an ER integral membrane protein implicated in polysome assembly and, therefore, protein synthesis [108]. RRPB1 has been suggested as essential in regulation of UPR signaling molecules and autophagy [109]. Finally, the F-box family SCF-E3 ubiquitin ligase, FBXO46, had a single 3’UTR SNP.
A previous report showed that expression of genes involved in the actin cytoskeleton and cytoskeletal organization is positively correlated with drip loss in pig [74]. In this study, a total of 10 SNPs were associated with moisture content were identified in seven genes engaged in cytoskeleton regulation (Table 5). These genes are encoding serum response factor (SRF), kinesin-1 heavy chain (KIF5B), inositol-trisphosphate 3-kinase C (IP3KC), supervillain (SVIL), calcineurin subunit B type 1 (PPP3R1), eIF4A and EIF4B. Briefly, SRF is a master regulator of the actin cytoskeleton [110]. Mutations in KIF5A caused cytoskeletal defects in humans [111]. Two, 3’UTR SNPs were identified in IP3KC and PPP3R1. IP3K and calcium/calcineurin signaling play critical roles in maintaining Ca2+ homeostasis that has a profound effect on the cytoskeleton [88, 112, 113]. Supervillin (SVIL) is one of the first components of the costameric membrane skeleton to assemble during muscle formation. It establishes a high-affinity connection between the membranes and actin cytoskeleton [114]. Translation initiation factors, including eIF4A and EIF4B, associate with the actin cytoskeleton, which affects protein synthesis [115].
In our previous work, we profiled transcriptome expression of fish families (YC 2010) showing contrasting phenotypes in fat content, which revealed only 17 differentially expressed transcripts associated with fat content [31]. About 90% of the genetic variation among individuals comes from SNPs [116], and therefore, identifying SNP markers associated with complex traits is most suitable for genetic evaluation in selection programs. Few previous GWA studies identified a small number of SNPs responsible for the additive variance for fat content in Atlantic salmon and Common Carp [11, 34, 38]. The current GWA analysis identified a total of 137 SNPs in windows explaining at least 2% of the additive genetic variance for fat content, suggesting a better characterization of the genetic basis underlying variation in fat content. The discrepancies among the different GWA studies might be due to; 1) usage of different algorithms in the GWA studies, 2) variation in population size, 3) substantial difference in the capacity of the SNP arrays, 4) polygenic nature of intramuscular fat content, 5) different thresholds in each study including sliding window size [35].
Compared to our WssGBLUP analysis, the single marker GWA analysis revealed a smaller number of SNP markers associated with variation in intramuscular fat and moisture content. Besides, these two GWA approaches revealed different significant peaks associated with traits of interest. Aguilar et al. [117] showed that the highest peak based on the p-value was not the same based on the proportion of variance explained, and this is because the latter depends on allele frequency, i.e., high effect but low frequency decreases the variance explained. This result is consistent with other studied traits, such as fillet firmness, protein content [39], and bodyweight gain [40] in rainbow trout. The potential factors associated with observed heterogeneity between the two approaches are different algorithms, thresholds, and windows size used in each approach. For instance, WssGBLUP uses a flexible default HWE threshold as it assumes selection may have caused a departure from equilibrium, and therefore only extreme outliers would be excluded in order to keep the information content of haplotypes. The WssGBLUB was more effective than the single marker GWA in examining the genetic architecture of studied traits and identifying common QTL between traits. This method has proven to be optimal for breeding populations given the data structure: phenotyped individuals may not have genotypes, and there is a long history of pedigree recording [117]. Common QTL identified in this study may explain the high negative correlation between fat and moisture content. The recombinational progression of QTL and nearby markers determines the information content of haplotypes [118]. However, SNP-harboring genes identified by the two approaches had similar biological functions and were involved in lipid metabolism, protein turnover, and cytoskeletal remodeling. Routine use of single-SNP and multi-makers for GWA analysis was previously recommended to take advantage of the complete information content of the genotypes [118].
Taken together, controlling muscular fat content can help the aquaculture industry to produce a final product of expected quality, including moisture content, drip loss, firmness, and shelf-life. Dietary lipids are used to increase fillet fat to improve fillet sensory characteristics [119]. However, this can also elevate the feeding costs, increase visceral fat, and accelerate lipid oxidation, which increases fillet degradation. Alternatively, genetic/genomic selection can be used to control fillet fat content. The findings of the current study can help breeders where GEBV for muscular fat and moisture content can be added to multi-trait selection indices that reflect the various needs of producers and consumers [120].
Conclusions
The current GWA analyses identified novel genomic regions associated with additive genetic variance for fat and moisture content in rainbow trout. SNP-harboring genes encode proteins with a role in lipid metabolism, actin cytoskeleton remodeling, and protein synthesis/degradation. This work reveals significant QTL associated with fat content, which appears to be a polygenic trait. The top common windows affecting additive genetic variance for fat and moisture content are mainly on chromosome 19. These findings provide a genetic basis for description of the molecular mechanisms underlying fat and moisture content in teleost fish. Variation in moisture content is likely to be driven by changes in fat content in an antagonistic fashion. This work provides putative markers that could be prioritized when estimating genomic breeding values for fat and moisture content. GEBV for muscular fat and moisture content can be included in multi-trait selection indices that meet the various producers’ and consumers’ demands [121].
Methods
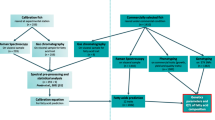
Fish population, tissue sampling, and phenotypic traits
The fish population used in the current GWA analyses was previously described in [122]. Briefly, a five-generation selective breeding program was established at NCCCWA in 2002 by intercrossing seven domesticated strains of rainbow trout; this fish population was selected for improved growth performance [30]. Phenotypic data for muscle fat and moisture content were obtained from 789 fish representing 197 full-sib families produced from two consecutive generations (YC 2010 and 2012). Single-sire×single-dam matings occurred over 6 weeks to produce full-sib families. Individuals from each family were reared together in a 200-L tank in order to keep the pedigree information. Tagging fish at ~ 5-months post-hatch allowed to rear different fish families together in 800-L communal tanks until ~ 13 months post-hatch. Fish were starved for 5 days before harvest to facilitate viscera removal.
Over five weeks, a single fish/family was collected and randomly assigned to a harvest group as described in [40]. Fish from YC 2010 were younger than those from YC 2012. Fish were euthanized (300 mg of MS-222 per liter), harvested, and eviscerated. Afterwards, the carcasses were hand-made into skinless fillets.
Proximate analyses, including crude lipid and moisture content (water content), were previously described [123]. Crude lipid content was determined using Soxhlet extraction with petroleum ether, whereas moisture content was assessed by the loss on drying method. When muscle fat and moisture content were regressed on body weight, coefficient of determination (R2) values of 0.23 and 0.38 were observed, respectively. Heritability was estimated for fat and moisture content using a genomic relationship matrix (GRM) [100].
SNP genotyping and quality control
A 50 K, transcribed gene SNP-chip was recently developed and used in identifying genomic loci responsible for additive genetic variance in fillet yield [36]. SNPs utilized to construct the SNP chip were reported in our previous study [122]. The chip included ~ 5 K nonsynonymous SNPs and ~ 21 K SNPs exhibiting potential allelic imbalances with economic traits [36, 122]. Other SNPs were added to the chip to reach a total of 50,006 SNPs with a minimum of 2 SNPs per SNP-harboring gene.
In total, 1728 rainbow trout fish were used for genotyping and quality assessment of the SNP chip. Genotyped samples were filtered by the SNPolisher software using a call rate threshold of 0.97 and Dish QC cutoff of 0.82 [36]. Genotyped fish with records for intramuscular fat and moisture content (789 fish), were used for the current GWA studies.
Fifty-SNP window GWA analysis
Estimates of SNP effects from WssGBLUP were utilized to conduct the current GWA analyses, as described in [36]. The WssGBLUP combines phenotypes, genotypes, and pedigree information into a single evaluation. GRM was created based on VanRaden equation [124]. SNPs used to create the GRM were weighted according to the proportion of additive variance they explain. The following single-trait model was used:
where y is the vector of phenotypes (fat or moisture content), b, a and w are vectors of fixed and random effects, and e is the vector of residual effects. Random and fixed effects were determined according to [35, 125]. X, Z1, and Z2 are incidence matrices for fixed and random effects in vectors b, a, and w, respectively. Whereas the family and residual random effects were considered uncorrelated, the animal effect was correlated. The covariance structure for the animal effect was given by \( \mathbf{H}{\upsigma}_{\mathrm{a}}^2 \), where H is a matrix that combines pedigree- and genomic-based relationships [126] and \( {\upsigma}_{\mathrm{a}}^2 \) is the additive variance.
The variance components were estimated using AIREMLF90 [127]. The inbreeding coefficient was calculated by INBUPGF90 [128], as described in [36]. Genomic data were edited using PREGSF90 [127], and samples or SNPs were kept according to the following parameters: minor allele frequency (MAF) > 0.05, a default value of Hardy-Weinberg equilibrium (HWE) < 0.15, and call rate > 0.90. In total, 35,322 SNPs (70.6%) passed the QC and were used for the WssGBLUP analyses.
The filtered SNPs (~ 35 K) were subjected to a two-iteration WssGBLUP analysis. All SNPs were assigned weight = 1.0 in the first iteration. SNP effects (\( \hat{u} \)) were determined using POSTGSf90 (part of GBLUPf90 software family) according to:
Where q is a weight factor based on SNPs frequency, D is a weight matrix of SNPs, Z is a matrix of gene content adjusted for allele frequencies, and â is Genomic Breading Values of genotyped animals [129].
Using \( \hat{u} \) and the allele frequency (p), SNP weights were determined as \( {\hat{u}}^22p\left(1-p\right) \) in the second iteration. SNP effects and updated weights were computed by POSTGSF90 [128] using genomic sliding windows of 50 contiguous SNPs. The proportion of additive variance explained by an i-th region was computed according to [129]:
Where i is the i-th region that consists of 50 adjacent SNPs, ai is the genetic value of the contiguous 50 SNP region, σ2a is the total genetic variance, j is the j-th SNP within the 50 SNP region, Zj is a vector of the gene content of an SNP for all fish, and ûj is the effect of an SNP within the contiguous 50 SNP region.
The qqman package [130] in R was used to generate Manhattan plots showing the percentage of genetic variance explained.
Single marker GWA analysis
PLINK [131] was used to filter the genomic data before performing a single marker association analysis. The filtering criteria included MAF > 0.05 and a default value of HWE < 0.001. PLINK was used to retrieve R-squared values (R2) of association between the quantitative traits and genotypes. R2 is the proportion of variance in the phenotype explained by the genetic factors in a linear regression model. ONETOOL [100] was used to perform a family-based association analysis where it allows for incorporating multiple covariates and accounts for family structure. Covariates were incorporated in the linear model to account for fixed effects (harvest group and hatch-year) and population structure. Bonferroni corrected p-values were calculated as (α/total number of variants), where α = 0.05 was used for a genome-wide significance level, and α = 0.3 was used for the suggestive significance level. Manhattan plots showing single markers associated with variation in intramuscular fat and moisture content were generated using qqman package [130].
Gene annotation
The genome annotation file was used to annotate the SNP-harboring genes by Bedtools [132]. SNPs were categorized as genic or intergenic depending on their physical location relative to the body of the gene. Genic SNPs exist in coding DNA sequence (CDS), introns, or untranslated regions (5’UTR and 3’UTR). Intergenic SNPs are defined as SNPs located in the region between genes.
Availability of data and materials
All datasets generated for this study are included in the manuscript and/or the Additional Files. The genotypes (ped and .map files) are available in our previous publication [39].
Abbreviations
- BCWD:
-
Bacterial Cold Water Disease
- GO:
-
Gene ontology
- GWA:
-
Genome-wide association
- HWE:
-
Hardy–Weinberg equilibrium
- MAF:
-
Minor allele frequency
- NCCCWA:
-
USDA National Center of Cool and Cold Water Aquaculture
- QC:
-
Quality control
- QTL:
-
Quantitative trait loci
- SNP:
-
Single nucleotide polymorphism
- UTR:
-
Untranslated region
- WssGBLUP:
-
Weighted single-step GBLUP
- YC:
-
Year class
References
FAO. Livestock's long shadow: environmental issues and options. Rome; 2006.
Kankainen M, Setälä J, Kause A, Quinton C, Airaksinen S, Koskela J. Economic values of supply chain productivity and quality traits calculated for a farmed European whitefish breeding program. Aquaculture Econ Manage. 2016;20(2):131–64.
Conte F, Passantino A, Longo S, Voslarova E. Consumers' attitude towards fish meat. Ital J Food Saf. 2014;3(3):1983.
Kenney PB, DJW. Growth Performance, Fillet Quality, and Reproductive Maturity of Rainbow Trout (Oncorhynchus mykiss) Cultured to 5 Kilograms within Freshwater Recirculating Systems. J Aquaculture Res Dev. 2014;05(04).
Chaiyapechara S, Liu KKM, Barrows FT, Hardy RW, Dong FM. Proximate composition, lipid oxidation, and sensory characteristics of fillets from rainbow trout Oncorhynchus mykiss fed diets containing 10 to 30% lipid. J World Aquacult Soc. 2003;34(3):266–77.
Robb DHF, Kestin SC, Warriss PD, Nute GR. Muscle lipid content determines the eating quality of smoked and cooked Atlantic salmon (Salmo salar). Aquaculture. 2002;205(3–4):345–58.
Morkore T, Vallet JL, Cardinal M, Gomez-Guillen MC, Montero P, Torrissen OJ, Nortvedt R, Sigurgisladottir S, Thomassen MS. Fat content and fillet shape of Atlantic Salmon: relevance for processing yield and quality of raw and smoked products. J Food Sci. 2001;66(9):1348–54.
Manor M, Weber G, Salem M, Yao J, Aussanasuwannakul A, et al. Effect of sexual maturation and triploidy on chemical composition and fatty acid content of energy stores in female rainbow trout, Oncorhynchus mykiss. Aquaculture. 2012;364:312–21.
Florence L, Mireille C, Jérôme B, Laurent L, Françoise M, Edwige Q. Selection for muscle fat content and triploidy affect flesh quality in pan-size rainbow trout, Oncorhynchus mykiss. Aquaculture. 2015;448:569–77.
Kause A, Kiessling A, Martin SA, Houlihan D, Ruohonen K. Genetic improvement of feed conversion ratio via indirect selection against lipid deposition in farmed rainbow trout (Oncorhynchus mykiss Walbaum). Br J Nutr. 2016;116(9):1656–65.
Sodeland M, Gaarder M, Moen T, Thomassen M, Kjøglum S, Kent M, Lien S. Genome-wide association testing reveals quantitative trait loci for fillet texture and fat content in Atlantic salmon. Aquaculture. 2013;408–409:169–74.
Rye M, Gjerde B. Phenotypic and genetic parameters of body composition traits and flesh colour in Atlantic salmon, Salmo salar L. Aquac Res. 1996;27(2):121–33.
Gjedrem T. Flesh quality improvement in fish through breeding. Aquac Int. 1997;5(3):197–206.
Jensen C, Birk E, Jokumsen A, Skibsted LH, Bertelsen G. Effect of dietary levels of fat, α-tocopherol and astaxanthin on colour and lipid oxidation during storage of frozen rainbow trout ( Oncorhynchus mykiss ) and during chill storage of smoked trout. Zeitschrift für Lebensmitteluntersuchung und -Forschung A. 1998;207(3):189–96.
Chaiyapechara S, Casten MT, Hardy RW, Dong FM. Fish performance, fillet characteristics, and health assessment index of rainbow trout (Oncorhynchus mykiss) fed diets containing adequate and high concentrations of lipid and vitamin E. Aquaculture. 2003;219(1–4):715–38.
Kiessling A, Pickova J, Johansson L, Åsgård T, Storebakken T, Kiessling KH. Changes in fatty acid composition in muscle and adipose tissue of farmed rainbow trout (Oncorhynchus mykiss) in relation to ration and age. Food Chem. 2001;73(3):271–84.
Regost C, Arzel J, Cardinal M, Laroche M, Kaushik SJ. Fat deposition and flesh quality in seawater reared, triploid brown trout (Salmo trutta) as affected by dietary fat levels and starvation. Aquaculture. 2001;193(3–4):325–45.
Paneru B, Ali A, Al-Tobasei R, Kenney B, Salem M. Crosstalk among lncRNAs, microRNAs and mRNAs in the muscle 'degradome' of rainbow trout. Sci Rep. 2018;8(1):8416.
Binsi PK, Viji P, Visnuvinayagam S, Ninan G, Sangeeta G, Triveni A, Ravishankar CN. Microbiological and shelf life characteristics of eviscerated and vacuum packed freshwater catfish (Ompok pabda) during chill storage. J Food Sci Technol. 2015;52(3):1424–33.
Özogul F, Polat A, Özogul Y. The effects of modified atmosphere packaging and vacuum packaging on chemical, sensory and microbiological changes of sardines (Sardina pilchardus). Food Chem. 2004;85(1):49–57.
Shahidi F, Jones YM, Kitts D. Seafood safety, processing, and biotechnology. UK: Taylor & Francis; 1997.
Ang CYW, Young LL, Wilson R. Interrelationships of protein, fat and moisture content of broiler meat. J Food Sci. 1984;49(2):359–62.
Watanabe G, Motoyama M, Nakajima I, Sasaki K. Relationship between water-holding capacity and intramuscular fat content in Japanese commercial pork loin. Asian-Australas J Anim Sci. 2018;31(6):914–8.
Li YX, Cabling MM, Kang HS, Kim TS, Yeom SC, Sohn YG, Kim SH, Nam KC, Seo KS. Comparison and correlation analysis of different swine breeds meat quality. Asian-Australas J Anim Sci. 2013;26(7):905–10.
Salem M, Kenney PB, Rexroad CE, Yao J. Molecular characterization of muscle atrophy and proteolysis associated with spawning in rainbow trout. Comp Biochem Physiol Part D Genomics Proteomics. 2006;1(2):227–37.
Salem M, Manor ML, Aussanasuwannakul A, Kenney PB, Weber GM, Yao J. Effect of sexual maturation on muscle gene expression of rainbow trout: RNA-Seq approach. Physiol Rep. 2013;1(5):e00120.
Kause A, Ritola O, Paananen T, Mäntysaari E, Eskelinen U. Coupling body weight and its composition: a quantitative genetic analysis in rainbow trout. Aquaculture. 2002;211(1–4):65–79.
Kause A, Ritola O, Paananen T, Wahlroos H, Mäntysaari EA. Genetic trends in growth, sexual maturity and skeletal deformations, and rate of inbreeding in a breeding programme for rainbow trout (Oncorhynchus mykiss). Aquaculture. 2005;247(1–4):177–87.
Quillet E, Le Guillou S, Aubin J, Labbé L, Fauconneau B, Médale F. Response of a lean muscle and a fat muscle rainbow trout (Oncorhynchus mykiss) line on growth, nutrient utilization, body composition and carcass traits when fed two different diets. Aquaculture. 2007;269(1–4):220–31.
Leeds TD, Vallejo RL, Weber GM, Pena DG, Silverstein JS. Response to five generations of selection for growth performance traits in rainbow trout (Oncorhynchus mykiss). Aquaculture. 2016;465:341–51.
Ali A, Al-Tobasei R, Kenney B, Leeds TD, Salem M. Integrated analysis of lncRNA and mRNA expression in rainbow trout families showing variation in muscle growth and fillet quality traits. Sci Rep. 2018;8(1):12111.
García-Celdrán M, Ramis G, Manchado M, Estévez A, Navarro A. Estimates of heritabilities and genetic correlations of raw flesh quality traits in a reared gilthead sea bream ( Sparus aurata L.) population sourced from broodstocks along the Spanish coasts. Aquaculture. 2015;446:181–6.
Leeds T, Kenney P, Manor M. Genetic parameter estimates for feed intake, body composition, and fillet quality traits in a rainbow trout population selected for improved growth. In: International Symposium on Genetics in Aquaculture. Auburn, AL: Auburn University; 2012. p. 259.
Horn SS, Ruyter B, Meuwissen THE, Moghadam H, Hillestad B, Sonesson AK. GWAS identifies genetic variants associated with omega-3 fatty acid composition of Atlantic salmon fillets. Aquaculture. 2020;514:734494.
Gonzalez-Pena D, Gao G, Baranski M, Moen T, Cleveland BM, Kenney PB, Vallejo RL, Palti Y, Leeds TD. Genome-wide association study for identifying loci that affect fillet yield, carcass, and body weight traits in rainbow trout (Oncorhynchus mykiss). Front Genet. 2016;7:203.
Salem M, Al-Tobasei R, Ali A, Lourenco D, Gao G, Palti Y, Kenney B, Leeds TD. Genome-wide association analysis with a 50K transcribed gene SNP-Chip identifies QTL affecting muscle yield in rainbow trout. Front Genet. 2018;9(387):387.
Vallejo RL, Liu S, Gao G, Fragomeni BO, Hernandez AG, Leeds TD, Parsons JE, Martin KE, Evenhuis JP, Welch TJ, et al. Similar genetic architecture with shared and unique quantitative trait loci for bacterial cold water disease resistance in two rainbow trout breeding populations. Front Genet. 2017;8:156.
Zheng X, Kuang Y, Lv W, Cao D, Sun Z, Sun X. Genome-wide association study for muscle fat content and abdominal fat traits in common carp (Cyprinus carpio). PLoS One. 2016;11(12):e0169127.
Ali A, Al-Tobasei R, Lourenco D, Leeds T, Kenney B, Salem M. Genome-wide association study identifies genomic loci affecting filet firmness and protein content in rainbow trout. Front Genet. 2019;10(386):386.
Ali A, Al-Tobasei R, Lourenco D, Leeds T, Kenney B, Salem M. Genome-wide identification of loci associated with growth in rainbow trout. BMC Genomics. 2020;21(1):209.
Johansson L, Kiessling A, Kiessling KH, Berglund L. Effects of altered ration levels on sensory characteristics, lipid content and fatty acid composition of rainbow trout (Oncorhynchus mykiss). Food Qual Prefer. 2000;11(3):247–54.
Aussanasuwannakul A, Kenney PB, Weber GM, Yao J, Slider SD, Manor ML, Salem M. Effect of sexual maturation on growth, fillet composition, and texture of female rainbow trout (Oncorhynchus mykiss) on a high nutritional plane. Aquaculture. 2011;317(1–4):79–88.
Lu D, Sargolzaei M, Kelly M, Vander Voort G, Wang Z, Mandell I, Moore S, Plastow G, Miller SP. Genome-wide association analyses for carcass quality in crossbred beef cattle. BMC Genet. 2013;14:80.
Melody JL, Lonergan SM, Rowe LJ, Huiatt TW, Mayes MS, Huff-Lonergan E. Early postmortem biochemical factors influence tenderness and water-holding capacity of three porcine muscles. J Anim Sci. 2004;82(4):1195–205.
Zhang WG, Lonergan SM, Gardner MA, Huff-Lonergan E. Contribution of postmortem changes of integrin, desmin and mu-calpain to variation in water holding capacity of pork. Meat Sci. 2006;74(3):578–85.
Claussen IC, Gullsvåg PE, Bantle M, Tolstorebrov I, Kvalsvik K. Superchilling of organic food: Part 2: Storage test with superchilled organic salmon and pork chops. In: SINTEF Energy Research, Thermal Energy; 2017.
Rotabakk BT, Melberg GL, Lerfall J. Effect of season, location, filleting regime and storage on water-holding properties of farmed Atlantic Salmon (Salmo salar L.). Food Technol Biotechnol. 2018;56(2):238–46.
Garcia-Ruiz A, Cole JB, VanRaden PM, Wiggans GR, Ruiz-Lopez FJ, Van Tassell CP. Changes in genetic selection differentials and generation intervals in US Holstein dairy cattle as a result of genomic selection. Proc Natl Acad Sci U S A. 2016;113(28):E3995–4004.
Mørkøre T, Rødbotten M, Vogt G, Fjæra SO, Kristiansen IØ, Manseth E. Relevance of season and nucleotide catabolism on changes in fillet quality during chilled storage of raw Atlantic salmon (Salmo salar L.). Food Chem. 2010;119(4):1417–25.
Sheridan MA. Exposure to seawater stimulates lipid mobilization from depot tissues of juvenile coho (Oncorhynchus kisutch) and Chinook (O. tshawytscha) salmon. Fish Physiol Biochem. 1988;5(4):173–80.
Suppes C, Tiemeier OW, Deyoe CW. Seasonal Variations of Fat, Protein, and Moisture in Channel Catfish. Trans Kansas Acad Sci (1903). 1967;70(3):349.
Thibeaux S, Siddiqi S, Zhelyabovska O, Moinuddin F, Masternak MM, Siddiqi SA. Cathepsin B regulates hepatic lipid metabolism by cleaving liver fatty acid-binding protein. J Biol Chem. 2018;293(6):1910–23.
Matsuo Y, Irie K, Kiyonari H, Okuyama H, Nakamura H, Son A, Lopez-Ramos DA, Tian H, Oka S, Okawa K, et al. The protective role of the transmembrane thioredoxin-related protein TMX in inflammatory liver injury. Antioxid Redox Signal. 2013;18(11):1263–72.
Heinonen S, Saarinen L, Naukkarinen J, Rodriguez A, Fruhbeck G, Hakkarainen A, Lundbom J, Lundbom N, Vuolteenaho K, Moilanen E, et al. Adipocyte morphology and implications for metabolic derangements in acquired obesity. Int J Obes. 2014;38(11):1423–31.
Joo JI, Yun JW. Gene expression profiling of adipose tissues in obesity susceptible and resistant rats under a high fat diet. Cell Physiol Biochem. 2011;27(3–4):327–40.
Flor AC, Wolfgeher D, Wu D, Kron SJ. A signature of enhanced lipid metabolism, lipid peroxidation and aldehyde stress in therapy-induced senescence. Cell Death Discov. 2017;3:17075.
Lemos MV, Chiaia HL, Berton MP, Feitosa FL, Aboujaoud C, Camargo GM, Pereira AS, Albuquerque LG, Ferrinho AM, Mueller LF, et al. Genome-wide association between single nucleotide polymorphisms with beef fatty acid profile in Nellore cattle using the single step procedure. BMC Genomics. 2016;17:213.
Sohle J, Machuy N, Smailbegovic E, Holtzmann U, Gronniger E, Wenck H, Stab F, Winnefeld M. Identification of new genes involved in human adipogenesis and fat storage. PLoS One. 2012;7(2):e31193.
Gaudet P, Livstone MS, Lewis SE, Thomas PD. Phylogenetic-based propagation of functional annotations within the gene ontology consortium. Brief Bioinform. 2011;12(5):449–62.
Lin NY, Lin TY, Yang WH, Wang SC, Wang KT, Su YL, Jiang YW, Chang GD, Chang CJ. Differential expression and functional analysis of the tristetraprolin family during early differentiation of 3T3-L1 preadipocytes. Int J Biol Sci. 2012;8(5):761–77.
Wahl S, Drong A, Lehne B, Loh M, Scott WR, Kunze S, Tsai PC, Ried JS, Zhang W, Yang Y, et al. Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity. Nature. 2017;541(7635):81–6.
Wu YH, Ko TP, Guo RT, Hu SM, Chuang LM, Wang AH. Structural basis for catalytic and inhibitory mechanisms of human prostaglandin reductase PTGR2. Structure. 2008;16(11):1714–23.
Langner M, Repasky EA, Hui SW. Relationship between membrane lipid mobility and spectrin distribution in lymphocytes. FEBS Lett. 1992;305(3):197–202.
Sweet C, Zull JE. Interaction of the erythrocyte--membrane protein, spectrin, with model membrane systems. Biochem Biophys Res Commun. 1970;41(1):135–41.
Machnicka B, Czogalla A, Hryniewicz-Jankowska A, Boguslawska DM, Grochowalska R, Heger E, Sikorski AF. Spectrins: a structural platform for stabilization and activation of membrane channels, receptors and transporters. Biochim Biophys Acta. 2014;1838(2):620–34.
Finocchiaro G, Ito M, Tanaka K. Purification and properties of short chain acyl-CoA, medium chain acyl-CoA, and isovaleryl-CoA dehydrogenases from human liver. J Biol Chem. 1987;262(17):7982–9.
Patwari P, Emilsson V, Schadt EE, Chutkow WA, Lee S, Marsili A, Zhang Y, Dobrin R, Cohen DE, Larsen PR, et al. The arrestin domain-containing 3 protein regulates body mass and energy expenditure. Cell Metab. 2011;14(5):671–83.
Lin JC. RBM4-MEF2C network constitutes a feed-forward circuit that facilitates the differentiation of brown adipocytes. RNA Biol. 2015;12(2):208–20.
Mora S, Yang C, Ryder JW, Boeglin D, Pessin JE. The MEF2A and MEF2D isoforms are differentially regulated in muscle and adipose tissue during states of insulin deficiency. Endocrinology. 2001;142(5):1999–2004.
Novokhatska O, Dergai M, Tsyba L, Skrypkina I, Filonenko V, Moreau J, Rynditch A. Adaptor proteins intersectin 1 and 2 bind similar proline-rich ligands but are differentially recognized by SH2 domain-containing proteins. PLoS One. 2013;8(7):e70546.
Porat-Shliom N, Milberg O, Masedunskas A, Weigert R. Multiple roles for the actin cytoskeleton during regulated exocytosis. Cell Mol Life Sci. 2013;70(12):2099–121.
Komarova Y, De Groot CO, Grigoriev I, Gouveia SM, Munteanu EL, Schober JM, Honnappa S, Buey RM, Hoogenraad CC, Dogterom M, et al. Mammalian end binding proteins control persistent microtubule growth. J Cell Biol. 2009;184(5):691–706.
Wayne R, Tazawa M. The Actin Cytoskeleton and Polar Water Permeability in Characean Cells. In: Tazawa M, editor. Cell Dynamics: Molecular Aspects of Cell Motility Cytoskeleton in Cellular Structure and Activity. Vienna: Springer Vienna; 1989. p. 116–30.
Ponsuksili S, Jonas E, Murani E, Phatsara C, Srikanchai T, Walz C, Schwerin M, Schellander K, Wimmers K. Trait correlated expression combined with expression QTL analysis reveals biological pathways and candidate genes affecting water holding capacity of muscle. BMC Genomics. 2008;9:367.
Johnson JA, Hemnes AR, Perrien DS, Schuster M, Robinson LJ, Gladson S, Loibner H, Bai S, Blackwell TR, Tada Y, et al. Cytoskeletal defects in Bmpr2-associated pulmonary arterial hypertension. Am J Physiol Lung Cell Mol Physiol. 2012;302(5):L474–84.
Tan-Sindhunata MB, Mathijssen IB, Smit M, Baas F, de Vries JI, van der Voorn JP, Kluijt I, Hagen MA, Blom EW, Sistermans E, et al. Identification of a Dutch founder mutation in MUSK causing fetal akinesia deformation sequence. Eur J Hum Genet. 2015;23(9):1151–7.
Cayrol C, Lacroix C, Mathe C, Ecochard V, Ceribelli M, Loreau E, Lazar V, Dessen P, Mantovani R, Aguilar L, et al. The THAP-zinc finger protein THAP1 regulates endothelial cell proliferation through modulation of pRB/E2F cell-cycle target genes. Blood. 2007;109(2):584–94.
Diamond G, Cedar H, Marcus M. A temperature-sensitive mutation in asparaginyl-tRNA synthetase causes cell-cycle arrest in early S phase. Exp Cell Res. 1989;184(1):53–60.
Nakaseko Y, Yanagida M. Cell biology. Cytoskeleton in the cell cycle. Nature. 2001;412(6844):291–2.
Bendris N, Lemmers B, Blanchard JM. Cell cycle, cytoskeleton dynamics and beyond: the many functions of cyclins and CDK inhibitors. Cell Cycle. 2015;14(12):1786–98.
Sontag E, Nunbhakdi-Craig V, Lee G, Bloom GS, Mumby MC. Regulation of the phosphorylation state and microtubule-binding activity of tau by protein phosphatase 2A. Neuron. 1996;17(6):1201–7.
Qi H, Liu S, Guo C, Wang J, Greenaway FT, Sun MZ. Role of annexin A6 in cancer. Oncol Lett. 2015;10(4):1947–52.
Sugiyama M, Takenaga F, Kitani Y, Yamamoto G, Okamoto H, Masaoka T, Araki K, Nagoya H, Mori T. Homozygous and heterozygous GH transgenesis alters fatty acid composition and content in the liver of Amago salmon (Oncorhynchus masou ishikawae). Biol Open. 2012;1(10):1035–42.
Sferra A, Baillat G, Rizza T, Barresi S, Flex E, Tasca G, D'Amico A, Bellacchio E, Ciolfi A, Caputo V, et al. TBCE mutations cause early-onset progressive encephalopathy with distal spinal muscular atrophy. Am J Hum Genet. 2016;99(4):974–83.
Greenberg DL, Mize GJ, Takayama TK. Protease-activated receptor mediated RhoA signaling and cytoskeletal reorganization in LNCaP cells. Biochemistry. 2003;42(3):702–9.
Lu JP, Wang Y, Sliter DA, Pearce MM, Wojcikiewicz RJ. RNF170 protein, an endoplasmic reticulum membrane ubiquitin ligase, mediates inositol 1,4,5-trisphosphate receptor ubiquitination and degradation. J Biol Chem. 2011;286(27):24426–33.
Huang L, Chardon JW, Carter MT, Friend KL, Dudding TE, Schwartzentruber J, Zou R, Schofield PW, Douglas S, Bulman DE, et al. Missense mutations in ITPR1 cause autosomal dominant congenital nonprogressive spinocerebellar ataxia. Orphanet J Rare Dis. 2012;7:67.
Hepler PK. The cytoskeleton and its regulation by calcium and protons. Plant Physiol. 2016;170(1):3–22.
Andersen KM, Madsen L, Prag S, Johnsen AH, Semple CA, Hendil KB, Hartmann-Petersen R. Thioredoxin Txnl1/TRP32 is a redox-active cofactor of the 26 S proteasome. J Biol Chem. 2009;284(22):15246–54.
David Y, Ziv T, Admon A, Navon A. The E2 ubiquitin-conjugating enzymes direct polyubiquitination to preferred lysines. J Biol Chem. 2010;285(12):8595–604.
Hatakeyama S, Yada M, Matsumoto M, Ishida N, Nakayama KI. U box proteins as a new family of ubiquitin-protein ligases. J Biol Chem. 2001;276(35):33111–20.
Ferreira V, van Dijk KW, Groen AK, Vos RM, van der Kaa J, Gijbels MJ, Havekes LM, Pannekoek H. Macrophage-specific inhibition of NF-kappaB activation reduces foam-cell formation. Atherosclerosis. 2007;192(2):283–90.
Cahir-McFarland ED, Davidson DM, Schauer SL, Duong J, Kieff E. NF-kappa B inhibition causes spontaneous apoptosis in Epstein-Barr virus-transformed lymphoblastoid cells. Proc Natl Acad Sci U S A. 2000;97(11):6055–60.
Lin WJ, Yang CY, Li LL, Yi YH, Chen KW, Lin YC, Liu CC, Lin CH. Lysosomal targeting of phafin1 mediated by Rab7 induces autophagosome formation. Biochem Biophys Res Commun. 2012;417(1):35–42.
Caillet M, Janvier K, Pelchen-Matthews A, Delcroix-Genete D, Camus G, Marsh M, Berlioz-Torrent C. Rab7A is required for efficient production of infectious HIV-1. PLoS Pathog. 2011;7(11):e1002347.
Trombetta ES, Ebersold M, Garrett W, Pypaert M, Mellman I. Activation of lysosomal function during dendritic cell maturation. Science. 2003;299(5611):1400–3.
Jung J, Bohn G, Allroth A, Boztug K, Brandes G, Sandrock I, Schaffer AA, Rathinam C, Kollner I, Beger C, et al. Identification of a homozygous deletion in the AP3B1 gene causing Hermansky-Pudlak syndrome, type 2. Blood. 2006;108(1):362–9.
Lawson MA. The role of integrin degradation in post-mortem drip loss in pork. Meat Sci. 2004;68(4):559–66.
Cong J, Goll DE, Peterson AM, Kapprell HP. The role of autolysis in activity of the Ca2+−dependent proteinases (mu-calpain and m-calpain). J Biol Chem. 1989;264(17):10096–103.
Song YE, Lee S, Park K, Elston RC, Yang HJ, Won S. ONETOOL for the analysis of family-based big data. Bioinformatics. 2018;34(16):2851–3.
Zhu G, Ye R, Jung DY, Barron E, Friedline RH, Benoit VM, Hinton DR, Kim JK, Lee AS. GRP78 plays an essential role in adipogenesis and postnatal growth in mice. FASEB J. 2013;27(3):955–64.
Zhu ZL, Yang QM, Li C, Chen J, Xiang M, Chen MM, Yan M, Zhu ZG. Identification of biomarkers for childhood obesity based on expressional correlation and functional similarity. Mol Med Rep. 2018;17(1):109–16.
Hwangbo C, Wu J, Papangeli I, Adachi T, Sharma B, Park S, Zhao L, Ju H, Go GW, Cui G, et al. Endothelial APLNR regulates tissue fatty acid uptake and is essential for apelin's glucose-lowering effects. Sci Transl Med. 2017;9(407).
Yang Z, Bowles NE, Scherer SE, Taylor MD, Kearney DL, Ge S, Nadvoretskiy VV, DeFreitas G, Carabello B, Brandon LI, et al. Desmosomal dysfunction due to mutations in desmoplakin causes arrhythmogenic right ventricular dysplasia/cardiomyopathy. Circ Res. 2006;99(6):646–55.
Merline R, Schaefer RM, Schaefer L. The matricellular functions of small leucine-rich proteoglycans (SLRPs). J Cell Commun Signal. 2009;3(3–4):323–35.
Urano T, Shiraki M, Sasaki N, Ouchi Y, Inoue S. SLC25A24 as a novel susceptibility gene for low fat mass in humans and mice. J Clin Endocrinol Metab. 2015;100(4):E655–63.
Wei B, Yang Z, Cheng Y, Zhou J, Yang H, Zhang L, Yang X. Proteomic analysis of the Hepatopancreas of Chinese mitten crabs (Eriocheir sinensis) fed with a linoleic acid or α-Linolenic acid diet. Front Physiol. 2018;9:1430.
Ueno T, Kaneko K, Sata T, Hattori S, Ogawa-Goto K. Regulation of polysome assembly on the endoplasmic reticulum by a coiled-coil protein, p180. Nucleic Acids Res. 2012;40(7):3006–17.
Pan Y, Cao F, Guo A, Chang W, Chen X, Ma W, Gao X, Guo S, Fu C, Zhu J. Endoplasmic reticulum ribosome-binding protein 1, RRBP1, promotes progression of colorectal cancer and predicts an unfavourable prognosis. Br J Cancer. 2015;113(5):763–72.
Miano JM, Long X, Fujiwara K. Serum response factor: master regulator of the actin cytoskeleton and contractile apparatus. Am J Physiol Cell Physiol. 2007;292(1):C70–81.
Nicolas A, Kenna KP, Renton AE, Ticozzi N, Faghri F, Chia R, Dominov JA, Kenna BJ, Nalls MA, Keagle P, et al. Genome-wide analyses identify KIF5A as a novel ALS gene. Neuron. 2018;97(6):1268–83 e1266.
Wu J, Zheng C, Wang X, Yun S, Zhao Y, Liu L, Lu Y, Ye Y, Zhu X, Zhang C, et al. MicroRNA-30 family members regulate calcium/calcineurin signaling in podocytes. J Clin Invest. 2015;125(11):4091–106.
Xia HJ, Yang G. Inositol 1,4,5-trisphosphate 3-kinases: functions and regulations. Cell Res. 2005;15(2):83–91.
Oh SW, Pope RK, Smith KP, Crowley JL, Nebl T, Lawrence JB, Luna EJ. Archvillin, a muscle-specific isoform of supervillin, is an early expressed component of the costameric membrane skeleton. J Cell Sci. 2003;116(Pt 11):2261–75.
Howe JG, Hershey JW. Translational initiation factor and ribosome association with the cytoskeletal framework fraction from HeLa cells. Cell. 1984;37(1):85–93.
Collins FS, Brooks LD, Chakravarti A. A DNA polymorphism discovery resource for research on human genetic variation. Genome Res. 1998;8(12):1229–31.
Aguilar I, Legarra A, Cardoso F, Masuda Y, Lourenco D, Misztal I. Frequentist p-values for large-scale-single step genome-wide association, with an application to birth weight in American Angus cattle. Genet Sel Evol. 2019;51(1):28.
Lorenz AJ, Hamblin MT, Jannink JL. Performance of single nucleotide polymorphisms versus haplotypes for genome-wide association analysis in barley. PLoS One. 2010;5(11):e14079.
Rasmussen RS. Quality of farmed salmonids with emphasis on proximate composition, yield and sensory characteristics. Aquac Res. 2001;32(10):767–86.
Cole JB, VanRaden PM. Symposium review: possibilities in an age of genomics: the future of selection indices. J Dairy Sci. 2018;101(4):3686–701.
Karaman E, Lund MS, Su G. Correction: Multi-trait single-step genomic prediction accounting for heterogeneous (co)variances over the genome. Heredity (Edinb). 2020;124(4):618.
Al-Tobasei R, Ali A, Leeds TD, Liu S, Palti Y, Kenney B, Salem M. Identification of SNPs associated with muscle yield and quality traits using allelic-imbalance analyses of pooled RNA-Seq samples in rainbow trout. BMC Genomics. 2017;18(1):582.
Manor ML, Cleveland BM, Kenney PB, Yao J, Leeds T. Differences in growth, fillet quality, and fatty acid metabolism-related gene expression between juvenile male and female rainbow trout. Fish Physiol Biochem. 2015;41(2):533–47.
VanRaden PM. Efficient methods to compute genomic predictions. J Dairy Sci. 2008;91(11):4414–23.
Fox GA, Negrete-Yankelevich S, Sosa VJ. Ecological statistics: contemporary theory and application. UK: Oxford University Press; 2015.
Legarra A, Aguilar I, Misztal I. A relationship matrix including full pedigree and genomic information. J Dairy Sci. 2009;92(9):4656–63.
Misztal I, Tsuruta S, Lourenco D, Aguilar I. style="mso-bidi-font-style: Li, normal"> a, Vitezica Z: manual for BLUPF90 family of programs. Athens, USA: Univ. Georg.; 2014.
BLUPF90 and related programs (BGF90) [WWW Document], in Proceeding of 7th World Congress on Genetics Applied to Livestock Production (Montpellier) [http://nce.ads.uga.edu/wiki/lib/exe/fetch.php?media=28-07.pdf]. Accessed 24 July 2020.
Wang H, Misztal I, Aguilar I, Legarra A, Fernando RL, Vitezica Z, Okimoto R, Wing T, Hawken R, Muir WM. Genome-wide association mapping including phenotypes from relatives without genotypes in a single-step (ssGWAS) for 6-week body weight in broiler chickens. Front Genet. 2014;5:134.
Turner SD. qqman: an R package for visualizing GWAS results using Q-Q and manhattan plots. bioRxiv. 2014.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75.
Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–2.
Acknowledgments
The authors acknowledge J. Everson, M. Hostuttler, K. Jenkins, J. Kretzer, J. McGowan, K. Melody, T. Moreland, and D. Payne for technical assistance.
Funding
This study was supported by a competitive grant No. 2014–67015-21602 from the United States Department of Agriculture, National Institute of Food and Agriculture (MS), and by the USDA, Agricultural Research Service CRIS Project 1930–31000-010 “Utilizing Genetics and Physiology for Enhancing Cool and Cold Water Aquaculture Production”. The content is solely the responsibility of the authors and does not necessarily represent the official views of any of the funding agents.
Author information
Authors and Affiliations
Contributions
MS, TL, and BK conceived and designed the experiments. RA-T, MS, TL, and BK performed the experiments. RA-T, AA, DL, BK, and MS analyzed the data. AA and MS wrote the manuscript. The authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Institutional Animal Care and Use Committee of the United States Department of Agriculture, National Center for Cool and Cold Water Aquaculture (Leetown, WV) specifically reviewed and approved all husbandry practices used in this study (IACUC protocol #056).
Consent for publication
Not applicable.
Competing interests
Mohamed Salem is a member of the editorial board of BMC Genomics. Otherwise, the authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Table S1
. Genetic variance for fat content explained by all SNP markers in genomic sliding windows. Table S2. SNP markers in genomic sliding windows explaining at least 2% of the genetic variance for fat content. Table S3. Genetic variance for moisture content explained by all SNP markers in genomic sliding windows. Table S4. SNP markers in genomic sliding windows explaining at least 2% of the genetic variance for moisture content. Table S5. SNP markers significantly associated with variability in fat content (highlighted in yellow) using family-based association analysis. Table S6. SNP markers significantly associated with variability in moisture content (highlighted in yellow) using family-based association analysis.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Ali, A., Al-Tobasei, R., Lourenco, D. et al. Genome-wide scan for common variants associated with intramuscular fat and moisture content in rainbow trout. BMC Genomics 21, 529 (2020). https://doi.org/10.1186/s12864-020-06932-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-020-06932-0