Abstract
Background
Cotton (Gossypium spp.) is the most important natural fiber crop worldwide, and cottonseed oil is its most important byproduct. Phospholipid: diacylglycerol acyltransferase (PDAT) is important in TAG biosynthesis, as it catalyzes the transfer of a fatty acyl moiety from the sn-2 position of a phospholipid to the sn-3 position of sn-1, 2-diacylglyerol to form triacylglycerol (TAG) and a lysophospholipid. However, little is known about the genes encoding PDATs involved in cottonseed oil biosynthesis.
Results
A comprehensive genome-wide analysis of G. hirsutum, G. barbadense, G. arboreum, and G. raimondii herein identified 12, 11, 6 and 6 PDATs, respectively. These genes were divided into 3 subfamilies, and a PDAT-like subfamily was identified in comparison with dicotyledonous Arabidopsis. All GhPDATs contained one or two LCAT domains at the C-terminus, while most GhPDATs contained a preserved single transmembrane region at the N-terminus. A chromosomal distribution analysis showed that the 12 GhPDAT genes in G. hirsutum were distributed in 10 chromosomes. However, none of the GhPDATs was co-localized with quantitative trait loci (QTL) for cottonseed oil content, suggesting that their sequence variations are not genetically associated with the natural variation in cottonseed oil content. Most GhPDATs were expressed during the cottonseed oil accumulation stage. Ectopic expression of GhPDAT1d increased Arabidopsis seed oil content.
Conclusions
Our comprehensive genome-wide analysis of the cotton PDAT gene family provides a foundation for further studies into the use of PDAT genes to increase cottonseed oil content through biotechnology.
Similar content being viewed by others
Background
Cotton, especially upland cotton, is the world’s most important fiber crop, and oil is extracted from its oil-rich seeds. Indeed, cotton ranks sixth among the world’s oil crops. Cottonseed oil makes up approximately 16% of the seed weight [1], and is the most valuable product derived from cotton seed. Cottonseed oil is typically composed of approximately 26% saturated palmitic acid (C16:0), 15% monounsaturated oleic acid (C18:1), and 58% polyunsaturated linoleic acid (C18:2) [2]. From 1999 to 2009, the world-wide consumption of vegetable oils increased by > 50% [3]. Therefore, research into the molecular mechanisms of oil biosynthesis and the development of new high-seed oil content cotton varieties using classical breeding techniques and biotechnological approaches is becoming increasingly important.
Triacylglycerols (TAGs) are major components of vegetable oils. The 3 pathways of DAG /TAG production with different FA compositions have previously been reviewed [4]. These pathways are de novo DAG/TAG synthesis (Kennedy pathway), acyl editing to provide PC-modified FA for de novo DAG/TAG synthesis, and PC-derived DAG/TAG synthesis. Phospholipid: diacylglycerol acyltransferase (PDAT) in the second pathway catalyzes the transfer of a fatty acyl moiety from the sn-2 position of a phospholipid to the sn-3 position of sn-1, 2-diacylglyerol, thus forming TAG and a lysophospholipid. PDAT enzyme activity was first identified in the use of phospholipids as acyl donors and DAG as an acceptor for TAG biosynthesis in yeast and plants [5].
Arabidopsis contains two PDAT genes, AtPDAT1 (At5g13640) and AtPDAT2 (At3g44830) [6]. No significant differences were found in total acyl composition or TAG content between 17-day-old AtPDAT-overexpressing and wild-type (WT) seedlings [6]. Additionally, the fatty acid content and composition of seeds also showed no significant difference in the pdat mutant versus WT [7]. However, in 5-week-old developing Arabidopsis leaves, the overexpression or knockout of AtPDAT1 in led to significant changes in fatty acid and TAG synthesis [8]. AtPDAT2 is highly expressed in seeds, but plays no role in TAG biosynthesis [6, 9]. In castor bean, 3 PDAT genes have been identified [10]. The endoplasmic reticulum-located PDAT1–2 enhances hydroxy fatty acid accumulation in transgenic castor bean plants [11]. In flax (Linum usitatissimum), 6 PDATs have been identified (LuPDAT1, LuPDAT2, LuPDAT3, LuPDAT4, LuPDAT5, and LuPDAT6) [12]. LuPDAT1/LuPDAT5 and LuPDAT2/LusPDAT4, but not LusPDAT3 or LusPDAT6, have the unique ability to preferentially channel a-linolenic acid into TAG. Recently, the PDAT gene Lro1 was shown to be responsible for hepatitis C virus core-induced lipid droplet formation in a yeast model system [13]. PDAT genes were also found in the unicellular green alga Chlamydomonas reinhardtii [14] and the bacterium Streptomyces coelicolor [15]. However, no mammalian counterpart has yet been found.
Previously, a genome-wide analysis of eudicots found 6 PDATs in Gossypium raimondii (two each in clades V, VI, and VII) [16]. To further understand the complexity of PDATs and TAG biosynthetic mechanisms in cotton, we performed a comprehensive genome-wide analysis of the PDAT gene family in cotton in the present study.
Results
Genome-wide identification and phylogenetic tree analysis of PDAT genes
Allotetraploid cotton G. hirsutum and G. barbadense contain two ancestral genomes: the At and Dt subgenomes. To identify all PDAT proteins in G. hirsutum (AD1), G. barbadense (AD2), and its two diploid ancestors G. arboreum (AA genome) and G. raimondii (DD genome), we used Arabidopsis PDAT protein sequences (AtPDAT1/At5g13640 and AtPDAT2/At3g44830) to query the four reference genomes to screen out candidate PDAT-like proteins in cotton. Combined with the previously identified PDATs from G. raimondii [16], 12 deduced PDATs were identified in G. hirsutum [17], 11 in G. barbadense [18], 6 in G. arboreum [19] and 6 in G. raimondii [20].
To interpret the relationship between AtPDAT1, AtPDAT2, and cotton PDAT proteins, we constructed a phylogenetic tree (Fig. 1). This classified PDAT genes into 3 subfamilies; PDAT1, PDAT1-like, and PDAT2, corresponding to clades VI, V, and VII, respectively [16]. The sequence similarity between GhPDAT1-like and GhPDAT1 was higher than that of GhPDAT2 (Fig. 1). Based on the phylogenetic tree and sequence similarity analysis, we also analyzed orthologous PDAT gene pairs in G. hirsutum, G. barbadense, and their corresponding diploid ancestors (Table 1). Only one gene, GbPDAT1b-like, was not found or lost in G. barbadense. The PDAT gene name, gene identifier, gene pairs, and predicted properties of PDAT proteins are listed in Table 1.
Phylogenetic tree and gene structure of the PDAT gene family in Arabidopsis and Gossypium. The phylogenetic tree of all PDAT proteins in Arabidopsis and four Gossypium species (Additional file 3) was constructed using Neighbor-Joining method. The exon/intron structure of PDAT genes in Arabidopsis and four Gossypium species. Black boxes show exons and lines show introns
Gene structure analysis and chromosomal distribution of PDAT genes in cotton
Generic Feature Format files of the four Gossypium species were used to analyze the exon-intron structure of putative PDAT genes. Figure 1 shows the exon-intron structure of each gene. Although the locations of introns differed, most PDAT genes contained 5 introns and 6 exons. For example, in the PDAT1 subfamily, AtPDAT1, GbPDAT1a (Gbscaffold24182.2.0), and the counterparts from G. hirsutum, G. arboreum and G. raimondii included 5 introns and 6 exons. However, the other 3 PDAT1 genes GbPDAT1b (Gbscaffold14656.14.0), GbPDAT1c (Gbscaffold1227.2.0), and GbPDAT1d (Gbscaffold10824.9.0), contained 9 introns and 10 exons, 6 introns and 7 exons and 6 introns and 7 exons, respectively. Interestingly, only 3 of 11 PDAT genes in G. barbadense had the same gene structure.
Based on the sequenced genome sequence, cotton PDAT genes were physically mapped to chromosomes (Fig. 2; Table 1). In G. hirsutum and G. barbadense, PDAT genes were uniformly distributed on the At and Dt chromosome, excluding one lost in G. barbadense. In G. hirsutum, 12 PDAT genes were located on 5 Dt chromosomes (D6, D7, D8, D9 and D13) and 5 At chromosomes (A6, A7, A8, A9 and A13). Two PDAT genes were located on both chromosome A6 and D6. Chromosomal localization data are listed in Fig. 2 and Table 1.
Localization of PDAT genes in the four cotton species. Thirty-five PDAT genes were mapped on different chromosomes in Gossypium raimondii (a), Gossypium arboreum (b), Gossypium hirsutum (c), and Gossypium barbadense (d). Only the chromosomes where PDAT genes were mapped are shown. The scale represents the megabases (Mb)
Protein domain analysis of PDATs in Gossypium hirsutum
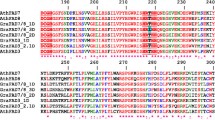
To improve the comparison of protein domains among GhPDATs, the putative protein domains of 12 GhPDATs were predicted using the SMART database (http://smart.embl-heidelberg.de/). As shown in Fig. 3, a single transmembrane region in the N-terminus has been preserved in most GhPDATs, while all GhPDATs contain one or two LCAT domains at their C-termini.
Protein domain prediction for the GhPDATs. The potential transmembrane regions and functional motifs of GhPDAT proteins were identified using SMART database (http://smart.embl-heidelberg.de/)
Adaptive evolution analysis of the PDAT gene family
To explore which type of Darwinian selection determined the process of PDAT gene divergence after duplication, the Ka/Ks substitution ratio was used to assess the coding sequences of 12 pairs of PDAT gene family orthologs between G. hirsutum/G. barbadense and G. arboreum/ G. raimondii (Table 1). A Ka/Ks ratio > 1 represents positive selection, a ratio of 1 represents neutral evolution and a ratio < 1 represents purifying selection [21]. The Ka/Ks ratios of PDAT genes ranged from 0.575 to ∞ (Table 2), indicating that the PDAT gene family had undergone purifying selection and positive selection in cotton. As shown in Table 2, the majority of PDAT genes had undergone positive selection, especially GbPDAT1b, GhPDAT1d, GbPDAT1d and GhPDAT2d. Only four PDAT genes GhPDAT1a, GbPDAT1a, GhPDAT1c and GbPDAT2b had undergone purifying selection.
Phylogenetic tree analysis showed that each AtPDAT gene corresponded to four PDAT genes in tetraploid cotton and two genes in diploid cotton. Therefore, the 12 GhPDATs were divided into 6 pair of duplicates, and the Ka/Ks ratio for each pair was calculated (Table 3). All Ka/Ks ratios were < 1, suggesting that the PDAT genes from G. hirsutum have mainly experienced purifying selection pressure.
Expression profiles of PDAT genes in Gossypium hirsutum
To reveal the gene expression pattern for the GhPDAT genes identified, we analyzed the transcript profiles of PDAT genes in 22 cotton tissues (Fig. 4) based on published TM-1 data [17]. GhPDAT1a and GhPDAT1b maintained a low expression level in 22 cotton tissues. GhPDAT1c and GhPDAT1d were highly expressed in the stem, leaf, and torus, and were also expressed in the ovule and fiber. GhPDAT1-like genes were expressed in 22 cotton tissues. AtPDAT2 was highly expressed in seeds, but plays no role in TAG biosynthesis [6, 9]. GhPDAT2 was also highly expressed in 20 days post anthesis (DPA)-35 DPA ovules and 25 DPA fibers, and only marginally in other organs. This suggested that GhPDAT2 plays no role in TAG biosynthesis. Cottonseed oil mainly accumulates in the ovules after 15 DPA-20 DPA, at which stage, most of the GhPDATs were expressed. Therefore, GhPDATs may play a role in the biosynthesis of TAGs in developing cotton seeds.
Expression analysis of GhPDAT genes in Gossypium hirsutum acc TM-1 across 22 tissues. The RNA-seq expression profiles of G. hirsutum acc. TM-1 [17] were used to identify the expression levels of GhPDAT genes. FPKM represents fragments per kilobase of exon model per million mapped reads
To reveal the gene expression pattern for the GhPDAT genes identified, we analyzed their transcript profiles in our unpublished RNA-seq datasets. This was based on transcriptomic information for two upland BILs, i.e., 3012 vs. 3008 (with Gossypium barbadense germplasm introgression), with differing seed kernel oil contents of 25.88 and 33.52% (Additional file 1: Figure S1). There was no significant difference in the expression levels of GhPDAT genes between the two BIL genotypes.
Co-localization of PDAT genes with quantitative trait loci (QTLs) for cottonseed oil
To determine if any GhPDATs were genetically associated with the cottonseed oil content, we performed co-localization analysis of GhPDATs with QTLs for seed oil content. QTLs were downloaded from the CottonQTL database [22]. However, no PDAT gene was localized in the cottonseed oil QTL interval (data not shown).
Ectopic expression of GhPDAT1d increased the oil content of Arabidopsis seeds
In PDAT1 clade, the expression level of GhPDAT1c and GhPDAT1d (gene pairs from the corresponding At and Dt subgenome) was higher in 15–20 DPA ovules than that of GhPDAT1a and GhPDAT1b (Figs. 4 and 5a). GhPDAT1d was thus selected for further functional analysis. Transgenic Arabidopsis plants overexpressing GhPDAT1d were generated and used to characterize its biological functions in oil content. Relative expression levels of GhPDAT1d analyzed by qRT-PCR in transgenic Arabidopsis and WT plants showed that GhPDAT1d was highly expressed in the transgenic plants (Fig. 5b). No visible difference between transgenic Arabidopsis and WT plants was observed at different developmental stages (data not shown).
Improved oil content of GhPDAT1d transgenic plants. a Tissue-specific expression profile of GhPDAT1d in different tissues of G. hirsutum accession TM-1. The ΔCt value of GhPDAT1d in root was set as the control. The data presented are the means ± SD of three replicates. b Relative expression level of GhPDAT1d in four transgenic Arabidopsis lines (L1, L2, L3, and L4). The ΔCt value of GhPDAT1d in transgenic line L1 was set as the control. The data presented are the means ± SD of three replicates. c Seed oil content of GhPDAT1d transgenic lines (L1, L2, L3, and L4) and WT. The data presented are the means ± SD of three replicates; *, P < 0.05 (Student’s t-test)
In order to determine whether GhPDAT1d could increase the oil content, the oil contents of transgenic and WT plants were compared using an NMI20-Analyst nuclear magnetic resonance spectrometer (Niumag, Shanghai, China). Significantly increased oil content, 6.55 to 17.61% higher, was observed in transgenic line L2-L4 (Fig. 5c). There is no significant change in fatty acid compositions of WT and GhPDAT1d transgenic Arabidopsis seeds (Table 4).
Discussion
Despite the fact that many previous studies have revealed a crucial role for PDAT encoded products in TAG biosynthesis, our knowledge of PDATs in cotton remains limited. Therefore, this study aimed to present an overall picture of Gossypium PDATs, including their sequence variation, adaptive evolutionary analysis, protein domains, expression profiles and co-localization with QTLs.
The PDAT gene family in Gossypium
PDAT genes exist in all plants, including algae, lowland plants (mosses and lycophytes) and highland plants (monocots and eudicots) [16]. This study revealed the details of 12 deduced PDATs from G. hirsutum, 11 deduced PDATs from G. barbadense, 6 deduced PDATs in G. arboretum and 6 deduced PDATs in G. raimondii. Evolutionary analysis previously showed that the PDAT gene family can be clearly divided into 7 major clades [16]. In the present study, Gossypium PDAT amino acid sequences were clustered into 3 clades (subfamilies), and the additional clade, PDAT1-like, was found in cotton. Clades I-IV were not found in cotton. This compares with Arabidopsis, in which only two PDAT genes (AtPDAT1 and AtPDAT2) have been identified [6].
We observed that each AtPDAT gene corresponded to four PDAT genes in tetraploid cotton and two genes in diploid cotton. This suggested that PDAT gene duplication events occurred in diploid cotton before the emergence of tetraploid cotton, which is consistent with a previously reported eudicot-wide PDAT gene expansion [16]. Additionally, a single transmembrane region in the N-terminus has been preserved in most GhPDATs, and one or two LCAT domains were located at the C-terminus of all GhPDATs.
PDATs in relation to seed oil content
Cottonseed oil accumulates in ovules after 15–20 DPA. At this stage, most of the GhPDATs were expressed (Fig. 3), indicating that they play a role in the biosynthesis of TAGs in developing cotton seeds. Additionally, we found GhPDATs were expressed in developing fibers (Fig. 3), suggesting they are also involved in this stage of development. However, no PDAT gene was localized in the cottonseed oil QTL interval (data not shown).
In 5-week-old developing Arabidopsis leaves, the overexpression or knockout of AtPDAT1 led to significant changes in fatty acid and TAG synthesis [8]. Cottonseed oil was widely believed to accumulate in ovules after 15 DPA. At this stage, most GhPDATs were found to be expressed (Fig. 4). In this study, we proved that ectopic expression of GhPDAT1d could increase the oil content of Arabidopsis seeds. Any fatty acid in the seed oil was found to be significantly changed as previously reported Arabidopsis pdat-ko mutant [7]. Together, these results implied that PDATs are conserved in upland cotton cultivars.
Conclusion
In conclusion, we performed a comprehensive genome-wide analysis of the PDAT gene family in cotton. A total of 35 PDAT genes were identified in four sequenced Gossypium species and grouped into 3 distinct clades. Ectopic expression of GhPDAT1d increased Arabidopsis seed oil content. Our detailed analysis of sequence variation, adaptive evolutionary analysis, protein domains, expression profiles, and QTL co-localization provides an important lead for further studies of PDAT genes in cotton.
Methods
Sequence retrieval, multiple sequence alignment, and phylogenetic analysis
The cotton genome sequences of G. arboreum (A2, BGI_V1.0) [19], G. raimondii (D5, BGI_V1.0) [20], G. hirsutum (AD1, NBI_V1.1) [17] and G. barbadense (AD2, SGI_V1.0) [18] were downloaded from the CottonGen database (https://www.cottongen.org). AtPDAT1 (At5g13640) and AtPDAT2 (At3g44830) were acquired from TAIR 10 (http://www.arabidopsis.org). To identify PDAT genes, AthPDAT1 and AthPDAT2 protein sequences were used as queries against cotton genome sequences. Multiple sequence alignments of all identified PDATs in this study were performed using Clustal X2 (http://www.clustal.org/). A phylogenetic tree was constructed using the neighbor-joining algorithm with default parameters and 1000 bootstrap replicates in MEGA 6 (http://www.megasoftware.net/). The sequence length, molecular weight, and isoelectric point of PDAT proteins were calculated using ExPasy (http://web.expasy.org).
In-silico mapping and genetic structure analysis of PDAT genes
Mapping of PDAT genes was performed using MapChart (https://www.wur.nl/en/show/Mapchart.htm) [23]. QTLs in this paper were downloaded from CottonQTLdb (http://www.cottonqtldb.org) [22]. The structures of PDAT genes were generated using the GSDS (Gene Structure Display Server) algorithm (http://gsds.cbi.pku.edu.cn/).
Detection of protein domains
Potential transmembrane regions and functional motifs of GhPDAT proteins were identified using the SMART database (http://smart.embl-heidelberg.de/).
Ka and Ks calculations
PDAT gene pairs were used to calculate Ka and Ks using the DnaSP software of phylogenetic analysis by the maximum likelihood method.
Analysis of PDAT genes in RNA-seq data
RNA-seq data of 22 cotton tissues were previously published (accession codes, SRA: PRJNA248163) [17]. Unpublished RNA-seq datasets were generated in our own laboratory using transcriptomic information for two upland BILs, i.e., 3012 vs. 3008 (with Gossypium barbadense germplasm introgression), with differing seed kernel oil contents of 25.88 and 33.52%. The expression of PDAT genes was analyzed based on these data.
Transgenic plant generation and expression analysis
Transgenic plant generation and expression analysis were performed as previously reported [24]. Briefly, complete coding sequence of GhPDAT1d (Additional file 4) was amplified with gene specific primers from G. hirsutum acc. TM-1. The resulting PCR product was cloned into a digested pBI121 vector with BamH I and Sac I using ClonExpress R II One Step Cloning Kit (Vazyme, Nanjing, China). Agrobacterium tumefaciens strain GV3101 containing the binary construct was used to transform Arabidopsis plants. We performed quantitative real-time PCR (qRT-PCR) to determine the expression pattern of GhPDAT1d, with t2-ΔΔCt method used to quantify the expression level of GhPDAT1d relative to the 18S rRNA endogenous control. Primers are listed in Additional file 2: Table S1.
Oil content analysis
Total oil content was determined with about 0.3 g seeds per sample using an NMI20-Analyst nuclear magnetic resonance spectrometer (Niumag, Shanghai, China) as previously reported [24].
Fatty acid composition analysis
A gas chromatography/mass spectrometry GC/MS analysis was performed to determine the fatty acid compsitions using a gas chromatograph (7890A, Agilent Technologies, USA) equipped with a flame ionization detector (FID) and an HP-FFAP capillary column (30 m × 250 μm × 0.25 μm). WT and GhPDAT1d transgenic Arabidopsis seeds (about 100 seeds) were performed to determine the fatty acid components.
Abbreviations
- DAG:
-
Diacylglycerol
- DPA:
-
Days post anthesis
- PDAT:
-
Phospholipid: diacylglycerol acyltransferase
- QTL:
-
Quantitative trait loci
- TAG:
-
Triacylglycerol
- WT:
-
Wild-type
References
Liu Q, Surinder S, Chapman K, Green A. Bridging traditional and molecular genetics in modifying cottonseed oil. London: Springer Science + Business Media; 2009. p. 353–82.
Liu Q, Singh SP, Green AG. High-stearic and high-oleic cottonseed oils produced by hairpin RNA-mediated post-transcriptional gene silencing. Plant Physiol. 2002;129(4):1732–43.
Lu C, Napier JA, Clemente TE, Cahoon EB. New frontiers in oilseed biotechnology: meeting the global demand for vegetable oils for food, feed, biofuel, and industrial applications. Curr Opin Biotechnol. 2011;22(2):252–9.
Bates PD, Browse J. The significance of different diacylgycerol synthesis pathways on plant oil composition and bioengineering. Front Plant Sci. 2012;3:147.
Dahlqvist A, Stahl U, Lenman M, Banas A, Lee M, Sandager L, Ronne H, Stymne S. Phospholipid:diacylglycerol acyltransferase: an enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc Natl Acad Sci U S A. 2000;97(12):6487–92.
Stahl U, Carlsson AS, Lenman M, Dahlqvist A, Huang BQ, Banas W, Banas A, Stymne S. Cloning and functional characterization of a phospholipid : diacylglycerol acyltransferase from Arabidopsis. Plant Physiol. 2004;135(3):1324–35.
Mhaske V, Beldjilali K, Ohlrogge J, Pollard M. Isolation and characterization of an Arabidopsis thaliana knockout line for phospholipid: diacylglycerol transacylase gene (At5g13640). Plant Physiol Biochem. 2005;43(4):413–7.
Fan J, Yan C, Zhang X, Xu C. Dual role for phospholipid:diacylglycerol acyltransferase: enhancing fatty acid synthesis and diverting fatty acids from membrane lipids to triacylglycerol in Arabidopsis leaves. Plant Cell. 2013;25(9):3506–18.
Zhang M, Fan JL, Taylor DC, Ohlrogge JB. DGAT1 and PDAT1 acyltransferases have overlapping functions in Arabidopsis triacylglycerol biosynthesis and are essential for Normal pollen and seed development. Plant Cell. 2009;21(12):3885–901.
van Erp H, Bates PD, Burgal J, Shockey J, Browse J. Castor phospholipid:diacylglycerol acyltransferase facilitates efficient metabolism of hydroxy fatty acids in transgenic Arabidopsis. Plant Physiol. 2011;155(2):683–93.
Kim HU, Lee KR, Go YS, Jung JH, Suh MC, Kim JB. Endoplasmic reticulum-located PDAT1-2 from castor bean enhances hydroxy fatty acid accumulation in transgenic plants. Plant Cell Physiol. 2011;52(6):983–93.
Pan X, Siloto RMP, Wickramarathna AD, Mietkiewska E, Weselake RJ. Identification of a pair of phospholipid: diacylglycerol acyltransferases from developing flax (Linum usitatissimum L.) seed catalyzing the selective production of Trilinolenin. J Biol Chem. 2013;288(33):24173–88.
Iwasa S, Sato N, Wang CW, Cheng YH, Irokawa H, Hwang GW, Naganuma A, Kuge S. The phospholipid:diacylglycerol acyltransferase Lro1 is responsible for hepatitis C virus core-induced lipid droplet formation in a yeast model system. PLoS One. 2016;11(7):e0159324.
Yoon K, Han D, Li Y, Sommerfeld M, Hu Q. Phospholipid: diacylglycerol acyltransferase is a multifunctional enzyme involved in membrane lipid turnover and degradation while synthesizing triacylglycerol in the unicellular green microalga Chlamydomonas reinhardtii. Plant Cell. 2012;24(9):3708–24.
Arabolaza A, Rodriguez E, Altabe S, Alvarez H, Gramajo H. Multiple pathways for triacylglycerol biosynthesis in Streptomyces coelicolor. Appl Environ Microbiol. 2008;74(9):2573–82.
Pan X, Peng FY, Weselake RJ. Genome-wide analysis of PHOSPHOLIPID:DIACYLGLYCEROL ACYLTRANSFERASE (PDAT) genes in plants reveals the eudicot-wide PDAT gene expansion and altered selective pressures acting on the core eudicot PDAT paralogs. Plant Physiol. 2015;167(3):887–904.
Zhang TZ, Hu Y, Jiang WK, Fang L, Guan XY, Chen JD, Zhang JB, Saski CA, Scheffler BE, Stelly DM, et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat Biotechnol. 2015;33(5):531–U252.
Yuan D, Tang Z, Wang M, Gao W, Tu L, Jin X, Chen L, He Y, Zhang L, Zhu L, et al. The genome sequence of Sea-Island cotton (Gossypium barbadense) provides insights into the allopolyploidization and development of superior spinnable fibres. Sci Rep. 2015;5:17662.
Li FG, Fan GY, Wang KB, Sun FM, Yuan YL, Song GL, Li Q, Ma ZY, Lu CR, Zou CS, et al. Genome sequence of the cultivated cotton Gossypium arboreum. Nat Genet. 2014;46(6):567–72.
Wang KB, Wang ZW, Li FG, Ye WW, Wang JY, Song GL, Yue Z, Cong L, Shang HH, Zhu SL, et al. The draft genome of a diploid cotton Gossypium raimondii. Nat Genet. 2012;44(10):1098.
Akhunov ED, Sehgal S, Liang HQ, Wang SC, Akhunova AR, Kaur G, Li WL, Forrest KL, See D, Simkova H, et al. Comparative analysis of syntenic genes in grass genomes reveals accelerated rates of gene structure and coding sequence evolution in polyploid wheat. Plant Physiol. 2013;161(1):252–65.
Said JI, Knapka JA, Song MZ, Zhang JF. Cotton QTLdb: a cotton QTL database for QTL analysis, visualization, and comparison between Gossypium hirsutum and G-hirsutum x G-barbadense populations. Mol Gen Genomics. 2015;290(4):1615–25.
Voorrips RE. MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered. 2002;93(1):77–8.
Zang XS, Pei WF, Wu M, Geng YH, Wang NH, Liu GY, Ma JJ, Li D, Cui YP, Li XL, et al. Genome-scale analysis of the WRI-like family in Gossypium and functional characterization of GhWRI1a controlling triacylglycerol content. Front Plant Sci. 2018;9:1516.
Acknowledgements
We thank Sarah Williams, PhD, from Liwen Bianji, Edanz Group China (www.liwenbianji.cn), for editing the English text of a draft of this manuscript.
Funding
The research was mainly supported by grants from the National Natural Science Foundation of China (grant No. 31801415 and 31621005). The National Natural Science Foundation of China (grant No. 31801415 and 31621005) supported us to design of the study, analysis, interpretation of data and measure the oil content and fatty acid compositions. The National Key Research and Development Program of China (grant No. 2016YFD0101400) and the National Research and Development Project of Transgenic Crops of China (grant No. 2016ZX08005005) supported us to edit the English text of a draft of this manuscript and pay publication fees.
Availability of data and materials
AtPDAT1 (accession number At5g13640) and AtPDAT2 (accession number At3g44830) can be found in TAIR 10 (http://www.arabidopsis.org). All of the cotton PDAT genes with the accession number in Table 1 can be found in cottonGen database (https://www.cottongen.org). The data and materials supporting the results are included in the manuscript and additional files. The other data and materials are available from the corresponding author on reasonable request.
Author information
Authors and Affiliations
Contributions
JY, JZ and XZ designed and directed the experiments. XZ conceived the study, performed most of the experiments and wrote the manuscript. XG and NW acquired and analyzed the data. LM performed a gas chromatography/mass spectrometry GC/MS analysis to determine the fatty acid compsitions. WP and MW performed transgenic Arabidopsis plant generation and measured total oil content. JY and JZ revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
Although JZ is one of the associate editors of BMC Genomics, all the authors including JZ declare that they have no competing interests and review process is transparent.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Figure S1. Expression analysis of GhPDAT genes in our unpublished RNA-seq datasets: with transcriptomic information for two Upland BILs, i.e., 3012 vs. 3008 (with Gossypium barbadense germplasm introgression), with differing seed kernel oil content 25.88 and 33.52%. FPKM represents fragments per kilobase of exon model per million mapped reads. (JPG 286 kb)
Additional file 2:
Table S1. Primers used in this paper. (DOCX 18 kb)
Additional file 3:
Phylogenetic data of Fig. 1. (DOCX 21 kb)
Additional file 4:
Coding sequence of GhPDAT1d. (DOCX 16 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Zang, X., Geng, X., Ma, L. et al. A genome-wide analysis of the phospholipid: diacylglycerol acyltransferase gene family in Gossypium. BMC Genomics 20, 402 (2019). https://doi.org/10.1186/s12864-019-5728-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-019-5728-8