Abstract
Background
Acute hepatopancreatic necrosis disease (AHPND) has emerged as a major debilitating disease that causes massive shrimp death resulting in substantial economic losses in shrimp aquaculture. Given that several diseases and infections have been associated with microRNAs (miRNAs), we conducted a comparative transcriptomic analysis using the AHPND (VA) and non-AHPND (VN) strains of Vibrio parahemolyticus to identify miRNAs potentially involved in AHPND pathogenesis in Litopenaeus vannamei.
Results
A total of 83 miRNAs (47 upregulated and 36 downregulated) were significantly differentially expressed between the VA and VN challenged groups, while 222 target genes of these miRNAs were predicted. Functional enrichment analysis revealed that the miRNAs target genes were involved in multiple biological processes including metabolic pathways, amoebiasis, Vibrio cholerae infection etc. Finally, interaction network and qPCR (Real-time Quantitative PCR) analysis of 12 potential key AHPND-related miRNAs and their predicted target genes, revealed their possible involvement in modulating several immune-related processes in the pathogenesis of AHPND.
Conclusions
We have shown using comparative transcriptomic analysis, miRNAs and their target genes that are responsive to AHPND V. parahemolyticus infection in shrimp, therefore suggesting their possible role in defense response to AHPND V. parahemolyticus infection.
Similar content being viewed by others
Background
MicroRNAs (miRNAs) are endogenous non-coding small RNA (sRNA) [1], which act by negatively modulating gene expression at the post-transcription level [2], via repression of messenger RNAs (mRNA) translation, destabilization and degradation of transcript by binding to the 3′ UTR (Untranslated Region) of the target mRNAs [3]. These molecules act as important regulators in diverse life processes, including cellular proliferation, apoptosis and immunity [4].
Litopenaeus vannamei is one of the largest aquaculture shrimp species in the world with huge production and economic benefits [5]. The shrimp aquaculture industry has however been saddled with diseases and infections, which cause significant losses in shrimp production. A number of studies in mammals have suggested that host miRNAs are key regulators of virus-host interactions during an infection [6,7,8], while in different crustaceans miRNAs have been identified to be involved in the response to some diseases and infections. For instance, Li et al. identified some microRNAs involved in the response of Chinese shrimp Fenneropenaeus chinensis to white spot syndrome virus (WSSV) infection [9]. Similarly, Zhu et al. screened 55 host miRNAs which responded to Vibrio alginolyticus infection in shrimp Marsupenaeus japonicas [10], while Ou et al. identified 33 dysregulated miRNAs in crayfish Procambarus clarkii upon challenge with Spirolasma eriocheiris [11]. In recent years, an emerging shrimp disease called acute hepatopancreatic necrosis disease (AHPND), brought about significant losses in aquaculture shrimp production, resulting in great economic losses. The pathogenesis of AHPND was first though to be related to the environment, wherein a multiple and complex interplay of factors including bacteria [12, 13], virus [14, 15] as well as other environmental factors coexist [16, 17]. Of these factors, the V. parahemolyticus with photorhabdus insect-related (Pir) toxins was finally implicated as the most probable major pathogenic factor [18],as this bacteria is capable of producing the characteristic symptoms of AHPND [19]. However, it is still largely unknown how shrimp miRNAs respond to AHPND V. parahaemolyticus infection.
Our unpublished shrimp hemocytes comparative transcriptome data (GenBank accession number: PRJNA385392) revealed a large number of differentially expressed genes following challenge of shrimps with AHPND and non-AHPND V. parahaemolyticus. Based on this, we sought to explore in this study the responsive miRNAs that might regulate the expression of these genes in shrimps infected with AHPND and non-AHPND V. parahaemolyticus. This study therefore provides a transcriptome-based analysis of miRNAs and their target genes in L. vannamei challenged with AHPND V. parahaemolyticus, thereby laying the framework for further exploring miRNAs and their roles in regulating host responses to AHPND.
Methods
Animal challenge and hemocytes collection
Healthy adult shrimps (L. vannamei) (weighing about 10~ 12 g) were obtained from Shantou Huaxun Aaquatic Product Corporation (Shantou, Guangdong, China) and acclimatized to laboratory conditions for 3–5 days before challenge experiments. Bacteria (non-AHPND and AHPND V. parahaemolyticus, kind gift from Prof. Lo, National Cheng Kung University, Taiwan, China) at the logarithmic growth phase were cultured in Luria-Bertani media (tryptone: 10 g/L, yeast extract: 5 g/L, NaCl 30 g/L) at 28 °C with shaking, counted and diluted to the appropriate colony forming units (CFU) with 0.65% normal saline. After the period of laboratory acclimatization, shrimps were randomly divided into three experimental groups with at least 50 shrimps per group per container. The VN group denotes shrimps injected with 50 μl of non-AHPND V. parahaemolyticus (1.0 × 105 CFU/ml), VA group represent shrimps injected with 50 μl of AHPND V. parahaemolyticus (1.0 × 105 CFU/ml), while the NS group denotes shrimps injected with 50 μl of normal saline (0.65% NaCl, Xilong Scientific, Shantou, China). Hemolymph from ten randomly selected shrimps per group was drawn at 24 h post infection (hpi) through the pericardial sinus into an equal volume of cold anti-coagulant buffer (NaCl 450 mM; KCl 10 mM; EDTA-Na2 10 mM; HEPES 10 mM. pH 7.0), centrifuged immediately at 800 g for 10 min at 4 °C to collect the hemocytes for RNA extraction. Three biological replicates per treatment group were prepared for the sRNA library construction.
sRNA library and high-throughput sequencing
Total RNA including sRNA was extracted from hemocytes using the mirVana miRNA Isolation Kit (Ambion, Austin, TX, USA) according to the manufacturer’s instructions. The concentration and purity of the total RNA was quantified with a NanoDrop 2000 spectrophotometer (Nano-drop Technologies, Wilmington, DE), while the total RNA integrity was evaluated using an Agilent 2100 Bioanalyzer system (Agilent Technologies, Palo Alto, California.) with a RNA integrity number (RIN) > 8.5. Only high quality RNA samples were used for constructing the cDNA libraries. Before use, all samples were diluted to a final concentration of 200 ng/μL. Sequencing libraries were generated with the TruSeq® smallRNA Sample Preparation kit (Illumina, San Diego, USA). The concentration of the cDNA libraries was determined with the KAPA Library Quantification Kit (KAPA Biosystems) followed by sequencing on an Illumina HiSeq2000 sequencer (Illumina, San Diego, USA) employing single-end sequencing strategy with 1 × 50-bp read length. Sequencing work was commissioned by Beijing Genomics Institute (BGI, Wuhan, China). The raw sequencing data from this study have been deposited at GenBank with accession number PRJNA414944.
Bioinformatic analysis of miRNA data
The raw miRNA data was first cleaned by removing 5′ primer contaminants, no insert tags, oversized insertions, low quality reads and polyA tags. Length distribution analysis of the clean reads was then carried out before miRNA prediction. To remove rRNA-, scRNA-, snoRNA-, snRNA-, and tRNA-associated tags, reads were aligned with GenBank and Rfam reference databases using Blast or Bowtie. The obtained sequences were then mapped against our L. vannamei hemocyte transcriptome (GenBank accession: PRJNA385392) to remove degraded mRNA and reads of unannotated transcripts. Following this, reads were then mapped against the miRNA database (miRBase 21), to obtain matching miRNA sequences conserved in L. vannamei. The obtained miRNAs sequences were used for family classification and annotation with the miRClassify web server [20]. Finally, the sRNAs that did not map to other miRNA sequences were regarded as novel miRNAs [21]. Differential expression of the identified miRNAs was computed according to the DESeq method [22], while the Bayesian approach was applied for the statistical analysis of the data [23].
Target genes prediction and miRNA-mRNA interaction network analysis
To test for significant associations between genes, miRNAs, pathways and GO terms, the prediction software TargetScan and RNAhybrid, together with the differentially expressed genes in the L. vannamei hemocytes transcriptome data (Genebank accession numbers: PRJNA385392) were used. The relatedness or anticorrelation of the expression profiles of both the target genes and the associated miRNAs (downregulated miRNAs – upregulated mRNAs and upregulated miRNAs – downregulated mRNAs) based on the output of TargetScan or/and RNAhybrid were considered. Next, all the target genes obtained were subjected to GO and KEGG analysis. Finally, Cytoscape (version 3.5.1), an open source software platform was used to construct the interaction network between the miRNAs and mRNAs.
Real-time quantitative PCR (qPCR) analysis
To validate the miRNA-seq data, 12 VA versus VN significant differentially expressed miRNAs were selected for qPCR analysis. One microgram (1 μg) of the high quality total RNA samples, extracted using the mirVana miRNA Isolation Kit (Ambion, Austin, TX, USA), from each treatment were polyadenylated with poly (A) followed by first-strand cDNA synthesis with the TransScript miRNA First-Strand cDNA Synthesis SuperMix (TransGen Biotech, Beijing, China) kit following the manufacturer’s instruction. The forward miRNA primers (Table 1), designed based on the mature miRNA sequence, were used with a universal primer (supplied with TransScript miRNA First-Strand cDNA Synthesis SuperMix Kit, TransGen Biotech, Beijing, China) to amplify the mature miRNAs. The qPCR assay was performed at the following cycling conditions: one cycle at 95 °C for 10 min and 45 cycles of 95 °C for 15 s and 60 °C for 30 s, with the TransStart Tip Green qPCR SuperMix (TransGen Biotech, Beijing, China) kits on Roche LightCycler 480 detection system (Roche, Basel, Switzerland). U6 snRNA (small nuclear RNA) was used as an internal control (see Table 1 primers sequence). All the reactions were performed in replicates and with three biological replicates. The relative miRNA expression levels were calculated using the 2-∆∆CT method. Statistical analysis was performed using a two-tailed unpaired Student’s t-test with p < 0.05 considered statistically significant.
Results
Overview of miRNA transcriptome data
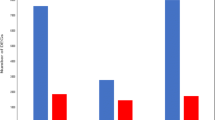
To characterize the AHPND related miRNAs in shrimp, nine sRNA libraries of shrimp hemocytes from three independent experiments were constructed, with three treatment groups denoted NS (normal saline), VN (non-AHPND V. parahemolyticus), and VA (AHPND V. parahemolyticus). High-throughput Illumina sequencing was performed on the sRNA libraries generating a total of 107,159,619 raw sequencing reads. Further filtering to remove rRNA (ribosomal RNA), scRNA (small cytoplasmic RNA), snoRNA(small nucleolar RNA), snRNA, tRNA (transfer RNA), and degraded mRNA fragments, was carried out by aligning with GenBank and Rfam reference databases as well as with our own hemocytes transcriptome data. After data filtering, 98,629,518 clean reads were left representing 92.04% of total raw reads (Table 2). The resulting reads were then mapped against the miRBase 21.0 database to obtain the matching shrimp miRNA homologs. A total of 4952, 5105 and 5122 known miRNA homologs from NS, VN and VA respectively were obtained, giving a grand total of 5990 identified known miRNAs (Fig. 1a). Of the total identified known miRNAs, 4235 were common to all treatment groups while 376, 378 and 282 miRNAs were unique to NS, VN and VA respectively (Fig. 1a). MiRNA family cluster analysis with other species yielded 70, 58 and 59 miRNAs as probable novel miRNAs from NS, VN and VA respectively, giving a total of 162 predicted novel miRNAs (Fig. 1b). Size distribution analysis of the 5990 identified known miRNAs (Fig. 1c) showed that 96.57% were between 20 and 24 nt in length, which is typical of miRNAs [24].
Characteristics of identified miRNA from L. vannamei. Summary of known (a) and predicted novel (b) miRNAs from hemocyte miRNA libraries, with samples from AHPND V. parahaemolyticus (VA), non-AHPND V. parahaemolyticus (VN) and normal saline (NS) challenged shrimps. c Mature miRNAs from sRNAs matched to miRBase. d MiRNA family analysis of conserved miRNAs. The Y-axis indicated the number of conserved miRNA families of L. vannamei aligned in arthropods. e Diagram summarizing differentially expressed miRNAs (known and novel) and their predicted target genes. Target genes were obtained from in-house hemocyte transcriptome data. Boxed shows data comparison between VN and VA
Next, the identified miRNAs were classified into 103 families (Additional file 1: Table S1), with most of the miRNA families highly conserved in arthropods. For instance, 71 were found in Drosophila melabgaster, 67 in Drosophila simulans, 65 in Bombyx mori, 62 in Drosophila pseudoobscura, but only 35 found in crustaceans (i.e., 34 found in Daphnia pulex and 1 in Marsupenaeus japonicas). Figure 1d shows the family distribution of the identified miRNAs in arthropods.
To identify the V. parahemolyticus responsive miRNAs, the DESeq method [22], which requires biological replicate samples, with a fold change > 2 for upregulated miRNAs and fold change ≤ 0.5 for downregulated miRNAs were used [22]. With these fold change cutoffs, 71 dysregulated miRNAs were identified upon AHPND V. parahemolyticus challenge while 64 dysregulated miRNAs were identified upon non-AHPND V. parahemolyticus challenge. When the non-AHPND (VA) and AHPND (VN) infection groups were compared, 47 upregulated and 36 downregulated miRNAs were identified (Fig. 1e).
Target genes prediction and functional analysis of differentially expressed miRNAs
Given that miRNAs negatively regulate mRNA expression [25], we went on to combine this miRNA transcriptome data with our our previous shrimp L. vannamei mRNA transcriptome data (GenBank accession number: PRJNA385392) so as to identify the miRNA target genes. Two target gene prediction software, i.e., Targetscan and RNAhybrid were used to obtain all the differentially expressed miRNA target genes. Next, target genes whose expressions were inversely correlated or with reciprocal expression pattern with their corresponding miRNAs were extracted. Using these target prediction criteria, 125 and 50 target genes were identified for the AHPND and non-AHPND V. parahemolyticus infected groups, respectively. When the predicted target genes for VA and VN were compared, 208 upregulated and 14 downregulated targets genes were identified to be candidate AHPND-related miRNA target genes (Fig. 1e and Additional file 2: Table S2).
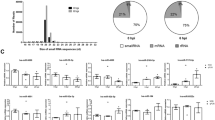
To gain an insight into how miRNAs affect AHPND, the AHPND-related miRNA target genes identified were categorized by GO and KEGG analyses. For the highly represented terms in each GO category, 77 DEGs in the VN versus VA treated groups were enriched in cellular component (89), molecular function (80), and biological process (137). In terms of GO function annotation, most of the DEGs were enriched in cell (26), cell part (26), catalytic (49), binding (28), cellular process (33), and metabolic process (29) (Fig. 2a). KEGG analysis was further performed to so as to identify the pathways involved in shrimps’ response to AHPND. As shown in Fig. 2b, the five most highly represented pathways include metabolic pathways (43), amoebiasis (15), Vibrio cholerae infection (11), drug metabolism - other enzymes (8), starch and sucrose metabolism (8).
Among the 222 predicted AHPND-related miRNA target genes, we excluded uncharacterized or unannotated genes, leaving with 124 genes, after which 38 immune related genes and their corresponding miRNAs (17 miRNAs) were chosen for further evaluation. Next, we went on to construct network interaction between these 17 miRNAs and their putative target genes (Fig. 2c and Table 3). It was observed that 14 downregulated miRNAs had 35 upregulated target genes while 3 upregulated miRNAs had 3 downregulated target genes (Fig. 2c). Broadly, these 38 immune-related genes encode for eight categories of proteins including 4 apoptosis related proteins, 1 heat shock protein, and 6 immunoproteins (Fig. 2c and Table 3). The network interaction analysis showed some possible interaction between the AHPND-related miRNAs and HSP70 (Fig. 2c).
Validation of potential key AHPND-related miRNAs using qPCR
To further substantiate the miRNA data obtained, 12 key putative AHPND-related miRNAs were chosen from the network analysis (Fig. 2c) for validation using qPCR, with U6 snRNA as the internal control. Three miRNAs, mja-miR-6489-5p, dre-miR-107b, and bta-miR-4286 were significantly upregulated in VA as compared to VN, while nine miRNAs including tch-miR-210, sha-miR-10b, and dpu-miR-193 were significantly downregulated (Fig. 3). The expression levels of all the miRNAs were expressed relative the NS group. The qPCR results were generally consistent with the miRNA transcriptome data, as shown in Table 3, where data for upregulated and downregulated miRNAs showed a similar trend.
Validation of differentially expressed miRNAs between VN and VA treated group by qPCR. Real-time qPCR was used to analyze the relative expression of 12 miRNAs and normalized to U6 snoRNA. Data presented is average of three independent experiments. The error bar represents the S.D. of the mean (n = 3). The statistical significance was calculated using Student’s t-test with p < 0.05 considered statistically significant compared with NS control
Discussion
Acute Hepatopancreatic Necrosis Syndrome (AHPNS) or Early Mortality Syndrome (EMS), which has now been shown to have bacterial aetiology [26], has caused significant losses in shrimp aquaculture in recent years. Given their role as one of the main post-transcriptional regulatory factors, miRNAs are implicated in various physiological and pathophysiological processes including differentiation, metabolism, signal transduction, stress responses, and immunity [27]. In the last couple of years, multiple functions of some crustaceans’ miRNAs have been reported. For instance, it has been shown that shrimp miR-1000 could target the 3′-untranslated region (3’UTR) of the p53 gene to regulate apoptosis and virus infection via the p53 pathway [28]. Similarly, miR-9 was shown to be upregulated or downregulated when shrimp apoptosis was induced or inhibited, suggesting that miR-9 participated in the apoptosis pathway by acting as a modulator [29]. Similarly, shrimp miR-1959 mediates a feedback loop, which could directly regulate the NFκB/IκB complex by targeting the 3’-UTR of cactus gene [30].
Although L. vannamei is the most widely cultured shrimp species in the world, and miRNAs have been extensively studied in the last couple of years, there is still limited information on shrimp miRNAs. Ruan et al. were the first to isolate 35 miRNAs from Marsupenaeus japonicas, and showed that the expression levels of 22 miRNAs changed upon WSSV infection [31]. Similarly, Huang et al. identified 63 shrimp miRNAs in WSSV-infected shrimp at different time points post-infection [32]. Since AHPND has had devastating effects on shrimp aquaculture couple with the conflicting accounts of the disease etiology and pathogenesis, we sought to gain a better understanding of the pathogenesis and progression of AHPND. To do this, we went about to determine the miRNAs that might be differentially expressed following infection of shrimps with AHPND and non-AHPND strains of V. parahaemolyticus. MiRNA transcriptome analysis showed 47 upregulated and 36 downregulated miRNAs between VA and VN. Twelve differentially expressed and potential AHPND-related miRNAs including miR-210, miR-10b and miR-193 were selected and validated by qPCR. Interestingly, miR-210 had previously been reported in eel, Branchiostoma belcheri, and was shown to increase in expression upon V. parahaemolyticus infection [33], while miR-10b was shown to be upregulated upon lipopolysaccharide (LPS) treatment [34]. It thus seems miR-210 and miR-10b are typical bacterial induced miRNAs, as our KEGG analysis showed that the predicted target genes of the 12 potential AHPND-related miRNAs, were mainly enriched in the Vibrio cholerae subcategory. For miR-193, it had previously been reported to be associated with WSSV infection [35], and was shown to be upregulated upon low virulent WSSV infection [36]. While miR-210, miR-10b and miR-193 were shown in previous studies to be upregulated, their expression in this study was observed to be downregulated (VA relative to VN), suggesting that probably the expression of these miRNAs was suppressed so as to increase the expression or function of their associated target genes; a possible implication of their importance in shrimp immune defense.
Since the complete genome sequence of L. vannamei is currently unavailable, we used our previous V. parahaemolyticus infected shrimp mRNA transcriptome data (GenBank accession number: PRJNA385392) to help us identify the AHPND V. parahaemolyticus specific responsive genes, which are the target genes of the identified miRNAs. Based on this in-house database, p53 was predicted as the target gene of miRNA-210 and miRNA-210-5p (downregulated in virulent V. parahaemolyticus infected shrimp). Indeed, miRNA-210 has been reported to suppress p53 in cancer cell line [37]. Thus, the upregulation of p53 expression seems to suggest that apoptosis was induced in shrimp hemocytes upon AHPND V. parahaemolyticus infection [28]. At the same time, caspase 2, a miRNA-22-5p predicted target gene, was upregulated in AHPND V. parahaemolyticus infected shrimp. Since caspase 2 is anti-apoptosis or prosurvival related, its upregulation might be a response to counter the apoptosis induction [38, 39]. The strong modulation of apoptosis seems to induce a higher metabolic rate and consequently a stronger immune responses in AHPND V. parahaemolyticus infected shrimp. For example, miRNA-278-5p, miRNA-22-5p, miRNA-211-5p, and miRNA-9c-5p whose predicted target genes are cytochrome P450, 2 L1, 49a1, and 3A2, were downregulated in AHPND V. parahaemolyticus infected shrimp, suggesting the involvement of energy metabolism [40] in AHPND V. parahaemolyticus infected shrimp. Since cytochrome P450 is involved in the metabolism of insecticides [41], the dysregulation by these miRNAs suggests some response to the bacterial Pir toxin [42, 43]. In response to the AHPND V. parahaemolyticus infection, shrimps mounted a strong immune response, as there was downregulation of miRNA-127, miRNA-193, and miRNA-10b, whose target genes are C-type lectin, Chymotrypsin BII, and cytokine receptor-like isoform 1, which are immune-related. There was also the downregulation of miR-211, which was predicted to target the Mucin2 gene. MiR-211 has been reported as a potential suppressor of MUC4 and shown to inhibit cervical cancer cell invasion and epithelial-to-mesenchymal transition (EMT) [44]. Given that Mucin glycoproteins are among the main components of the first barrier that bacteria encounter in an invasion [45, 46], it thus suggest the downregulation of miR-211 in AHPND V. parahaemolyticus infected shrimp might be an immune defense mechanism employed by hemocytes by enhancing the expression of Mucin. Apart from these, network interaction analysis revealed some possible interaction between the AHPND-related miRNAs and HSP70, which is in tandem with a recent study which have reported that HSP70 and HSP90 are involved in the tolerance of shrimp to AHPND-causing strain of V. parahaemolyticus [47]. Collectively, these results suggest that the dysregulation of the 12 miRNAs identified following the infection of shrimps with AHPND V. parahaemolyticus was an immune defense mechanism, since this was to enhance the level of metabolism, increase the expression of immune related genes, as well as modulate cellular processes such as apoptosis.
Conclusions
In this study, we observed the expression of 5990 shrimp miRNAs following V. parahaemolyticus infection, among which 12 potential AHPND-related miRNAs were identified. Furthermore, the predicted target genes of these miRNAs are involved in several immune-related processes. This work provides useful information for future research on AHPND and shrimp immune defense in terms of the relevant related miRNAs and their target genes.
Abbreviations
- AHPND:
-
Acute Hepatopancreatic Necrosis Disease
- EMS:
-
Early Mortality Syndrome
- hpi:
-
Hours post infection
- LPS:
-
Lipopolysaccharide
- miRNAs:
-
microRNAs
- mRNA:
-
Messenger RNAs
- NS:
-
Normal Saline
- Pir toxin:
-
Photorhabdus insect-related toxin
- qPCR:
-
Real-time Quantitative PCR
- rRNA:
-
Ribosomal RNA
- scRNA:
-
Small cytoplasmic RNA
- snoRNA:
-
Small nucleolar RNA
- snRNA:
-
Small nuclear RNA
- sRNA:
-
Small RNA
- tRNA:
-
Transfer RNA
- UTR:
-
Untranslated Region
- VA:
-
AHPND Vibrio parahaemolyticus
- VN:
-
Non-AHPND Vibrio parahaemolyticus
- WSSV:
-
White Spot Syndrome Virus
References
Bader AG, Brown D, Stoudemire J, Lammers P. Developing therapeutic microRNAs for cancer. Gene Ther. 2011;18(12):1121–6.
Cheng C, Bhardwaj N, Gerstein M. The relationship between the evolution of microRNA targets and the length of their UTRs. BMC Genomics. 2009;10(1):431.
Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310(5755):1817–21.
Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148(6):1172–87.
FAO Yearbook: Fishery and Aquaculture Statistics. 2014. http://www.fao.org/3/a-i5716t.pdf. Accessed 18 Dec 2017.
Umbach JL, Cullen BR. The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev. 2009;23(10):1151–64.
Triboulet R, Mari B, Lin YL, Chable-Bessia C, Bennasser Y, Lebrigand K, Cardinaud B, Maurin T, Barbry P, Baillat V, et al. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science. 2007;315(5818):1579–82.
Wang FZ, Weber F, Croce C, Liu CG, Liao XD, Pellett PE. Human cytomegalovirus infection alters the expression of cellular microRNA species that affect its replication. J Virol. 2008;82(18):9065–74.
Li X, Meng X, Luo K, Luan S, Shi X, Cao B, Kong J. The identification of microRNAs involved in the response of Chinese shrimp Fenneropenaeus chinensis to white spot syndrome virus infection. Fish Shellfish Immunol. 2017;68:220–31.
Zhu F, Wang Z, Sun BZ. Differential expression of microRNAs in shrimp Marsupenaeus japonicus in response to Vibrio alginolyticus infection. Dev Comp Immunol. 2016;55:76–9.
Ou J, Li Y, Ding Z, Xiu Y, Wu T, Du J, Li W, Zhu H, Ren Q, Gu W, et al. Transcriptome-wide identification and characterization of the Procambarus clarkii microRNAs potentially related to immunity against Spiroplasma eriocheiris infection. Fish Shellfish Immunol. 2013;35(2):607–17.
Xiao JZ, Liu LY, Ke YY, Li XF, Liu YF, Pan YJ, Yan SL, Wang YJ. Shrimp AHPND-causing plasmids encoding the PirAB toxins as mediated by pirAB-Tn903 are prevalent in various Vibrio species. Sci Rep-Uk. 2017;7:42177.
Han JE, Tang KFJ, Aranguren LF, Piamsomboon P. Characterization and pathogenicity of acute hepatopancreatic necrosis disease natural mutants, pir AB vp (−) V. parahaemolyticus, and pir AB vp (+) V . campbellii strains. Aquaculture. 2017;470:84–90.
Liu Q, Huang J, Yang HL, Yang B, Liu S, Wang HL, Wang QT, Liu F, Zhang QL. Detection of a new genotype of yellow-head virus in farmed shrimp suspicious of EMS/AHPNS infection. Oceanologia Et Limnologia Sinica. 2014;45:703–9.
Zhang QL, Wang X, Liang Y, Wang QT, Liu Q, Liu S, Huang J, Yang H, Liu S, Zhu L. A new nodavirus (CMNV) is associated withcovert mortality disease of shrimp. J Gen Virol. 2014;95(Pt 12):2700.
Lin YC, Chen JC. Acute toxicity of ammonia on Litopenaeus vannamei Boone juveniles at different salinity levels. J Exp Mar Biol Ecol. 2001;259(1):109–19.
Lin YC, Chen JC. Acute toxicity of nitrite on Litopenaeus vannamei (Boone) juveniles at different salinity levels. Aquaculture. 2003;224(1–4):193–201.
Yang Y-T, Chen I-T, Lee C-T, Chen C-Y, Lin S-S, Hor L-I, Tseng T-C, Huang Y-T, Sritunyalucksana K, Thitamadee S, Wang H-C, Lo C-F. 2014. Draft genome sequences of four strains of Vibrio parahaemolyticus, three of which cause early mortality syndrome/acute hepatopancreatic necrosis disease in shrimp in China and Thailand. Genome Announc. 2(5):e00816–14. https://doi.org/10.1128/genomeA.00816-14.
Lee CT, Chen IT, Yang YT, Ko TP, Huang YT, Huang JY, Huang MF, Lin SJ, Chen CY, Lin SS. The opportunistic marine pathogen Vibrio parahaemolyticus becomes virulent by acquiring a plasmid that expresses a deadly toxin. Proc Natl Acad Sci U S A. 2015;112(39):10798–803.
Zou Q, Mao Y, Hu L, Wu Y, Ji Z. miRClassify: an advanced web server for miRNA family classification and annotation. Comput Biol Med. 2014;45(1):157–60.
Friedlander MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40(1):37–52.
Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106.
Audic S, Claverie JM. The significance of digital gene expression profiles. Genome Res. 1997;7(10):986–95.
Sha ZX, Gong GY, Wang SL, Lu Y, Wang L, Wang QL, Chen SL. Identification and characterization of Cynoglossus semilaevis microRNA response to Vibrio anguillarum infection through high-throughput sequencing. Dev Comp Immunol. 2014;44(1):59–69.
He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–31.
Tran L, Nunan L, Redman RM, Mohney LL, Pantoja CR, Fitzsimmons K, Lightner DV. Determination of the infectious nature of the agent of acute hepatopancreatic necrosis syndrome affecting penaeid shrimp. Dis Aquat Org. 2013;105(1):45–55.
Huang Y, Shen XJ, Zou Q, Wang SP, Tang SM, Zhang GZ. Biological functions of microRNAs: a review. J Physiol Biochem. 2011;67(1):129–39.
Gong Y, Ju C, Zhang X. The miR-1000-p53 pathway regulates apoptosis and virus infection in shrimp. Fish Shellfish Immunol. 2015;46(2):516–22.
Yang G, Yang L, Zhao Z, Wang J, Zhang X. Signature miRNAs involved in the innate immunity of invertebrates. PLoS One. 2012;7(6):e39015.
Zuo H, Yuan J, Chen Y, Li S, Su Z, Wei E, Li C, Weng S, Xu X, He J. A MicroRNA-mediated positive feedback regulatory loop of the NF-kappaB pathway in Litopenaeus vannamei. J Immunol. 2016;196(9):3842–53.
Ruan L, Bian X, Ji Y, Li M, Li F, Yan X. Isolation and identification of novel microRNAs from Marsupenaeus japonicus. Fish Shellfish Immunol. 2011;31(2):334–40.
Huang TZ, Xu DD, Zhang XB. Characterization of host microRNAs that respond to DNA virus infection in a crustacean. BMC Genomics. 2012;13(1):1–10.
Jin P, Li SJ, Sun LJ, Lv CY, Ma F. Transcriptome-wide analysis of microRNAs in Branchiostoma belcheri upon Vibrio parahemolyticus infection. Dev Comp Immunol. 2017;74:243–52.
Liao X, Yang L, Zhang Q, Chen J. microRNA expression changes after lipopolysaccharide treatment in gills of amphioxus Branchiostoma belcheri. Dev Comp Immunol. 2017;70:39–44.
Du ZQ, Leng XY, Shen XL, Jin YH, Li XC. Identification and characterization of lymph organ microRNAs in red swamp crayfish, Procambarus clarkii infected with white spot syndrome virus. Fish Shellfish Immunol. 2017;69:78–84.
Sun BZ, Wang Z, Zhu F. Molecular cloning of kuruma shrimp Marsupenaeus japonicus phosphopyruvate hydratase and its role in infection by white spot syndrome virus and Vibrio alginolyticus. Aquaculture. 2016;455:87–96.
Amponsah PS, Fan P, Bauer N, Zhao Z, Gladkich J, Fellenberg J, Herr I. microRNA-210 overexpression inhibits tumor growth and potentially reverses gemcitabine resistance in pancreatic cancer. Cancer Lett. 2017;388:107–17.
Bergeron L, Perez GI, Macdonald G, Shi L, Sun Y, Jurisicova A, Varmuza S, Latham KE, Flaws JA, Salter JCM. Defects in regulation of apoptosis in caspase-2-deficient mice. Genes Dev. 1998;12(9):1304–14.
Terry MR, Arya R, Mukhopadhyay A, Berrett KC, Clair PM, Witt B, Salama ME, Bhutkar A, Oliver TG. Caspase-2 impacts lung tumorigenesis and chemotherapy response in vivo. Cell Death Differ. 2015;22(5):719–30.
Koop DR. Oxidative and reductive metabolism by cytochrome P450 2E1. FASEB J. 1992;6(2):724–30.
Yan L, Yang P, Jiang F, Cui N, Ma E, Qiao C, Cui F. Transcriptomic and phylogenetic analysis of Culex pipiens quinquefasciatus for three detoxification gene families. BMC Genomics. 2012;13(1):609.
Bowen D, Blackburn M, Rocheleau TA, Andreev O, Golubeva E, Ffrench-Constant RH. Insecticidal toxins from the bacterium Photorhabdus luminescens: gene cloning and toxin histopathology. Pest Manag Sci. 2015;55(6):666–8.
Ffrenchconstant RH, Dowling A, Waterfield NR. Insecticidal toxins from Photorhabdus bacteria and their potential use in agriculture. Toxicon. 2007;49(4):436–51.
Xu D, Liu S, Zhang L, Song L. MiR-211 inhibits invasion and epithelial-to-mesenchymal transition (EMT) of cervical cancer cells via targeting MUC4. Biochem Biophys Res Commun. 2017;485(2):556–62.
Parsons CL, Greenspan C, Moore SW, Mulholland SG. Role of surface mucin in primary antibacterial defense of bladder. Urology. 1977;9(1):48–52.
Moncada DM, Kammanadiminti SJ, Chadee K. Mucin and toll-like receptors in host defense against intestinal parasites. Trends Parasitol. 2003;19(7):305–11.
Junprung W, Supungul P, Tassanakajon A. HSP70 and HSP90 are involved in shrimp Penaeus vannamei tolerance to AHPND-causing strain of Vibrio parahaemolyticus after non-lethal heat shock. Fish Shellfish Immunol. 2017;60:237–46.
Acknowledgements
We acknowledge Professor Chu Fang Lo of National Cheng Kung University, Taiwan, China, for provision of the non-AHPND and AHPND V. parahaemolyticus bacteria.
Funding
This work was funded by National Natural Science Foundation of China (Nos. 31672689 & 31372558) and Natural Science Foundation of Guangdong Province (No. 2017A030311032). These funding bodies had no role in study design, analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
The sequence datasets associated with this study have been deposited at GenBank under the accession number PRJNA414944 (miRNA-seq) and PRJNA385392 (mRNA-seq). All other data generated or analyzed during this study are included in this published article or found in the additional data file.
Author information
Authors and Affiliations
Contributions
YLZ conceived and coordinated the study. ZHZ designed, performed and analyzed the experimental data. DFY, JSL, SKL and HYM contributed reagents /materials and provided technical assistance. YLZ, FW, ZHZ and JJA wrote the paper with help from the other authors. All authors reviewed the results and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All animal experiments were carried out in accordance with the guidelines and approval of the Animal Research and Ethics Committees of Shantou University, China.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Table S1. Conserved miRNA families analysis of L. vannamei. (XLS 76 kb)
Additional file 2:
Table S2. The potential targets of AHPND related miRNAs. (XLS 63 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Zheng, Z., Aweya, J.J., Wang, F. et al. Acute Hepatopancreatic Necrosis Disease (AHPND) related microRNAs in Litopenaeus vannamei infected with AHPND-causing strain of Vibrio parahemolyticus. BMC Genomics 19, 335 (2018). https://doi.org/10.1186/s12864-018-4728-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-018-4728-4