Abstract
Background
Euplotes, a ciliated protozoan, is a useful unicellular model organism. Studies on Euplotes have provided excellent insights into various basic biological principles. We have recently sequenced the macronuclear genome of the common freshwater species Euplotes octocarinatus to provide novel insights into Euplotes genetics and molecular biology.
Results
In this study, we present the E. octocarinatus Genome Database (EOGD), a functional annotation and analysis platform for the global study of the Euplotes genome. EOGD includes macronuclear genomic and transcriptomic data, predicted gene models, coding sequences, protein sequences, and functional annotations. The GBrowser and BLAST tools are embedded in EOGD to enable the search, visualization and analysis of E. octocarinatus genomic and transcriptomic data.
Conclusions
EOGD is a useful resource for the research community, particularly for researchers who conduct genome-scale analysis and molecular biology studies of Euplotes or other ciliates. EOGD will be continuously updated to integrate more datasets and analytical tools. EOGD is freely available at http://ciliates.ihb.ac.cn/database/home/#eo.
Similar content being viewed by others
Background
Euplotes is a free-living unicellular eukaryote that belongs to the ciliate phylum. This phylum also includes the model organisms Tetrahymena, Paramecium and Oxytricha. Like most ciliates, Euplotes exhibits nuclear dimorphism, possessing a germline micronucleus (MIC) and a somatic macronucleus (MAC). MIC is diploid, transcriptionally silent, and enables the transmission of genetic information between generations by sexual reproduction. MAC is generated from post-conjugation MIC, which is transcriptionally active during vegetative growth. Similar to other spirotrichous ciliates, Euplotes’s macronuclear chromosomes are tiny (nanochromosome) and mostly encode single genes that are differentially amplified to thousands of copies each [1, 2]. Two fundamental differences distinguish Euplotes from other ciliates: (i) the UGA codon of Euplotes is reassigned as cysteine or selenocysteine [3, 4]; and (ii) programmed ribosomal frameshifting (PRF) is widespread in Euplotes [5, 6].
Given its easy collection and laboratory cultivation, Euplotes is an attractive experimental system for studying basic eukaryotic biological processes. This organism has contributed to the discovery of fundamental eukaryotic mechanisms, such as the key telomerase protein [7], genome reorganization [8], defensive changes in cellular architecture in response to predator-produced signals [9], pheromone signaling [10] and stop-codon reassignment [3, 4]. Moreover, recent studies showed that Euplotes exhibits frequent PRF at stop codons [5, 6]. The frequent occurrence of ribosomal frameshifting during translation makes Euplotes an outstanding model for studying this universal phenomenon.
Using high-throughput sequencing approaches, we recently sequenced the macronuclear genome and the transcriptome of the typical species E. octocarinatus. We developed the E. octocarinatus Genome Database (EOGD) to provide the scientific research community with easy access to the genomic resources and information of Euplotes. EOGD provides user-friendly functions to access Euplotes genomic and transcriptomic data, as well as introduces the biology, taxonomy and morphology of E. octocarinatus. EOGD will serve as an important platform for researchers to facilitate research on Euplotes or other ciliates.
Construction and content
EOGD integrates three major data sets of E. octocarinatus: (i) the macronuclear genome sequence data, (ii) the annotations of predicted genes, and (iii) the RNA-seq data of growth stage. Furthermore, EOGD provides a detailed description of the biology, taxonomy, and morphology of E. octocarinatus.
The E. octocarinatus macronuclear genome was assembled using a specialized meta-assembly pipeline based on Illumina sequences [5]. A total of 41,980 contigs with an average length of 2117 bp were obtained, and over 70% of these contigs were capped with telomeres on both ends [5]. Given that the existence of PRF genes will influence the accuracy of gene prediction, contigs known to contain frameshifted genes [5] were excluded from ab initio gene finding. AUGUSTUS, a de novo prediction software [11], was used to predict gene models on non-PRF contigs (38,615 in total). Of the 29,076 putative protein-coding genes that were obtained, 90% were supported by RNA-Seq reads [5].
The functional annotation of genomes, an important part of EOGD, is important in genomic studies. To obtain integrated functional genomic information, multiple annotations were performed. First, all putative proteins were functionally annotated with BLAST (Basic Local Alignment Search Tool) alignments against the NCBI non-redundant (nr) protein database. Second, conserved motifs and functional domains were predicted by InterProScan5 software [12] against the InterPro database [13]. The InterPro database provides comprehensive functional information on proteins from some partner databases, such as Pfam, PRINTS, PANTHER, Gene3D and InterPro. Gene Ontology (GO) annotations were performed by mapping of GO terms to Pfam entries [5]. This mapping was generated from data supplied by InterPro for the InterPro2GO [14] mapping. Finally, the clusters of orthologous groups across multiple ciliates, including Tetrahymena thermophile, Paramecium tetraurelia, Ichthyophthirius multifiliis, Oxytricha trifallax, and Stylonychia lemnae, were constructed by OrthoMCL [15].
Transcriptomic data are powerful resources for characterizing and validating gene models. EOGD stores the deeply sequenced RNA-seq data of E. octocarinatus growth stage. In brief, the E. octocarinatus transcriptome was sequenced with the Illumina deep RNA sequencing strategy. We obtained 39,478,354 short reads with a total length of more than 4.9 Gb [5]. Then the high-quality reads of RNA-seq data were mapped to the Euplotes macronuclear genome by Tophat [16]. The Bio::DB::SAM adaptor allows the Generic Genome Browser (GBrowse) to display the binary file as xyplot plots.
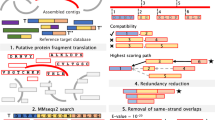
The schematic structure of EOGD is shown in Fig. 1. EOGD is built under the Linux system using a series of software packages, including Apache, PostgreSQL and Python. Genomic data and RNA-seq data were stored in the PostgreSQL database. The macronuclear genome sequences and gene sequences were formatted as a BLAST database. And we used Gbrowser [17], a widely used genome browser, to manipulate and visualize the genome annotations.
Schematic structure of EOGD. A flow diagram describing the database architecture is shown. Coding sequence, genome assembly data and protein sequence were formatted as BLAST database. Functional annotation information, RNA-seq data and genome data were stored in the PostqreSQL database. Keywords search and sequence search by BLAST allowed user to access the resources in EOGD easily. The EOGD also provides a description of E. octocarinatus
Utility and discussion
EOGD can be accessed through a user-friendly web interface. The EOGD website contains navigation tabs (Home, Genome browser, Blast, Morphology and Data download) and a search box on top of each page (Fig. 2a).
Web interface of EOGD. a Integrated searching box. The red arrow indicates the searching menu for selecting the “Gene ID”, “Genome ID” or “Keyword”. b Screenshot of search result interface of the EOGD website. (c–d) Gbrowse snapshots showing the search function interface (c) and search result interface (d) for Contig10891
EOGD provided two methods for users to rapidly search the genes of interest in the database: keywords and sequence search. If the user uses a “Keywords” to search, then any words (case insensitive) can be typed in the search box as search content. Scaffold ID and gene ID were also allowed to search specific genes. All searches will link to the result page (Fig. 2b). Furthermore, the user can search by a sequence. We implemented a server and an interface at our website for BLAST [18] search, which allows the user to search a query sequence against relevant Euplotes datasets (protein database, coding sequence database and genome database). Using BLAST tools, the user can obtain the detailed information of the sequence alignment for a region and download the BLAST results through the “DOWNLOAD” button.
EOGD graphically displays RNA-Seq and genome data and implements its search function through Gbrowse. Typically, the user can use “Gene ID”, “Scaffold ID” or a scaffold region to search the database (Fig. 2c). Three tracks, including a putative gene model track (linked to the gene sequence, annotation, and ortholog information), a RNA-Seq coverage plot track and a RPKM value track, are shown on the Gbrowse search result page (Fig. 2d). Through these tracks, the gene structure information and gene sequence can be obtained, which are important for exploring downstream biological functions. Any region of interest can be selected and the sequence can be downloaded in FASTA format by clicking the “Download Decorated FASTA File” in the pull-down menu or the “Export as FASTA sequence file” in the pull-down menu of the “File” button. Furthermore, the high-resolution image can be exported through Gbrowse.
By clicking the predicted gene model, users can obtain detailed information on the predicted gene, including annotations and sequence information (Fig. 3). Protein-coding gene models were annotated using BLAST alignments against the NCBI non-redundant (nr) protein database (E-value≤1e− 5). All predicted proteins were then functionally annotated (Fig. 3a). Functional domains and sites in all predicted protein models were identified by InterproScan (version Interproscan 5.2–45.0, run with default parameters) [12], which has combined signatures from a number of different database into one resource. All the predicted proteins were also annotated with GO for additional functional interpretation. In addition, OrthoMCL [15] was used to construct orthologous groups across multiple ciliates. The “Sequence” section includes the nucleotide and amino acid sequence of the predicted gene (fasta format) (Fig. 3b).
Case to illustrate the detailed search results. By clicking the predicted gene model on the Gbrowse search result page or searching the ID in the top search box, users can access the gene details page (taking the gene Contig10891.g834.t1 as an example). This gene details page will display all information for the gene of interest, including sequence name, scaffold ID, BLAST annotation, InterProScan domain information, annotated GO information, putative function annotation and ortholog information (a). Sequence information, including the nucleotide and amino acid sequence in FASTA format, will also be displayed (b)
On the “MORPHOLOGY” page (http://ciliates.ihb.ac.cn/database/morphology/#eo), the microscopic morphology and the detailed taxonomic classification of E. octocarinatus are described according to several previous studies [19,20,21]. Furthermore, EOGD provides a data download page (http://ciliates.ihb.ac.cn/database/download/#eo) for downloading E. octocarinatus genome sequences, protein sequences, coding sequences in FASTA format and annotation information. User can download these data by clicking the “download” button.
Further development of the EOGD
EOGD will be periodically updated. Additional Euplotes data sets, such as micronuclear genome, mitochondrial genome and proteome data, as well as gene expression profiles, will be uploaded to and integrated in EOGD in the future. Given the high-throughput and relatively low cost of next-generation sequencing, sequencing the genomes and transcriptomes of other Euplotes species in the future is possible.
Conclusions
EOGD allows researchers to access, browse, retrieve and analyze genomic and transcriptomic data and annotations. This hub will promote research on Euplotes biology, and will also enhance our understanding of eukaryotic molecular biology. We will periodically update EOGD by integrating more data. EOGD is an important and useful resource that is freely available to the research community.
Abbreviations
- BLAST:
-
Basic Local Alignment Search Tool
- EOGD:
-
Euplotes octocarinatus Genome Database
- GO:
-
Gene ontology
- MAC:
-
macronucleus
- MIC:
-
micronucleus
- NCBI:
-
National Center for Biotechnology Information
- RPKM:
-
Reads Per Kilobase Per Million Mapped Reads
References
Baird SE, Klobutcher LA. Differential DNA amplification and copy number control in the Hypotrichous ciliate Euplotes crassus. J Protozool. 1991;38(2):136–40.
Prescott DM. The DNA of ciliated protozoa. Microbiol Rev. 1994;58(2):233–67.
Turanov AA, Lobanov AV, Fomenko DE, Morrison HG, Sogin ML, Klobutcher LA, Hatfield DL, Gladyshev VN. Genetic code supports targeted insertion of two amino acids by one codon. Science. 2009;323(5911):259–61.
Meyer F, Schmidt HJ, Plümper E, Hasilik A, Mersmann G, Meyer HE, Engström A, Heckmann K. UGA is translated as Cysteine in pheromone-3 of Euplotes octocarinatus. Proc Natl Acad Sci U S A. 1991;88(9):3758–61.
Wang R, Xiong J, Wang W, Miao W, Liang A. High frequency of +1 programmed ribosomal frameshifting in Euplotes octocarinatus. Sci Rep. 2016;6:21139.
Lobanov AV, Heaphy SM, Turanov AA, Gerashchenko MV, Pucciarelli S, Devaraj RR, Xie F, Petyuk VA, Smith RD, Klobutcher LA, et al. Position-dependent termination and widespread obligatory frameshifting in Euplotes translation. Nat Struct Mol Biol. 2016;24(1):61–8.
Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276(5312):561–7.
Roth M, Prescott DM. DNA intermediates and telomere addition during genome reorganization in Euplotes crassus. Cell. 1985;41(2):411–7.
Kuhlmann HW, Heckmann K. Interspecific morphogens regulating prey-predator relationships in protozoa. Science. 1985;227(4692):1347–9.
Luporini P, Pedrini B, Alimenti C, Vallesi A: Revisiting fifty years of research on pheromone signaling in ciliates. Eur J Protistol 2016, 55(Pt A):26-328.
Stanke M, Keller O, Gunduz I, Hayes A, Waack S, Morgenstern B. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 2006;34(Web Server issue):W435–9.
Zdobnov EM, Apweiler R. InterProScan--an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17(9):847–8.
Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Biswas M, Bradley P, Bork P, Bucher P, et al. InterPro: an integrated documentation resource for protein families, domains and functional sites. Brief Bioinform. 2002;3(3):225–35.
Mitchell A, Chang HY, Daugherty L, Fraser M, Hunter S, Lopez R, McAnulla C, McMenamin C, Nuka G, Pesseat S, et al. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 2015;43(Database issue):D213–21.
Li L, Stoeckert CJ, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13(9):2178–89.
Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–11.
Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12(10):1599–610.
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–402.
Battistini R, Marcucci E, Verani M, Di Giuseppe G, Dini F, Carducci A. Ciliate-adenovirus interactions in experimental co-cultures of Euplotes octocarinatus and in wastewater environment. Eur J Protistol. 2013;49(3):381–8.
Lynn DH. The ciliated protozoa: characterization, classification and guide to the literature. 3rd ed. Dordrecht: Springer Verlag; 2008.
Gao F, Warren A, Zhang Q, Gong J, Miao M, Sun P, Xu D, Huang J, Yi Z, Song W. The all-data-based evolutionary hypothesis of ciliated Protists with a revised classification of the phylum Ciliophora (Eukaryota, Alveolata). Sci Rep. 2016;6:24874.
Acknowledgments
None.
Funding
This project is supported by grants from National Natural Science Foundation of China (No. 31372199) to A. Liang, National Natural Science Foundation of China (No. 31372168) to W. Miao, National Natural Science Foundation of China (No. 31471999) to W. Wang, Natural Science Foundation of China (No. 31301930) to J. Xiong and Youth Innovation Promotion Association, Chinese Academy of Sciences to J. Xiong.
Availability of data and materials
Database is available freely at http://ciliates.ihb.ac.cn/database/home/#eo.
Author information
Authors and Affiliations
Contributions
A.L. and W.M. conceived the work. W.M. and J.X. designed the database schema. R.W., J.X., W.M., W.W. and A.L. designed the Web interface. R.W. wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Wang, Rl., Miao, W., Wang, W. et al. EOGD: the Euplotes octocarinatus genome database. BMC Genomics 19, 63 (2018). https://doi.org/10.1186/s12864-018-4445-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-018-4445-z