Abstract
Background
Studies investigating associations between ACTN3 R577X and ACE I/D genotypes and endurance athletic status have been limited by small sample sizes from mixed sport disciplines and lack quantitative measures of performance. Aim: To examine the association between ACTN3 R577X and ACE I/D genotypes and best personal running times in a large homogeneous cohort of endurance runners.
Methods
We collected a total of 1064 personal best 1500, 3000, 5000 m and marathon running times of 698 male and female Caucasian endurance athletes from six countries (Australia, Greece, Italy, Poland, Russia and UK). Athletes were genotyped for ACTN3 R577X and ACE ID variants.
Results
There was no association between ACTN3 R577X or ACE I/D genotype and running performance at any distance in men or women. Mean (SD) marathon times (in s) were for men: ACTN3 RR 9149 (593), RX 9221 (582), XX 9129 (582) p = 0.94; ACE DD 9182 (665), ID 9214 (549), II 9155 (492) p = 0.85; for women: ACTN3 RR 10796 (818), RX 10667 (695), XX 10675 (553) p = 0.36; ACE DD 10604 (561), ID 10766 (740), II 10771 (708) p = 0.21. Furthermore, there were no associations between these variants and running time for any distance in a sub-analysis of athletes with personal records within 20% of world records.
Conclusions
Thus, consistent with most case-control studies, this multi-cohort quantitative analysis demonstrates it is unlikely that ACTN3 XX genotype provides an advantage in competitive endurance running performance. For ACE II genotype, some prior studies show an association but others do not. Our data indicate it is also unlikely that ACE II genotype provides an advantage in endurance running.
Similar content being viewed by others
Background
Although the likelihood of becoming an elite athlete is probably influenced by genetic variations across the human genome [1, 2], there is currently no evidence for a common genetic profile specific to elite endurance athletes, even when utilising a Genome-Wide Association Study (GWAS) approach [3]. However, there is considerable evidence suggesting that ACTN3 R577X and ACE I/D gene variants do influence muscle performance and metabolism in humans [4].
A common null polymorphism (rs1815739) was identified in the ACTN3 gene, which results in the replacement of an arginine (R) residue with a premature stop codon (X) at amino acid 577. Approximately 18% of the world population (~1.5 billion individuals) harbour the ACTN3 577XX genotype and consequently are completely deficient in α-actinin-3 protein. Importantly, α-actinin-3 deficiency does not cause any obvious muscle disease [5].
An association between the ACTN3 R577X genotype and athletic performance was initially found in a cohort of elite Australian athletes [6], with a very low proportion of elite sprint/power athletes harbouring the 577XX genotype. This genotype distribution pattern was quite consistent in other independent cohorts of elite athletes and has since been replicated in Finnish [7], Greek [8], Russian [9], Israeli [10], Polish [11] and Japanese [12] athletes.
A tendency for a higher proportion of elite athletes carrying the 577XX genotype was also found in Australian athletes excelling in aerobic activities [6], showing some evidence for association of this genotype with endurance performance. While this association was replicated in some cohorts of athletes [10, 13] other studies have shown no association between the ACTN3 R577X genotypes and endurance athletic status [7, 8]. Furthermore, a large study with Russian endurance athletes found that the frequency of the XX genotype was lower in endurance athletes than in controls [14], demonstrating the conflicting results between the association of this gene variant and endurance athletic performance. In line with this finding, an analysis comparing 50 elite male endurance cyclists and 52 Olympic-level endurance runners with 123 sedentary male controls [15] found no difference in genotype frequencies between controls and either of the two athlete groups. There was also no association between R577X genotypes and a common measure of endurance performance - maximal oxygen uptake (VO2max) - in either of the athlete groups. Cross-sectional studies showed no association of ACTN3 XX genotype with endurance performance [15, 16] as well, and debate is ongoing on whether the ACTN3 gene influences endurance performance. In a different human sporting context, namely the team sport of rugby union, the R allele has recently been associated with success in playing positions reliant on sprinting speed, while the X allele was associated with playing demands allowing relatively short recovery times [17].
Another candidate gene associated with elite performance is the ACE I/D polymorphism. The absence (deletion allele, D) rather than the presence (insertion allele, I) of a 287 base pair fragment is associated with higher tissue [18] and serum [19] ACE activity. While not directly functional [20], the ACE I/D polymorphism is related to ACE activity and accounts for up to 40% of the variation in circulating ACE activity in Caucasians [19]. An association between the ACE I/D polymorphism and athletic performance was initially found in a cohort of British mountaineers [21], with a very a low proportion of elite mountaineers harbouring the ACE DD genotype. This genotype distribution pattern was replicated in cohorts of elite endurance athletes [22, 23]. However, conflicting data also exist with ACE I/D genotype and endurance performance [24, 25] also found no association between the ACE I/D genotype with VO2max or its response to a 20-week endurance training programme in the HERITAGE Family study. Nevertheless, a more recent meta-analysis concluded that ACE II genotype was associated with superior endurance performance, with an odds ratio of 1.35 [4], however this was not replicated in the GAMES GWAS cohort analysis [3].
One of the limitations of most of the abovementioned studies investigating the association between the ACTN3 R577X and the ACE I/D genotypes and athletic status is the grouping of endurance athletes from mixed sport disciplines and events (e.g. middle distance runners, long distance runners, cyclists, swimmers), or analysing team sport athletes from a single sport yet with some variations in physiological demand according to playing position [17]. These approaches, while understandable given the very low number of World-class competitors in a single sport or event, reduce the consistency of the phenotype. Furthermore, those studies only used a simple case-control design based on athletic status without looking at measurable (quantitative) traits within the compared groups [26] and no studies have quantitatively linked those genotypes with endurance performance (e.g. running times) in elite athletes.
We sought to address these limitations by providing deeper insight into the possible association between the ACTN3 R577X and the ACE I/D variants and endurance performance. In the present study, we used the same quantitative approach previously introduced in elite sprinters [27] that showed both ACTN3 R577X and ACE I/D genotypes have a substantial association with sprint performance (100-400 m run) at the elite level. The aim of this study was to examine the association between the ACTN3 R577X and ACE I/D variants and personal best running times in 1500 m, 3000 m, 5000 m, 10,000 m and marathon in a large cohort of male and female Caucasian endurance runners.
Methods
The methodology been used in genotyping, data collection and statistical analysis has been previously described [17, 27].
Participants
A total of 1064 personal best 1500, 3000, 5000, 10,000 m and marathon running times of 698 Caucasian endurance athletes (441 males and 257 females) from Australia (n = 14), Greece (n = 16), Italy (n = 9), Poland (n = 60), Russia (n = 17) and the UK (n = 582), were analysed (Table 1). The endurance runners’ personal best times in official competitions were found online (www.iaaf.org and www.thepowerof10.info) or provided by coaches or the athletes themselves and independently corroborated.
We grouped the participants’ personal best times by event (1500, 3000, 5000, 10,000 m or marathon) as has been previously described [27]. First, we analysed the whole cohort of males and females separately. Then, we also performed a sub-analysis in males including only the endurance runners with times that were within 20% of the current world record of the examined events, following a similar approach to our recently published work [27]. We did not analyse the females in this sub-analysis because the sample size was too low (i.e. n < 5 for the XX genotype and n < 6 for the II genotype). We used the following world records as references:
-
Male endurance runners. 3:26.00 in the 1500 m (Hicham El Guerrouj, Morocco), 12:37.35 in the 5000 m (Kenenisa Bekele, Ethiopa, 26:17.53 in the 10,000 m (Kenenisa Bekele, Ethiopia), 2:02:57 in the Marathon Dennis Kipruto Kimetto, Kenya);
-
Female endurance runners. 3:50.07 in the 1500 m (Genzebe Dibaba, Ethiopia), 14:11.15 in the 5000 m (Tirunesh Dibaba, Ethiopia), 29:17.45 in the 10,000 m (Almaz Ayana, Ethiopia), 2:17:42 in the Marathon (Paula Radcliffe, UK).
Genotyping
In the UK ~70% of the samples were collected as whole blood, ~20% as buccal swabs, ~10% as saliva. As has been previously described [17] blood was drawn from a superficial forearm vein into an EDTA tube and stored in sterile tubes at −20 °C until processing. Saliva samples were collected into Oragene DNA OG-500 collection tubes (DNA Genotek, Ottawa, Ontario, Canada) according to the manufacturer’s protocol and stored at room temperature until processing. Sterile buccal swabs (Omni swab; Whatman, Springfield Mill, UK) were rubbed against the buccal mucosa of the cheek for 30 s. Tips were ejected into sterile tubes and stored at −20 °C until processing. Genomic DNA was isolated from buccal epithelium, or white blood cells. In the UK DNA isolation was performed with the QIAamp DNA Blood Mini kit and standard spin column protocol, following the manufacturer’s instructions (Qiagen, West Sussex, UK). In brief, 200 μL of whole blood/saliva, or one buccal swab, was lysed and incubated, the DNA washed, and the eluate containing isolated DNA stored at 4 °C.
The Australian, Greek and Italian endurance runners’ DNA samples were genotyped using the polymerase chain reaction (PCR)-restriction fragment length polymorphism (RFLP) method as previously described [5]. The DNA samples of the UK, Polish and Russian endurance runners were genotyped in duplicates using an allelic discrimination assay on a Step One Real-Time PCR instrument (Applied Biosystems, Carlsbad, California, USA) with TaqMan® probes. To discriminate ACTN3 R577X (rs1815739) and the ACE I/D alleles, a TaqMan® Pre-Designed SNP Genotyping Assay was used (assay ID: C_590093_1_ for rs1815739 (ACTN3 R577X) and C__29403047_10 for rs4341 (a tag SNP in perfect linkage disequilibrium with the 287-bp ACE I/D in Caucasians [28])), including appropriate primers and fluorescently labelled (FAM and VIC) MGB™ probes to detect the alleles. For the genotyping of the UK samples the StepOnePlus and Chromo4 (Bio-Rad, Hertfordshire, UK) were used.
Statistical analysis
To compare the endurance athletes’ running times between ACTN3 R577X or ACE I/D genotypes, we converted the running times to seconds and initially used the one-way analysis of variance (ANOVA). Then a simple linear regression with running time as the dependent variable and genotypes as the independent variable was also applied. We used two genetic models: the additive model where RR = 0, RX = 1 and XX = 2, or DD = 0, ID = 1 and II = 2, and the recessive genetic model where RR = RX = 0 and XX = 1 or DD = ID = 0 and II = 1 as has been previously described [27]. Males and females were analysed separately. The level of significance was set at 0.05. All data analyses were conducted with the R statistical software with the lme4 and lrtest packages.
Results
The mean (SD) personal best 1500 m, 3000 m, 5000 m, 10,000 m and marathon running times, according to the ACTN3 and ACE genotype and distribution, are presented in Tables 1 and 2, respectively.
ANOVA revealed no differences among the three genotypes (p > 0.05) of either ACTN3 or ACE and running performance at any distance. Similarly, linear regression analysis using an additive or recessive genetic model also showed no association between running time and genotype of either genetic variant at any running distance (p > 0.05).
No association between ACTN3 R577X or ACE I/D genotypes and personal best times in the whole cohort
In males and females alike and regardless of the chosen statistical analysis or genetic model, we found no association between either ACTN3 R577X or ACE I/D genotype and 1500 m, 3000 m, 5000 m, 10,000 m or marathon personal best times (Figs. 1 and 2, Tables 1 and 2).
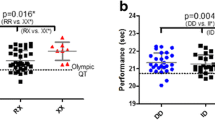
Individual 1500 m, 3000 m, 5000 m, 10,000 m personal best times in (a) male and (b) female endurance athletes according to their ACTN3 R577X genotype. Data are shown as boxplots and time is expressed in seconds. The red dashed line on each plot corresponds to the competition entry standard for the 2016 Olympic Games. 3000 m is not an Olympic event, so there is no red dashed line for this event
Individual 1500 m, 3000 m, 5000 m, 10,000 m personal best times in (a) male and (b) female endurance athletes according to their ACE I/D genotype. Data are shown as boxplots and time is expressed in seconds. The red dashed line on each plot corresponds to the competition entry standard for the 2016 Olympic Games. 3000 m is not an Olympic event, so there is no red dashed line for this event
No association between ACTN3 R577X or ACE I/D genotypes and personal best time in males within 20% of the world record
In males only, we conducted a sub-analysis of the athletes displaying times within 20% of the World record for the corresponding event, to see whether an association with the ACTN3 R577X or ACE I/D variants could be detected at the high end of the performance spectrum. Regardless of the chosen statistical analysis or genetic model, we found no association between ACTN3 R577X or ACE I/D genotypes and 1500 m, 3000 m, 5000 m, 10,000 m or marathon personal best time for those athletes within 20% of the world record (Table 3).
Discussion
Here, we have utilised similar approach as we previously did in elite sprinters [27] in a large cohort of elite Caucasian endurance runners. This quantitative assessment of genotype with qualifying time in 1064 personal best times of 698 elite endurance runners suggests that the potential association between ACTN3 R577X and ACE I/D genotypes and elite endurance running time is unproven.
In the present study, we examined whether a genotype association existed within athletes competing in particular endurance-running events (1500 m, 3000 m, 5000 m, 10,000 m and marathon) and in a subset of high-level athletes with personal-best times within 20% of the World record. Previous reports have grouped together endurance athletes from mixed endurance sports disciplines and events without quantifying measures of their actual endurance performance [6, 8,9,10,11,12]. Here, we have embraced a more stringent approach and included only endurance runners whose main sporting discipline was the 1500, 3000, 5000, 10,000 m or marathon, including their personal-best running performance. In this manner, we were able to differentiate between events that are estimated to have a different energy reliance on the aerobic system ranging from 77 to 86% (1500 m), 86-94% (3000 m) whereby it becomes increasingly dependent on aerobic metabolism up to the Marathon [29]. Despite addressing these subtle performance requirement differences, our results suggest that neither the ACTN3 R577X nor ACE I/D polymorphisms are likely to influence Caucasian endurance runners’ personal best times in 1500, 3000, 5000, 10,000 m and marathon, even at the high end of the performance spectrum.
The Actn3 KO mouse model attempted to mimic the ACTN3 R577X polymorphism in humans. Metabolically, the KO mice show a higher activity of oxidative enzymes and a lower activity of enzymes involved in the anaerobic pathway [30]. In addition, KO mice show enhanced glycogen accumulation due to lower glycogen phosphorylase activity [30, 31]. Their fast skeletal muscle fibre properties shift towards a slower metabolic profile, which has been linked to an increase in calcineurin signalling activity [32] and theoretically can favour endurance performance. Top-level endurance running performance is considered to be predominately based on the metabolic profile of slow-twitch (type I) and some recruitment of intermediate (type IIa) fibres due to the high reliance on the aerobic energy system, which has been hypothesized to favour the 577XX genotype. However, it may also depend on the endurance runner’s ability to recruit a greater number of type IIa myofibres during tactical surges (competitively critical phases requiring increase in pace) or finishing stages of a race (a sprint over a short distance), both of which require an increase in anaerobic energy/muscle recruitment (and may favour the 577RR genotype). While it is difficult to determine the relative contribution of muscle fibres in human competitive performance, it is well understood that murine muscle contains a significantly higher percentage of myofibres with faster twitch properties than human muscle and any association with the presence/absence of α-actinin-3 protein would probably be enhanced in this model. Therefore, any such association in humans might be extremely limited, thus offering no tangible or detectable advantage to a competitive α-actinin-3 deficient (ACTN3 577XX) endurance runner. In our study, we included only Caucasian endurance athletes because thus far we have recruited insufficient individuals of other ethnicities for effective analysis, so we cannot rule out the possibility that an association exists between ACTN3 R577X or ACE I/D and endurance running performance in elite runners with different geographic ancestry. However, most published associations on which our original hypothesis was based involved athletes or other individuals who were Caucasians.
Endurance performance is considered to be a complex trait effected by both genetic and environment (training) [33]. As recently shown, not only metabolism but also anthropometric and biomechanical factors are important in determining elite performance success [34]. Here we have not found evidence that supports the involvement of the two gene variants we studied in endurance running performance. Endurance running performance is dependent on extensive training and there is little evidence of either ACTN3 R577X or ACE I/D being associated with training responses of aerobic parameters [35]. Interestingly, in addition to this, it has been suggested that epigenetic modifications related to the ACE gene may also have a part to play in these discrepant findings [36]. Indeed, epigenetic factors such as CpG islands that modify the expression of genes without alteration to the DNA coding sequence have been identified in the ACE gene promoter [37]. Therefore, future studies on the ACE gene, in relation to human endurance, could benefit if these epigenetic factors that regulate ACE expression [37] were considered in addition to the I/D genotype [36]. These epigenetic factors reported to influence ACE activity cannot be controlled by our quantitative approach and sophisticated experimental designs are required to control more external factors and possibly explain some of the discrepant findings between ACTN3 R577X and ACE I/D genotype and endurance phenotypes.
The multi-centre cohort we used is larger than any previous such study regarding endurance performance, but a very small effect size could still go undetected using our sample size. It is increasingly recognised that the reality of complex human biology is that inter-individual differences in endurance performance are expected to be influenced by many common and perhaps rare genetic variations; none of which have been discovered at a genome-wide significance level or consistently replicated [3].
Our study of quantitative measures of endurance performance in a large homogeneous group of elite endurance runners suggests that the potential association between ACTN3 R577X and ACE I/D genotypes and elite endurance running performance should be regarded as unproven. Large population studies are needed to detect significant proportions of the underlying genetic profile and biology contributing to endurance performance.
Conclusions
In conclusion, this study has presented evidence that the ACTN3 and ACE polymorphisms are not associated with running performance in elite athletes, contrary to our hypotheses and again exposing the fallacy of products offered by numerous direct-to-consumer (DTC) genetic testing companies [38]. Our understanding of the genetic influences on human physical performance is evolving rapidly in the postgenomic era. Much more work remains to be done to answer a number of major questions in the field. One study utilising a genome-wide approach has been published recently [3] and future studies investigating the genomic contribution to elite endurance performance using genome-wide and targeted sequencing approaches are still needed to discover more genetic variants contributing to human physical performance capability. Understanding both genetic and environmental contributions, and how they interact, will be beneficial in understanding elite performance and muscle function in sport, health and disease.
Abbreviations
- DTC:
-
Genetic testing companies: Direct-to-Consumer genetic testing companies
- GWAS:
-
Genome-Wide Association Study
References
Pitsiladis Y, Wang G, Wolfarth B, Scott R, Fuku N, Mikami E, et al. Genomics of elite sporting performance: what little we know and necessary advances. Br J Sports Med. 2013;47:550–5.
Eynon N, Hanson ED, Lucia A, Houweling PJ, Garton F, North KN, et al. Genes for elite power and Sprint performance: ACTN3 leads the way. Sports Med. 2013;43:803–17.
Rankinen T, Fuku N, Wolfarth B, Wang G, Sarzynski MA, Alexeev DG, et al. No evidence of a common DNA variant profile specific to world class endurance athletes. PloS One U S A. 2016;11:e0147330.
Ma F, Yang Y, Li X, Zhou F, Gao C, Li M, et al. The Association of Sport Performance with ACE and ACTN3 genetic polymorphisms: a systematic review and meta-analysis. PLoS One. 2013;8:e54685.
North KN, Yang N, Wattanasirichaigoon D, Mills M, Easteal S, Beggs AH. A common nonsense mutation results in alpha-actinin-3 deficiency in the general population. Nat Genet U S A. 1999:353–4.
Yang N, MacArthur DG, Gulbin JP, Hahn AG, Beggs AH, Easteal S, et al. ACTN3 genotype is associated with human elite athletic performance. Am J Hum Genet. 2003;73:627–31.
Niemi A-K, Majamaa K. Mitochondrial DNA and ACTN3 genotypes in Finnish elite endurance and sprint athletes. Eur J Hum Genet. 2005;13:965–9.
Papadimitriou ID, Papadopoulos C, Kouvatsi A, Triantaphyllidis C. The ACTN3 gene in elite Greek track and field athletes. Int J Sports Med. 2008;29:352–5.
Druzhevskaya AM, Ahmetov I, Astratenkova IV, Rogozkin VA. Association of the ACTN3 R577X polymorphism with power athlete status in Russians. Eur J Appl Physiol. 2008;103:631–4.
Eynon N, Duarte JA, Oliveira J, Sagiv M, Yamin C, Meckel Y, et al. ACTN3 R577X polymorphism and Israeli top-level athletes. Int J Sports Med. 2009;30:695–8.
Cięszczyk P, Eider J, Ostanek M, Arczewska A, Leońska-Duniec A, Sawczyn S, et al. Association of the ACTN3 R577X polymorphism in polish power-orientated athletes. J Hum Kinet. 2011;28:55–61.
Mikami E, Fuku N, Murakami H, Tsuchie H, Takahashi H, Ohiwa N, et al. ACTN3 R577X genotype is associated with sprinting in elite Japanese athletes. Int J Sports Med. 2013;
Eynon N, Ruiz JR, Femia P, Pushkarev VP, Cieszczyk P, Maciejewska-Karlowska A, et al. The ACTN3 R577X polymorphism across three groups of elite male European athletes. PLoS One. 2012;7:e43132.
Ahmetov II, Druzhevskaya AM, Astratenkova IV, Popov DV, Vinogradova OL, Rogozkin VA. The ACTN3 R577X polymorphism in Russian endurance athletes. Br. J. Sports Med. 2010;44:649–52.
Lucia A, Gomez-Gallego F, Santiago C, Bandres F, Earnest C, Rabadan M, et al. ACTN3 genotype in professional endurance cyclists. Int J Sports Med Germany. 2006;27:880–4.
Paparini A, Ripani M, Giordano GD, Santoni D, Pigozzi F, Romano-Spica V. ACTN3 genotyping by real-time PCR in the Italian population and athletes. Med Sci Sports Exerc. 2007;39:810–5.
Heffernan SM, Kilduff LP, Erskine RM, Day SH, McPhee JS, McMahon GE, et al. Association of ACTN3 R577X but not ACE I/D gene variants with elite rugby union player status and playing position. Physiol Genomics. 2016;48:196–201.
Danser AHJ, Schalekamp MADH, Bax WA, van den Brink AM, Saxena PR, Riegger GAJ, et al. Angiotensin-converting enzyme in the human heart: effect of the deletion/insertion polymorphism. Circulation. 1995;92:1387–8.
Rigat B, Hubert C, Alhenc-Gelas F, Cambien F, Corvol P, Soubrier F. An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. J Clin Invest. 1990;86:1343–6.
Cox R, Bouzekri N, Martin S, Southam L, Hugill A, Golamaully M, et al. Angiotensin-1-converting enzyme (ACE) plasma concentration is influenced by multiple ACE-linked quantitative trait nucleotides. Hum Mol Genet. 2002;11:2969–77.
Montgomery HE, Marshall R, Hemingway H, Myerson S, Clarkson P, Dollery C, et al. Human gene for physical performance. Nature. 1998;393:221–2.
Gayagay G, Yu B, Hambly B, Boston T, Hahn A, Celermajer DS, et al. Elite endurance athletes and the ACE I allele--the role of genes in athletic performance. Hum Genet. 1998;103:48–50.
Myerson S, Hemingway H, Budget R, Martin J, Humphries S, Montgomery H. Human angiotensin I-converting enzyme gene and endurance performance. J Appl Physiol. 1999;87:1313–6.
Rankinen T, Wolfarth B, Simoneau JA, Maier-Lenz D, Rauramaa R, Rivera MA, et al. No association between the angiotensin-converting enzyme ID polymorphism and elite endurance athlete status. J Appl Physiol Bethesda Md 1985. 2000;88:1571–5.
Bouchard C, Rankinen T, Chagnon YC, Rice T, Pérusse L, Gagnon J, et al. Genomic scan for maximal oxygen uptake and its response to training in the HERITAGE family study. J Appl Physiol Bethesda Md 1985. 2000;88:551–9.
Wang G, Tanaka M, Eynon N, North KN, Williams AG, Collins M, et al. The future of genomic research in athletic performance and adaptation to training. Med Sport Sci Switzerland. 2016;61:55–67.
Papadimitriou ID, Lucia A, Pitsiladis YP, Pushkarev VP, Dyatlov DA, Orekhov EF, et al. ACTN3 R577X and ACE I/D gene variants influence performance in elite sprinters: a multi-cohort study. BMC Genomics. 2016;17:285.
Glenn KL, Du Z-Q, Eisenmann JC, Rothschild MF. An alternative method for genotyping of the ACE I/D polymorphism. Mol Biol Rep. 2009;36:1305–10.
Duffield R, Dawson B, Goodman C. Energy system contribution to 1500- and 3000-metre track running. J Sports Sci. 2005;23:993–1002.
Quinlan KGR, Seto JT, Turner N, Vandebrouck A, Floetenmeyer M, Macarthur DG, et al. Actinin-3 deficiency results in reduced glycogen phosphorylase activity and altered calcium handling in skeletal muscle. Hum Mol Genet. 2010;19:1335–46.
MacArthur DG, North KN. ACTN3: a genetic influence on muscle function and athletic performance. Exerc Sport Sci Rev. 2007;35:30–4.
Seto JT, Quinlan KGR, Lek M, Zheng XF, Garton F, MacArthur DG, et al. ACTN3 genotype influences muscle performance through the regulation of calcineurin signaling. J Clin Invest. 2013;123:4255–63.
Rankinen T, Bouchard C. Gene–physical activity interactions: overview of human studies. Obes Silver Spring Md. 2008;16:S47–50.
Stebbings GK, Williams AG, Herbert AJ, Lockey SJ, Heffernan SM, Erskine RM, et al. TTN genotype is associated with fascicle length and marathon running performance. Scand J Med Sci Sports. 2017. doi: https://doi.org/10.1111/sms.12927. [Epub ahead of print]
Rankinen T, Pérusse L, Gagnon J, Chagnon YC, Leon AS, Skinner JS, et al. Angiotensin-converting enzyme ID polymorphism and fitness phenotype in the HERITAGE family study. J Appl Physiol Bethesda Md 1985. 2000;88:1029–35.
Raleigh SM. Epigenetic regulation of the ACE gene might be more relevant to endurance physiology than the I/D polymorphism. J Appl Physiol Bethesda Md 1985. 2012;112:1082–3.
Rivière G, Lienhard D, Andrieu T, Vieau D, Frey BM, Frey FJ. Epigenetic regulation of somatic angiotensin-converting enzyme by DNA methylation and histone acetylation. Epigenetics. 2011;6:478–89.
Webborn N, Williams A, McNamee M, Bouchard C, Pitsiladis Y, Ahmetov I, et al. Direct-to-consumer genetic testing for predicting sports performance and talent identification: consensus statement. Br J Sports Med. 2015;49:1486–91.
Acknowledgments
IDP wishes to dedicate this paper to all athletes from all countries who participated in this multi-centre study.
Funding
No funding was received to assist in the preparation of this manuscript.
Availability of data and materials
The Authors declare that they have full control of the primary data and they agree to allow the journal to review their data if requested. The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Author information
Authors and Affiliations
Contributions
IDP: conceived the idea, done the initial analysis, wrote the paper and contributed in data collection and interpretation of the results. SJL, SV, AJH, FG, PJH, PC, AM, MS, MM, CMC, IVA, AK, AMD, MJ, IIA, GKS, SH, SHD, RE, CP, CK, KNN, AGW, NE: provided feedback on the initial draft, involved in data collection, data analysis, statistics, revised the manuscript and made substantial contributions in interpretation of the results. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Ethical approval was obtained from the Human Research Ethics Committees of the Children’s Hospital at Westmead (2003/086), the RCH Human Research Ethics Committee (35172), the ethics committee of the Manchester Metropolitan University, the Lithuanian National Committee of Biomedical Ethics, the Ethics Committee of Kazan State Medical University, the Ethics Committee of Gdansk University, the Ethics Committee of the University of Cagliari, and the UHWI/UWI/FMS Ethics Committee. All studies were conducted in accordance with the ethical standard laid down in the 1964 Declaration of Helsinki and its later amendments. From all athletes, informed consent to participate in the study was obtained and all participants signed a consent form.
Consent for publication
This manuscripts doesn’t include images/videos or any information relating to an individual person or participant.
Competing interests
All authors declare that they have no competing interests to declare that are directly relevant to the content of this manuscript. The corresponding author recently became a member of the editorial board of this journal.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Papadimitriou, I.D., Lockey, S.J., Voisin, S. et al. No association between ACTN3 R577X and ACE I/D polymorphisms and endurance running times in 698 Caucasian athletes. BMC Genomics 19, 13 (2018). https://doi.org/10.1186/s12864-017-4412-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-017-4412-0





