Abstract
Background
Multi-walled carbon nanotube (MWCNT) is one of the most momentous carbonaceous nanoparticles which is widely used for various applications such as electronics, vehicles, and therapeutics. However, their possible toxicity and adverse effects convert them into a major health threat for humans and animals.
Results
In this study, we employed weighted gene co-expression network analysis (WGCNA) to identify the co-expressed gene groups and dysregulated pathways due to the MWCNT exposure. For this purpose, three weighted gene co-expression networks for the microarray gene expression profiles of the mouse after 1, 6, and 12-month post-exposure to MWCNT were constructed. The module-trait analysis specified the significant modules related to different doses (1, 10, 40, and 80 µg) of MWCNT. Afterward, common genes between co-regulated and differentially expressed genes were determined. The further pathway analysis highlighted the enrichment of genes including Actb, Ube2b, Psme3, Ezh2, Alas2, S100a10, Ypel5, Rhoa, Rac1, Ube2l6, Prdx2, Ctsb, Bnip3l, Gp6, Myh9, Ube2k, Mbnl1, Kbtbd8, Riok3, Itgb1, Rap1a, and Atp5h in immune-, inflammation-, and protein metabolism-related pathways.
Conclusions
This study discloses the genotoxicity and cytotoxicity effects of various doses of MWCNT which also affect the metabolism system. The identified genes can serve as potential biomarkers and therapeutic candidates. However, further studies should be performed to validate them in human cells.
Similar content being viewed by others
Background
Multi-walled carbon nanotubes (MWCNTs) are fiber-shaped carbon nanomaterials that have been employed in various commercial products like home appliances, electronics, vehicles, and also biomedical applications [1]. Besides several advantages of using MWCNTs in industrial applications, the most challenging issues are their contaminations, toxicity, and hazardous effects. Therefore, it is unavoidable to survey the toxicity characteristics of carbon nanotubes. The toxicity assessment of materials on the gene expression in the animal models can be generalized to the human target organ [2, 3].
Predominantly, workers who work in an industrial factory and are in contact with nanomaterials like MWCNTs, as well as consumers, are the main vulnerable groups to nanomaterial [4]. MWVNTs are light and simply aerosolized. Therefore, the workplace is a considerable source of human exposure to carbon nanotubes through inhalation or dermal contact. Moreover, the respiratory tract and damaged skin are the possible ways of exposure due to the small size and low density of MWCNTs. They may deposit in the mice’s lungs by inhalation or pharyngeal aspiration which causes histologic alterations including fibrosis and inflammation [5, 6].
Carbon nanotubes can induce asbestos-like pathogenicity with carcinogenic risk [7]. The main mechanism for the MWCNTs toxicity is the induction of oxidative stress through the production of free radicals such as reactive oxygen species (ROS) or reactive nitrogen species (RNS) [8,9,10]. Oxidative stress causes the generation or boosting of inflammation which is a remarkable risk factor for pulmonary carcinogenicity [9, 11]. Genotoxicity can also be generated by direct interaction of carbon nanotubes with genetic contents or by indirect harm from the induction of ROS [10]. MWCNTs induce the increase of profibrotic inflammatory mediators as well as pleural mesothelioma, lung carcinoma, and DNA damage responses [12,13,14,15]. The thin and entangled MWCNTs induce pulmonary inflammation due to lymphocytic aggregates, granulomas, and macrophage infiltration [16].
Weighted gene co-expression network analysis (WGCNA) is a practical and data reduction approach to find the co-regulated genes and correlation patterns across various specimens. The potential biomarkers and possible therapeutic targets may be detected through constructing co-expression networks and then identification of hub genes based on their biological functions and connection with other genes [17, 18]. WGCNA has several advantages including the transformation of gene expression data into a small number of co-expression groups (modules), finding the hub nodes in each module, and the associations between modules with the external traits. The co-regulated modules assist the annotation of results and relevant signaling networks that might be liable for phenotypic traits of interest [19]. The disadvantages of WGCNA are likely simplicity and producing false positives for cascades [20]. However, the co-expressed genes are often functionally related, governed by a similar transcriptional regulatory program, or involved in similar pathways. Therefore, they are of biological interest. This method has been employed to determine the co-regulated genes to find the possible pathogenesis mechanisms and potential biomarkers for various cancers, pathogens-caused diseases, and nanomaterials-caused toxicity [21,22,23,24,25]. In this study, we aimed to find the genotoxic co-expressed genes due to MWCNTs. For this purpose, we constructed the weighted gene co-expression networks for the gene expression profiling of different mouse groups exposed to various doses of MWCNT in different periods [26]. Further analyses led to the identification of co-regulated genes which implicate various pathways related to cytotoxicity and carcinogenicity.
Results
Construction of weighted gene co-expression network
To build the co-expressed gene networks, the soft threshold powers β were determined as 4, 3, and 2 for MWCNT_1, MWCNT_6, and MWCNT_12, respectively. After calculating adjacency, TOM, dissTOM, hierarchical clustering, cutting branches and eventually merging close clusters, 39 modules for MWCNT_1, 48 modules for MWCNT_6, and 28 modules for MWCNT_12 were identified. Fig. 1a-c demonstrates the cluster dendrogram and modules before and after merging in which an inimitable color is ascribed to each module. In the dendrogram, the short vertical lines correspond to a gene. Moreover, branches of the dendrogram group show the densely interconnected and also highly co-expressed genes. The gene modules are related to the branches of the resulting dendrogram after merging the closed branches.
Identification of specific modules in the MWCNT-exposed samples
To detect the specific non-preserved modules of three MWCNT-exposed groups, the Zsummary score and medianRank for each module were computed. The specific modules were those that have Zsummary < 2 and medianRank ≥ 8, in which turquoise, violet, steelblue, paleturquoise, darkgrey, orange, skyblue3, ivory, white, lightyellow, darkred, lightcyan, lightsteelblue1, mediumpurple3, royalblue, darkorange, darkorange2, orangered4, yellowgreen, greenyellow, grey60, darkolivegreen, darkturquoise were specific in MWCNT_1m; lightcyan, lightgreen, darkslateblue, paleturquoise, thistle2, sienna3, violet, tan, brown, darkgrey, skyblue, darkorange2, brown4, orangered4, salmon, darkseagreen4, palevioletred3, lightsteelblue1, saddlebrown, honeydew1, skyblue3, darkolivegreen, darkred, thistle1, white in MWCNT_6m; and darkmagenta, steelblue, lightcyan, darkred, orange, darkturquoise, greenyellow, saddlebrown, royalblue, paleturquoise, darkgreen, lightyellow, darkolivegreen, white, pink, darkgrey, darkorange, cyan, grey60, purple, turquoise, gold, brown in MWCNT_12m (Supplementary data 1). Next, the correlations between the gene expression in each module and different doses of 1, 10, 40, 80 µg MWCNT were measured. Afterward, the module-trait analysis to find dose-related modules was performed. Fig S1, S2 and S3 represents the module-trait relationships in which p-value < 0.1 determines the modules that remarkably are correlated with individual traits [27]. The module-trait heatmap indicates the correlations of the module eigengenes with traits. The higher correlation shows the increasing eigengene with the increasing trait. In a signed network, it shows that genes of a module follow a similar pattern to increasing expression with increasing trait values. The modules that were significantly correlated to each dose of MWCNT are mentioned in Table 1. This table actually shows that which co-expressed genes as modules are mostly related to the exposure of MWCNT in various doses.
Protein–protein interactions (PPIs) and pathway enrichment analysis
The PPIs between the genes in each non-preserved module was determined through STRING database. In Table 1, the specific non-preserved and dose-related modules which also had connected PPI are specified as bold and mentioned in Supplementary data 2. These modules contain the genes that were co-expressed after the MWCNT exposure. In the next step, the differentially expressed genes (DEGs) were determined for each dose of MWCNT in different periods using Limma package considering adj.p.val < 0.05 and |logFC|> 3 (Supplementary data 3). Afterward, the common genes between DEGs and connected proteins in non-preserved modules were identified (Table 2). The further pathways analysis reveals that the common major genes between co-regulated genes and DEGs dysregulate the pathways related to the metabolism of proteins, immune system, and inflammation (Table 3). From this analysis, we found that although the increase of dose and the period of exposure to MWCNT may lead to dysregulation of more immune and sometimes cancer-related pathways, they affect similar pathways regardless of the MWCNT amount and time of exposure. However, it was found that pathways were disrupted by the function of different differentially expressed and co-expressed genes.
Discussion
One of the major challenging in the recent decade is the adverse effects of nanomaterials like MWCNT on the environment and human health. The increasing application of MWCNT in the industry makes the concern about the detrimental consequences of exposure. MWCNT has similar pathogenicity effects to asbestos fibers such as carcinogenic and profibrotic risk due to their resembling structures [28]. However, there is rare information about the dysregulated proteins and pathways due to MWCNT.
In this study, we explored toxicity pathways which are defined as the cellular response pathways that lead to a detrimental health effect when adequately perturbed. Systems toxicology provides a useful approach to determine the association between toxicity and the changed expression of a set of genes. One of the procedures to investigate the systems toxicology is finding co-expressed genes which contribute to a biological process. Moreover, a toxicity pathway can be determined through the identification of a set of co-expressed genes that are activated in response to the MWCNT exposure [29]. To determine these functional pathways, the weighted gene co-expression network and differentially expressed gene analyses were utilized.
The pathway enrichment analysis disclosed that different doses of MWCNT cause the activation of inflammation-, immune-, and carcinogenic-related pathways including Antigen processing: Ubiquitination & Proteasome degradation, Class I MHC mediated antigen processing & presentation, chemokine and cytokine signaling pathway, TGF-beta Signaling Pathway, MAPK family signaling cascades, apoptosis, immune system, oxidative stress, Transcriptional Regulation by TP53, VEGFA-VEGFR2 Signaling Pathway as well as metabolic pathways. In the following, we discuss the involvement of various proteins in the mentioned activated pathways due to MWCNT exposure.
There are several studies that reported the cytotoxicity effect of carbon nanotubes with disruption of the immune system [30,31,32,33]. The immune system is extensively affected by the MWCNT, which is concluded by the activation of Adaptive Immune System (UBE2B, KBTBD8), Innate Immune System (RHOA, CTSB, RAC1, ACTB, UBE2L6), and Cytokine Signaling in Immune system (RAP1A and ITGB1). Antigen processing: Ubiquitination & Proteasome degradation and Class I MHC mediated antigen processing & presentation are the pathways that were mainly activated by deregulation of ACTB, PSME3, UBE2B, UBE2K, and KBTBD8. It has been found that the designed nanomaterial may promote antigen processing and presentation utilizing a dual-trafficking route: cross-presentation and exogenous pathway [34]. Moreover, the proteasome system has a substantial efficacy in immune regulation through different mechanisms containing MHC class I antigen processing and regulation of inflammation/cytokine production [35].
It also has been disclosed that MWCNT and SWCNT induce the secretion of inflammatory factors, chemokines, and growth factors such as transforming growth factor (TGF)-b1 and tumor necrosis factor (TNF)-a in mouse macrophages [33, 36,37,38]. In this study, various inflammation pathways were also enriched with the involvement of several hub genes related to different doses of MWCNT including Inflammation mediated by chemokine and cytokine signaling pathway (RHOA, RAC1, ACTB, MYH9), TGF-beta Signaling Pathway (RHOA, RAC1, ITGB1), MAPK family signaling cascades (PSME3 and RAP1A), Oncogenic MAPK signaling (RAP1A), Transcriptional Regulation by TP53 (PRDX2 and BNIP3L), and Neutrophil degranulation (YPEL5). MWCNT induces inflammation and fibrosis through the liberalization of inflammatory cytokines from macrophages [39]. The MAPK pathway is a significant signal transduction pathway that governs a series of events that persuade gene expression relevant to inflammation, apoptosis, and fibrosis. This pathway may also be induced by carbon nanotube exposure [40]. MWCNT can also activate the TGF-β signaling pathway and induce the TGF- 1 production in macrophages, fibroblasts, and epithelial cells [41].
The carcinogenesis and genotoxic effects of CNTs such as oxidation of DNA base, breaking DNA strand, and also clastogenic and aneugenic effects have also been reported [42]. Herein, the dysregulation of pathways including DNA repair (PSME3 and ACTB), DNA Damage Bypass (UBE2B), and p53-Dependent G1 DNA Damage Response (PSME3) by the effect MWCNT were also observed. DNA damage induced by MWCNTs can be developed through downregulation of related genes which helps to the development of carcinogenicity [43]. Another enriched cancer-related pathway is VEGFA-VEGFR2 Signaling Pathway which was enriched by RAP1A, SH3BGRL3, and ITGB1. It is the main pathway that activates angiogenesis by inducing the survival, proliferation, and migration of endothelial cells. The recent studies also showed the modulation of CNT on the proliferation of various types of cells in animals [44].
Previous studies demonstrated that apoptosis could be induced in mitochondria by MWCNTs, possibly through two major mechanisms, including oxidative stress and mitochondrial membrane potential. Oxidative stress is implicated through the liberation of pro-inflammatory mediators [45]. The oxidative stress toxicity may also induce apoptosis by SWCNTs, leading to activating signals of p53-mediated DNA damage checkpoint and then apoptosis. Apoptosis is one of the enriched pathways by different doses of MWCNT through various pathway mechanisms including TNF-type receptor Fas induces apoptosis on ligand binding (S100A10), intrinsic apoptotic (BNIP3L), Regulation of Apoptosis (PSME3), PTEN dependent cell cycle arrest and apoptosis (ITGB1), and Apoptosis (CTSB and ACTB) [46, 47]. Oxidative stress may also be induced by the disturbed amino acids and the involved pathways. It has been found that nano-TiO2 can disrupt the metabolism of amino acids, inhibit the RNA and DNA synthesis, and damage energy production [48]. Similarly, the dysregulation of metabolism of proteins (ALAS2, UBE2B, ACTB), as well as various pathways related to degradation and modification of amino acids such as glycine, serine and threonine metabolic (ALAS2), Lysine degradation (EZH2), Metabolism of amino acids and derivatives (PSME3), and also adenosine ribonucleotides de novo biosynthesis (ATP5H) were observed. Moreover, the synthesis and ubiquitination of proteins are largely affected by changes in the expression levels of UBE2B, UBE2K, KBTBD8, PSME3, and ACTB [49, 50]. CNTs in the body can increase the level of free radicals leading to oxidative stress and oxidation of DNA, proteins, and lipids [51]. ROSs increase the oxidation of amino acids, inactivation of enzymes, and apoptosis [7, 51].
The nephrotoxicity of nanoparticles has also been reported. Some NPs may cause mitochondria and cell membrane perturbation as well as disturbance of the energy metabolism in rat kidneys [52]. Likewise, the dysregulation of pathways including glomerulonephritis and Genes controlling nephrogenesis (ITGB1), Nephrin/Neph1 signaling in the kidney podocyte (RAC1), and Nephrotic syndrome (MYH9) were also determined in this study due to the cytotoxicity effect of MWCNT.
Adipogenesis is the process by which adipocytes develop and accumulate as adipose tissue at different sites in the body. There are several studies that reported graphene and graphene oxide inhibited and enhanced adipogenesis, respectively [53]. Indeed, graphene oxide has a strong affinity towards insulin, which ultimately induces adipogenesis. We also found the dysregulation of adipogenesis pathway by deregulation of Mbnl1.
Moreover, the modulatory effect of carbon nanomaterials on stem cell differentiation has been reported [54]. The activation of Hematopoietic Stem Cell Differentiation pathway was also observed by deregulation of RIOK3. Hemostasis was also activated by the change in the expression levels of GP6, S100A10, RAP1A, and ITGB1 due to the MWCNT exposure. It is in agreement with the previous reports regarding the effect of nanomaterials in developing hemostasis [55, 56].
Generally, we did not find a comprehensive association between various doses of MWCNTs and the resulted dysregulated pathways. However, disparate genes participate in the dysregulation and activation of pathways. Our study has some limitations. The co-expressed gene groups were recognized by analysis of the mouse microarray dataset. Further in vitro studies must be carried out to validate the identified genes associated with MWCNT. Moreover, the analysis of more samples improves the reliability of WGCNA analysis and possibly finding the dose-related genes. The outcomes of WGCNA may technically be biased because of tissue contamination. However, this work partially clarifies the gene groups affected by MWCNT that should be considered for study the affected biological pathways.
Conclusion
The outcomes of this study reveal that regardless of the exposure dose, MWCNTs can induce genotoxic and carcinogenic effects. Although our study has some limitations it introduces several novel dysregulated genes that have important roles in the dysregulation of carcinogenic- and cytotoxic- related pathways. These genes should be further validated in the human cells in a large sample size.
Methods
Dataset and preprocessing
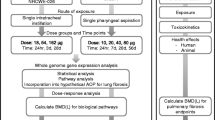
A gene expression microarray dataset with accession number GSE126959 [57] was downloaded from NCBI Gene Expression Omnibus (GEO) database. It contains the microarray gene expression profiles in mouse blood after 1-month (29 samples), 6-month (32 samples), and 12-month (35 samples) aspiration exposure to different doses (1, 10, 40, and 80 µg) of MWCNT. The gene expression profile of dispersion media (DM) control was also measured after one month (7 samples), six months (8 samples), and twelve months (9 samples). The MWCNT doses in mice were chosen to approximately be equivalent to the human occupational exposures [57]. For example, 10 µg MWCNT exposure in mice is almost equal to the deposition for a person doing work for about one month in a workplace with MWCNT aerosol of 400 µg/m3 or 9 months to 7.5 years with 4–40 µg/m3. Therefore, the employed MWCNT doses for mice approximately simulate the human occupational exposures to MWCNT. A total of 24 525 genes were firstly considered for analysis. The dataset was quantile normalized and log2 transformed. The flowchart containing the suggested procedure is depicted in Fig. 2.
Construction of weighted co-expression network and identification of modules
In order to construct the weighted co-expression networks, the WGCNA package in the R environment was employed [18]. Firstly, the “goodSamplesGenes” function was used to remove genes with exceeding missing values. After preprocessing with goodSamplesGenes function and removing unqualified genes, 24 378, 24 466, and 24 497 genes were considered in MWCNT_1m, MWCNT_6m, and MWCNT_12m groups for further analyses. Next, the outlier samples were explored using “hclust” function. Next, the “pickSoftThreshold” function was applied to find scale-free topology fitting indices (R2) versus various soft thresholding powers β. The threshold of R2 ≥ 0.8 was considered to choose the β value. Afterward, the pairwise correlation between genes was computed and then transformed into an adjacency matrix. Topological Overlap Matrix (TOM) and dissimilarity of TOM (1-TOM) were then determined. After that, the “hclust” function was employed to generate a hierarchical clustering tree (dendrogram) of genes based on dissTOM. The “cutreeDynamic” function was then used to cut the branches. Next, module eigengene (ME) that is the first principal component of a given module, was determined. The “mergeCloseModules” function was then utilized to merge close modules considering the height threshold of 0.25 [58].
Identification of non-preserved modules
Through module preservation analysis, the modules that were non-preserved in the DM control were determined. To this end, the “modulePreservation” function in WGCNA package and permutation-based statistics to determine Zsummary and medianRank scores were used. The Zsummary measures both aspects of density and connectivity preservation [59, 60]. Generally, a module with lower Zsummary and higher medianRank has a low tendency to be preserved. Here, a module with Zsummary < 2 and medianRank ≥ 8 was interpreted as non-preserved, a module with 2 < Zsummary ≤ 8 and medianRank < 8 was considered as semi-preserved, and a module with Zsummary > 10 and medianRank < 8 was defined as highly preserved [61]. For the modulePreservation function, some parameters (nPermutations = 200, maxModuleSize = 100) were set, whereas others were left as default.
Construction of module–trait association
The correlation between the traits (various doses of MWCNT) and module eigengenes (MEs) was determined as the module–trait association. To this end, “cor “ and “corPvalueStudent” functions in WGCNA package were employed. The p-value < 0.1 was considered to find meaningful associations.
Protein–protein interactions (PPIs) and enrichment analysis
The biological associations between proteins were identified utilizing the STRING database [62]. To this purpose, the proteins were submitted in STRING and the interactions.
with a combined score > 0.4 were considered as the cut-off criterion. In order to find the biological pathways enriched by hub genes in each module, ToppGene webtool was utilized [63]. To this end, ToppFun tools was used. It discovers the functional enrichment of input genes according to Transcriptome. Top pathway terms with a p-value < 0.05 were considered for further interpretations.
Availability of data and materials
The datasets used and/or analyzed during the current study are available at GEO database: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE126959. Programming language: R. Other requirements: R environment. R. Packages: GEOquery, Limma. Tested on R version 3.6.1.
References
X Zhao S Chang J Long J Li X Li YJF Cao 2019 The toxicity of multi-walled carbon nanotubes (MWCNTs) to human endothelial cells: The influence of diameters of MWCNTs Food Chem Toxicol 126 169 177
RK Newton M Aardema J Aubrecht 2004 The utility of DNA microarrays for characterizing genotoxicity Environ Health Perspect 112 4 420 422
J Aubrecht E Caba 2005 Gene expression profile analysis: an emerging approach to investigate mechanisms of genotoxicity Pharmacogenomics 6 4 419 428
Ghobadi MZ, Afsaneh E, Ghourchian HJTEHoNR, Safety, ELSI, Commercialization. Nanotechnology: Occupational Health Hazards of Nanoparticles and Legalization Challenges. 2020:113–34
LM Sargent DW Porter LM Staska AF Hubbs DT Lowry L Battelli 2014 Promotion of lung adenocarcinoma following inhalation exposure to multi-walled carbon nanotubes Particle and fibre toxicology 11 1 1 18
K Donaldson FA Murphy R Duffin CAJP Poland 2010 Asbestos, carbon nanotubes and the pleural mesothelium: a review of the hypothesis regarding the role of long fibre retention in the parietal pleura, inflammation and mesothelioma Particle and fibre toxicology 7 1 1 17
CA Poland R Duffin I Kinloch A Maynard WA Wallace A Seaton 2008 Carbon nanotubes introduced into the abdominal cavity of mice show asbestos-like pathogenicity in a pilot study Nat Nanotechnol 3 7 423 428
P Møller NR Jacobsen JK Folkmann PH Danielsen L Mikkelsen JG Hemmingsen 2010 Role of oxidative damage in toxicity of particulates Free radical research 44 1 1 46
KL Aillon Y Xie N El-Gendy CJ Berkland ML Forrest 2009 Effects of nanomaterial physicochemical properties on in vivo toxicity Advanced drug delivery reviews 61 6 457 466
J Nakanishi Y Morimoto I Ogura N Kobayashi M Naya M Ema 2015 Risk assessment of the carbon nanotube group Risk Analysis 35 10 1940 1956
AA Shvedova E Kisin AR Murray VJ Johnson O Gorelik S Arepalli 2008 Inhalation vs. aspiration of single-walled carbon nanotubes in C57BL/6 mice: inflammation, fibrosis, oxidative stress, and mutagenesis American Journal of Physiology-Lung Cellular and Molecular Physiology 295 4 L552 L65
ED Kuempel M-C Jaurand P Møller Y Morimoto N Kobayashi KE Pinkerton 2017 Evaluating the mechanistic evidence and key data gaps in assessing the potential carcinogenicity of carbon nanotubes and nanofibers in humans Crit Rev Toxicol 47 1 1 58
Fukushima S, Kasai T, Umeda Y, Ohnishi M, Sasaki T, Matsumoto MJjoOH. Carcinogenicity of multi-walled carbon nanotubes: challenging issue on hazard assessment. journal of Occupational Health. 2017:17–0102.
L Gaté KB Knudsen C Seidel T Berthing L Chézeau NR Jacobsen 2019 Pulmonary toxicity of two different multi-walled carbon nanotubes in rat: Comparison between intratracheal instillation and inhalation exposure Toxicol Appl Pharmacol 375 17 31
K Fujita S Obara J Maru S Endoh 2020 Cytotoxicity profiles of multi-walled carbon nanotubes with different physico-chemical properties Toxicol Mech Methods 30 7 477 489
KB Knudsen T Berthing P Jackson SS Poulsen A Mortensen NR Jacobsen 2019 Physicochemical predictors of Multi-Walled Carbon Nanotube–induced pulmonary histopathology and toxicity one year after pulmonary deposition of 11 different Multi-Walled Carbon Nanotubes in mice Basic Clin Pharmacol Toxicol 124 2 211 227
Y Guo J Ma L Xiao J Fang G Li L Zhang 2019 Identification of key pathways and genes in different types of chronic kidney disease based on WGCNA Mol Med Rep 20 3 2245 2257
P Langfelder S Horvath 2008 WGCNA: an R package for weighted correlation network analysis BMC Bioinformatics 9 559
M Wang L Wang L Pu K Li T Feng P Zheng 2020 LncRNAs related key pathways and genes in ischemic stroke by weighted gene co-expression network analysis (WGCNA) Genomics 112 3 2302 8
MM Saint-Antoine A Singh 2020 Network inference in systems biology: recent developments, challenges, and applications Current opinion in biotechnology 63 89 98
I Karkossa A Bannuscher B Hellack A Bahl S Buhs P Nollau 2019 An in-depth multi-omics analysis in RLE-6TN rat alveolar epithelial cells allows for nanomaterial categorization Particle and fibre toxicology 16 1 1 19
P Falagan-Lotsch CJJN Murphy 2020 Network-based analysis implies critical roles of microRNAs in the long-term cellular responses to gold nanoparticles Nanoscale 12 41 21172 21187
X Yin P Wang T Yang G Li X Teng W Huang 2021 Identification of key modules and genes associated with breast cancer prognosis using WGCNA and ceRNA network analysis Aging (Albany NY) 13 2 2519
Wang L, Liu X, Yue M, Liu Z, Zhang Y, Ma Y, et al. Identification of hub genes in bladder cancer based on weighted gene co-expression network analysis from TCGA database. 2021:e1557.
MZ Ghobadi S-H Mozhgani M Farzanehpour F Behzadian 2019 Identifying novel biomarkers of the pediatric influenza infection by weighted co-expression network analysis Virology journal 16 1 1 10
Weirauch MTJAsfnbmisb. Gene coexpression networks for the analysis of DNA microarray data. 2011;1:215–50.
AS Cesar LC Regitano JM Reecy MD Poleti PS Oliveira GB Oliveira de 2018 Identification of putative regulatory regions and transcription factors associated with intramuscular fat content traits BMC Genomics 19 1 499
LC Smith S Moreno S Robinson M Orandle DW Porter D Das 2019 Multi-walled carbon nanotubes inhibit estrogen receptor expression in vivo and in vitro through transforming growth factor beta1 NanoImpact 14 100152
P Schyman RL Printz SK Estes KL Boyd M Shiota A Wallqvist 2018 Identification of the toxicity pathways associated with thioacetamide-induced injuries in rat liver and kidney Frontiers in Pharmacology 9 1272
K Donaldson R Aitken L Tran V Stone R Duffin G Forrest 2006 Carbon nanotubes: a review of their properties in relation to pulmonary toxicology and workplace safety Toxicological sciences : an official journal of the Society of Toxicology 92 1 5 22
G Jia H Wang L Yan X Wang R Pei T Yan 2005 Cytotoxicity of carbon nanomaterials: single-wall nanotube, multi-wall nanotube, and fullerene Environ Sci Technol 39 5 1378 1383
HJ Johnston GR Hutchison FM Christensen S Peters S Hankin K Aschberger 2010 A critical review of the biological mechanisms underlying the in vivo and in vitro toxicity of carbon nanotubes: The contribution of physico-chemical characteristics Nanotoxicology 4 2 207 246
J Dong Q Ma 2015 Advances in mechanisms and signaling pathways of carbon nanotube toxicity Nanotoxicology 9 5 658 676
T Mocan C Matea F Tabaran C Iancu R Orasan L Mocan 2015 In vitro administration of gold nanoparticles functionalized with MUC-1 protein fragment generates anticancer vaccine response via macrophage activation and polarization mechanism Journal of Cancer 6 6 583
EJAM Sijts PM Kloetzel 2011 The role of the proteasome in the generation of MHC class I ligands and immune responses Cell Mol Life Sci 68 9 1491 1502
AA Shvedova ER Kisin R Mercer AR Murray VJ Johnson AI Potapovich 2005 Unusual inflammatory and fibrogenic pulmonary responses to single-walled carbon nanotubes in mice Am J Physiol Lung Cell Mol Physiol 289 5 L698 708
X He SH Young D Schwegler-Berry WP Chisholm JE Fernback Q Ma 2011 Multiwalled carbon nanotubes induce a fibrogenic response by stimulating reactive oxygen species production, activating NF-κB signaling, and promoting fibroblast-to-myofibroblast transformation Chem Res Toxicol 24 12 2237 2248
He X, Young S-H, Fernback JE, Ma Q. Single-Walled Carbon Nanotubes Induce Fibrogenic Effect by Disturbing Mitochondrial Oxidative Stress and Activating NF-κB Signaling. J Clin Toxicol. 2012;Suppl 5))5.
TA Brown JW Lee A Holian V Porter H Fredriksen M Kim 2016 Alterations in DNA methylation corresponding with lung inflammation and as a biomarker for disease development after MWCNT exposure Nanotoxicology 10 4 453 461
JW Lee H-M Kang E-J Won D-S Hwang D-H Kim S-J Lee 2016 Multi-walled carbon nanotubes (MWCNTs) lead to growth retardation, antioxidant depletion, and activation of the ERK signaling pathway but decrease copper bioavailability in the monogonont rotifer (Brachionus koreanus) Aquatic Toxicology 172 67 79
M Polimeni GR Gulino E Gazzano J Kopecka A Marucco I Fenoglio 2016 Multi-walled carbon nanotubes directly induce epithelial-mesenchymal transition in human bronchial epithelial cells via the TGF-β-mediated Akt/GSK-3β/SNAIL-1 signalling pathway Part Fibre Toxicol 13 1 27
D Berlo van MJ Clift C Albrecht RP Schins 2012 Carbon nanotubes: an insight into the mechanisms of their potential genotoxicity Swiss medical weekly 142 w13698
A Panek M Błażewicz A Frączek-Szczypta J Adamczyk J Wiltowska-Zuber CJAPPA Paluszkiewicz 2018 Applications of comet assay for the evaluation of genotoxicity and DNA repair efficiency in nanomaterials research Acta Phys. Pol. A 133 2 280 282
G Vietti S Ibouraadaten M Palmai-Pallag Y Yakoub C Bailly I Fenoglio 2013 Towards predicting the lung fibrogenic activity of nanomaterials: experimental validation of an in vitro fibroblast proliferation assay Particle and Fibre Toxicology 10 1 1 14
A Mandegary A Saeedi A Eftekhari V Montazeri E Sharif 2013 Hepatoprotective effect of silyamarin in individuals chronically exposed to hydrogen sulfide; modulating influence of TNF-α cytokine genetic polymorphism Daru 21 1 28
B Ren B Jia X Zhang J Wang Y Li H Liang 2021 Influence of multi-walled carbon nanotubes on enantioselective bioaccumulation and oxidative stress toxicity of indoxacarb in zebrafish (Danio rerio) Chemosphere 267 128872
J Yuan H Gao CB Ching 2011 Comparative protein profile of human hepatoma HepG2 cells treated with graphene and single-walled carbon nanotubes: an iTRAQ-coupled 2D LC–MS/MS proteome analysis Toxicology letters 207 3 213 21
Y Bo C Jin Y Liu W Yu H Kang 2014 Metabolomic analysis on the toxicological effects of TiO2 nanoparticles in mouse fibroblast cells: from the perspective of perturbations in amino acid metabolism Toxicology mechanisms and methods 24 7 461 9
SF Thai KA Wallace CP Jones H Ren E Grulke BT Castellon 2016 Differential genomic effects of six different TiO2 nanomaterials on human liver HepG2 cells Journal of biochemical and molecular toxicology 30 7 331 41
P Jin P Wei Y Zhang J Lin R Sha Y Hu 2016 Autophagy-mediated clearance of ubiquitinated mutant huntingtin by graphene oxide Nanoscale 8 44 18740 18750
D Mohanta S Patnaik S Sood N Das 2019 Carbon nanotubes: Evaluation of toxicity at biointerfaces Journal of pharmaceutical analysis 9 5 293 300
G Yan Y Huang Q Bu L Lv P Deng J Zhou 2012 Zinc oxide nanoparticles cause nephrotoxicity and kidney metabolism alterations in rats Journal of Environmental Science and Health 47 4 577 588
A Halim Q Luo Y Ju GJN Song 2018 A mini review focused on the recent applications of graphene oxide in stem cell growth and differentiation Nanomaterials 8 9 736
M Wei S Li W Le 2017 Nanomaterials modulate stem cell differentiation: biological interaction and underlying mechanisms Journal of Nanobiotechnology 15 1 75
F Salimi F Mohammadipanah 2021 Nanomaterials Versus The Microbial Compounds With Wound Healing Property Frontiers in Nanotechnology 2 21
T Platonova 2019 Silica Nanoparticles Effects on Hemostasis Jenny Stanford Publishing Immune Aspects of Biopharmaceuticals and Nanomedicines 731 752
BN Snyder-Talkington C Dong S Singh R Raese Y Qian DW Porter 2019 Multi-Walled Carbon Nanotube-Induced Gene Expression Biomarkers for Medical and Occupational Surveillance International journal of molecular sciences 20 11 2635
Zarei Ghobadi M, Mozhgani S-H, Erfani Y. Identification of dysregulated pathways underlying HTLV-1-associated myelopathy/tropical spastic paraparesis through co-expression network analysis. Journal of NeuroVirology. 2021.
P Langfelder R Luo MC Oldham S Horvath 2011 Is my network module preserved and reproducible? J PLoS Comput Biol 7 1 e1001057
M Zarei Ghobadi R Emamzadeh M Teymoori-Rad S-H Mozhgani 2021 Decoding pathogenesis factors involved in the progression of ATLL or HAM/TSP after infection by HTLV-1 through a systems virology study Virology Journal 18 1 175
MR Bakhtiarizadeh S Mirzaei M Norouzi N Sheybani MS Vafaei Sadi 2020 Identification of Gene Modules and Hub Genes Involved in Mastitis Development Using a Systems Biology Approach Front Genet 11 722
D Szklarczyk AL Gable D Lyon A Junge S Wyder J Huerta-Cepas 2019 STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets Nucleic acids research 47 D1 D607 D13
J Chen EE Bardes BJ Aronow AG Jegga 2009 ToppGene Suite for gene list enrichment analysis and candidate gene prioritization Nucleic acids research 37 suppl_2 W305 W11
Acknowledgements
Not applicable
Funding
There were no sources of funding for the research.
Author information
Authors and Affiliations
Contributions
SJS: conceptualization, investigation, editing, and revising the manuscript, MZG: supervision, project administration, formal analysis, investigation, writing, and editing the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Salih, S.J., Ghobadi, M.Z. Evaluating the cytotoxicity and pathogenicity of multi-walled carbon nanotube through weighted gene co-expression network analysis: a nanotoxicogenomics study. BMC Genom Data 23, 12 (2022). https://doi.org/10.1186/s12863-022-01031-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12863-022-01031-3