Abstract
Background
Ascites syndrome is a hypertensive, multifactorial, multigene trait affecting meat-type chickens imposing significant economic losses on the broiler industry. A region containing the CPQ gene has been previously identified as significantly affecting ascites phenotype. The region was discovered through whole genome resequencing focused on chicken chromosome 2. The association was confirmed through further genotyping in multiple broiler populations.
Results
The whole genome resequencing analyses have now been extended to the current chicken genome assembly. DNA samples were pooled according to gender and phenotype and the pools subjected to next generation sequencing. Loci were identified as clusters of single nucleotide polymorphisms where frequencies of the polymorphisms differed between resistant and susceptible chickens. The chickens are an unselected line descended from a commercial elite broiler line. Regions identified were specific to one or both genders. The data identify a total of 28 regions as potential quantitative trait loci for ascites. The genes from these regions have been associated with hypertensive-related traits in human association studies. One region on chicken chromosome 28 contains the LRRTM4 gene. Additional genotyping for the LRRTM4 region demonstrates an epistatic interaction with the CPQ region for ascites phenotype.
Conclusions
The 28 regions identified were not previously identified in a multi-generational genome wide association study using 60k Single Nucleotide Polymorphism panels. This work demonstrates the utility of whole genome resequencing as a cost effective, direct, and efficient method for identifying specific gene regions affecting complex traits. The approach is applicable to any organism with a genome assembly and requires no a priori assumptions.
Similar content being viewed by others
Background
Pulmonary hypertension syndrome (PHS) or ascites syndrome (AS) manifests as a complication of selection for rapid growth and high meat yield in broilers. AS is associated with accumulation of fluid in the abdominal cavity leading to death [1,2,3,4,5,6]. AS is a pathophysiological progression in which rapid growing birds are unable to meet the high oxygen (O2) demand for elevated metabolic rates [1,2,3, 6,7,8]. The hypoxic condition stimulates cardiac output leading to constriction of pulmonary arterioles causing hypertension in pulmonary circulation, right ventricular hypertrophy, and eventually right ventricular failure [3, 6, 9]. The primary cause of this disease is unknown, which makes it idiopathic in nature and makes AS an excellent model for human idiopathic arteriole hypertension [6, 10, 11]. AS incidence is influenced through environmental conditions including ventilation [12], lighting [13], temperature [14], feed restriction [1, 15], and diet composition [16,17,18]. However, world-wide economic losses are still estimated to be around 100 million dollars per year [6] (personal communication from Cobb-Vantress Inc. 2015). Many have suggested a genetic component to PHS, since AS related traits such as cardiac hypertrophy, and abdominal fluid, have moderate to high heritabilities [19,20,21,22,23,24,25,26,27]. Identification of quantitative trait loci (QTL) specifically affecting AS could lower economic losses in the broiler industry, with minimal impact on growth rate or meat yield.
Multiple Single Nucleotide Polymorphism (SNP) panel based genome wide association studies (GWAS) were used to identify potential genetic markers on chromosome 1, 2, 4, 9, and Z for this disease [28,29,30,31,32,33]. Unfortunately, further genotyping or marker assisted selection have only shown a marginal/minimal association between these regions and AS (unpublished). A whole-genome-resequencing (WGR) approach to further investigate potential regions on chromosomes 2 and 9 [29] did not support the previous regions but did identify a new region on chromosome 2 (127.65 – 127.75 Mbp; November 2017 assembly 5 coordinates) spanning part of the gene for plasma glutamate carboxypeptidase (PGCP or CPQ). GWAS data associates this gene with electrocardiogram, hypertension and blood pressure in humans (https://www.ncbi.nlm.nih.gov/gap/phegeni). Additional genotyping in a broiler research line, and three commercial broiler lines, demonstrated an association of homozygotes for the non-reference SNPs in this region with resistance in male birds. The CPQ gene region is the first demonstrated marker for resistance to AS in broilers [34]. The WGR investigation has now been extended to the rest of the genome to identify 27 additional regions as potential Quantitative Trait Loci (QTLs) for AS. Additional genotyping for one these new regions (LRRTM4) confirmed an association with AS. In addition, the data support a strong epistatic interaction between LRRTM4 and CPQ in contributing to AS phenotype.
Results
Whole Genome Resequencing and Templated Assembly
Next-generation WGR data was generated for eight pools of 10 individual DNA samples representing two biological replicates for each gender for both AS resistant and susceptible birds from the unselected, Relaxed (REL) line. The REL is descended from an elite commercial line [35]. Each pool was sequenced to >66x coverage using paired-end (2 x 125-bp) Illumina sequencing. FASTQ reads for each pool were mapped onto the May 2018 GRCg6a reference genome. The average read counts for all pools was 534,902,802±6,325,923, with 93.4±0.2% reads successfully assembled onto the reference genome. Potential SNPs identified by the read mapping totaled 12,024,469 in male pools and 11,933,041 in female pools. SNP counts were recorded, and SNP densities plotted for each chromosome according to gender (Fig. 1). The SNP density was observed to be higher for some of the microchromosomes in the assemblies for both genders. Whereas the average chromosomal SNP density is 1.36±0.61, SNP densities for microchromosomes 30, 31 and 33, are 2.3 to 3.6 SNPs per 100 bp. Most chromosomes have GC content of ≤ 47%. Chromosomes 30, 31 and 33 have a GC% of 58.7, 52.3, and 53.9, respectively. The current assembly of chromosome 30 is 1.82 Mbp, while chromosomes 31 and 33 are more complete at 6.15 and 7.82 Mbp, respectively. However, the assembly for chromosome 28 is 5.12 Mbp at 53% GC and the SNP density is 1.276/100 bp. This pattern of SNP density by chromosome is consistent with that observed for 3 commercial lines (manuscript in preparation) so it is not unique to the REL line. The SNP density is also not a function of read depth as read depth was roughly equivalent across all the chromosomes.
SNP density plots by chromosome in REL line birds. The SNP density computed per 100 bp is plotted for the entire genome (GRCg6a) or each individual chromosome for male (solid) and female (hatched). GRCg6a is for the entire genome and MT is mitochondrial genome. The insert is a plot of SNP density by GC% for individual chromosomes (excludes mitochondrial genome)
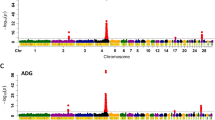
To identify potential QTLs for AS phenotype, the difference between the frequency of the SNP in resistant versus susceptible phenotypes were calculated (resistant SNP frequency - susceptible SNP frequency). The SNP frequency differences were visually inspected using the Integrative Genomics Viewer (IGViewer [36];) according to chromosome and chromosomal location for both genders to visually identify clusters of SNPs with frequencies skewed with respect to phenotype for either or both genders. Specifically, the SNP frequencies were scanned for clusters of SNPs showing frequency differences of >20%. Twenty eight regions on 13 chromosomes were detected showing clusters of SNPs differentially represented in the different phenotypes (Table 1). The identified regions included 15 that showed association in both genders, while 8 appeared to be specific to males and 5 to females. The majority of regions showed a higher frequency of non-reference SNPs in the ascites resistant birds, with 8 associated with resistance in both genders, 5 male specific and 3 female specific. The non-reference SNPs were associated with susceptibility (higher frequency in susceptible) for 1 region in both genders, 3 male specific and 2 female specific. There were two regions where the non-reference SNPs were associated with resistance in males and with susceptibility in females, while there were four regions where the reverse was found. The 64 genes identified in these regions (Table 1), were used to search the NCBI Phenotype-Genotype Integrator, (PheGenI; https://www.ncbi.nlm.nih.gov/gap/phegeni) for phenotypes that had been identified in human GWAS as associated with traits possibly contributing to PHS or AS. The most frequent traits from the PheGenI output are presented in Fig. 2. Traits that are of particular interest regarding AS in broilers include: platelet function tests, blood pressure, body mass index, echocardiography, mycocardial infarction, erythrocyte indices, and heart failure. Thus, the human GWAS data supports these 28 regions as potential candidate QTLs for AS.
Human phenotypic traits associated with genes found in the potential QTL regions. Chicken gene names from the regions listed in Table 1 were used to search the human PheGenI database at NCBI. Phenotypes identified were then used to total the number of regions which were associated with that trait in human GWAS studies. Only traits associated with 3 or more regions are listed
LRRTM4 Genotyping in Association with Ascites Syndrome
The region on chromosome 2 for CPQ has already been extensively analyzed for association with ascites phenotype and confirmed for association of the non-reference SNPs with ascites resistance in males [34]. The region on chromosome 22 associated with the LRRTM4 gene was selected for further investigation because an association was found in both genders, with a larger frequency difference in males than females. This region contains 711 successive, high-quality SNPs spanning the 4.400-4.455 Mbp region on Gga22 (Fig. 3). The frequency difference (resistant SNP frequency - susceptible SNP frequency) averages 27% in males and 7% in females. In males 641 SNPs have a positive SNP frequency difference while only 11 are negative. In females there were 261 positive, and 12 negative, for SNP frequency difference. Of these SNPs, 560 covered 45.1 kbp of the 3’end of the LRRTM4 gene which spans 211.6 kbp from 4,409,156 to 4,620,835 Mbp. The LRRTM4 gene encodes the leucine-rich repeat transmembrane neuronal protein 4 which has been suggested to play a role in the regulation of dendritic spine development in the nervous system [37, 38]. NCBI PheGenI associates the human LRRTM4 gene region with traits such as antihypertension, carotid artery disease, coronary heart disease, and pulmonary embolism, supporting a probable association with AS in broilers.
Integrative Genomics Viewer display of SNP frequency difference plots for a possible QTL for ascites syndrome on chromosome 22. Difference in SNP frequency for the alternative phenotypes are plotted according to position (Mbp). Resistant minus Susceptible SNP frequency (Res-Sus SNP frequency) are for the male and female data aligned with the gene annotations showing an association with the 3’ end of the LRRTM4 gene
An exonuclease assay was developed to target two SNPs (4,405,679, and 4,405,681 Mbp) which gave SNP frequency differences of approximately 50% in males and 15% in females. These SNPs are in the intergenic region, 3.5 kbp from the 3’ end of the LRRTM4 gene. The exonuclease assay was used to genotype more than 600 archived DNA samples from REL birds previously phenotyped for AS susceptibility in the hypobaric chamber [29, 33, 34]. The observed genotype frequencies were in agreement with calculated genotype frequencies (computed from allele frequencies) consistent with Hardy Weinberg Equilibrium (HWE) which implies that the quantitative Polymerase Chain Reaction (qPCR) genotyping is valid, there are no significant issues with null alleles, and that the samples of DNA utilized was non-biased. These same DNA samples had been used in the genotype associations with AS for the CPQ gene on chromosome 2 [34]. Based on the WGR SNP frequency plots the expectation was an association of resistance for males homozygous with the non-reference SNPs. Instead there was a significant association of the homozygous non-reference SNPs in females (AG genotype in females in Table 2; adjusted P-value=0.047). There was also a significant adjusted P-value (P=0.0083) for all samples, combining male and female data (Table 2). In both cases the homozygous non-reference SNPs were associated with increased susceptibility (AG genotype higher frequency of susceptible than RR or GA genotypes). The homozygous non-reference increases susceptibility by 19% in the females and 12 % in the entire population.
Male broilers with homozygous non-reference genotype for intron 6 SNPs in CPQ gene have shown approximately 20% higher overall resistance [34]. To test for potential interactions between CPQ and LRRTM4, the genotype data for LRRTM4 with the genotype data for the same samples for the CPQ gene [34] were combined. The combined genotype of heterozygous for LRRTM4 and homozygous non-reference for CPQ (RRCC) genotype was significantly associated with ascites resistance in males (P=0.033), contrary to the LRRTM4 genotype alone. The RRCC genotype was the most abundant genotype in the samples (n=131). Whereas, for most combined genotypes the males were nearly 50:50, 85% of the 61 RRCC males were resistant to AS (Table 3). For CPQ genotypes alone, 72% of male homozygous non-reference males were resistant, so the combination of the two loci may increase resistance over CPQ alone. Additionally, for homozygous reference LRRTM4 (GA), the addition of a heterozygous CPQ also seems to favor resistance, but the P-values are not significant. In summary, the CPQ non-reference homozygous genotype confers approximately 2:1 odds on ascites resistance in males, but the addition of one non-reference allele for LRRTM4 appears to improve the odds to approximately 6:1.
Tissue-specific Expression Evaluation of LRRTM4
LRRTM4 gene expression for each of the homozygous genotypes was measured by reverse transcriptase qPCR (RT-qPCR) for heart, lung, liver, brain, and testis samples. TATA-binding protein (TBP) served as the reference, as TBP is recommended across multiple human tissues [39], recommended for chicken cardio-pulmonary gene expression assays [40], and we have found it to show little variation across multiple chicken tissues [41]. No expression was detected in lung, liver, brain, or testis RNA samples, but expression was observed in the heart. When comparing the level of expression of LRRTM4 between homozygous reference (ΔΔCt ± sd = 6.8 ± 0.8) and homozygous non-reference (ΔΔCt ± sd = 5.7 ± 1.0) genotypes there was no difference in expression between genotypes (P=0.188). The expression data indicates that LRRTM4 in broiler heart is expressed at about 0.016 (0.56) the level as TBP. The RNAseq summary in NCBI for human LRRTM4 (Gene- Full Report) suggests that LRRTM4 expression is primarily restricted to the brain. Human RNAseq RPKM values for LRRTM4 are brain 5.6±1.1, heart 0.05±0.03, liver 0.033±0.012, lung 0.24±0.14, and testis 0.024±0.011. Based on the RPKM values for human TBP and LRRTM4 for these tissues, and an average ΔCt of 15.2±1.5 for TBP in the RT-qPCR analyses from REL broilers, amplification for all these tissues for LRRTM4 was expected within the 25 qPCR cycles employed. The relative expression in human heart of LRRTM4 to TBP is 0.0154 which is in agreement with the ΔΔCt values in heart for chickens. Thus, LRRTM4 expression in lung, liver, brain, and testis, is apparently much lower than the expression observed in human. Further, the two alternative alleles of LRRTM4, distinguished based on SNP differences distal to the 3’ end of the gene, do not appear to significantly differ in expression levels. Therefore, the contribution of LRRTM4 to AS more likely result from differences in the polypeptide sequences, or post-translational processes.
Discussion
The poultry industry has fueled the affordable production of chicken by selecting for economically important traits such as growth rate, feed conversion, and meat yield. This rapid growth and muscle deposition make chicken an excellent model for human idiopathic arteriole hypertension [10]. About 3% of the fast-growing broilers develop idiopathic arteriole hypertension and exposure to stressors such as elevated dietary sodium, cold temperature [8] or high altitude [42] can increase the incidence to 20%.
Multiple investigations have estimated high heritability of AS susceptibility which led our group to pursue the genetic basis of AS [19, 20, 22, 23, 27, 35, 43, 44]. This current study, used whole-genome resequencing in the ascites research line, REL. The REL is descended from a commercial elite line in 1996. In the current study, 28 candidate regions on 13 different chromosomes were identified (Table 1). Previous GWAS studies had identified candidate regions for ascites phenotype [29,30,31,32], however, extensive genotyping and marker-assisted selection revealed minimal, or no, association with the phenotype. Therefore, these earlier candidate regions were not informative QTLs for AS (unpublished). Conversely, the first region identified from an WGR approach contained the CPQ gene on chromosome 2. Additional genotyping confirmed this region as the first verified QTL for ascites in broilers [34]. Human CPQ gene encodes a carboxypeptidase with a function in protein hydrolysis and thyroxine synthesis [45]. Dey et al. [34] showed that the non-reference homozygous genotype for CPQ has a significant association with AS resistance in REL broilers as well as some commercial lines. This is consistent with human GWAS results, where CPQ was associated with traits such as electrocardiography, hypertension, blood pressure, and heart rate (PheGenI database at NCBI).
The CPQ work led to the expansion of the WGR analyses to the rest of the assembled genome. This identified 27 additional regions for ascites phenotype (Table 1). The 28 regions include 64 genes that either reside within or are proximal to the clusters of differentially represented SNPs. The PheGenI database reveals that many of these regions have some association with hypertension, body weight, fat deposition, heart failure, or cardiac hypertrophy in humans (Fig. 2).
Further investigation of one of these regions on chromosome 22 associated with the LRRTM4 gene supported an association with AS phenotype. LRRTM4 is a member of the leucine-rich-repeat (LRR) transmembrane protein family, whose members are involved in synapse development and maintenance of the nervous system [37]. The LRRTM4 non-reference homozygote shows an association with ascites susceptibility in REL line females. Surprisingly, the combined genotypic data for CPQ and LRRTM4 revealed a male specific, epistatic interaction between the LRRTM4 heterozygote and the intron-6 non-reference homozygote of CPQ. The interaction increased the resistance in males as compared to CPQ or LRRTM4 alone. In combination, these genes can be employed in marker-assisted selection for increasing ascites resistance in the broiler industry. However, the exact mechanisms by which these genes contribute to resistance in broilers are still unknown. Studies on understanding the gene networks, downstream regulation, or interactions between the CPQ and LRRTM4 protein products in chicken are warranted.
None of the current regions reported above have appeared in any previous GWAS studies on AS [30,31,32,33]. Identification of the CPQ gene [34] and now LRRTM4 demonstrates WGR to be an effective and robust method for high-resolution mapping of QTLs for complex traits. In traditional GWAS, the panel of SNPs utilized must be pre-specified and limited in numbers. Conversely, WGR identifies the SNPs segregating in a population and can detect tens of millions of SNPs. In the REL line WGR identified more than 12 million SNPs despite the population being closed for more than 18 generations. With decreasing costs in library construction and sequencing, coupled with increasing NGS output, the WGR approach is highly cost-effective and provides far higher resolution than conventional SNP-panel-based GWAS.
Conclusions
CPQ and LRRTM4 are gender-biased markers for ascites resistance in broilers. MAS studies are in progress to confirm the contributions of these two gene regions for ascites syndrome resistance. The additional 26 regions are available for further investigation as potential QTLs associated with ascites syndrome. The current findings may also be relevant to pursuit of the genetics underlying idiopathic pulmonary arteriole hypertension, as well as other forms of hypertension, in humans.
Methods
Reference genome
All genomic positions presented are relative to the May 2018 assembly of the Gallus gallus genome (RefSeq accession ID: GCA_000002315.5; GRCg6a).
Bird stocks and hypobaric chamber trials
All animal procedures were approved by the University of Arkansas Institutional Animal Care and Use Committee under protocols 15039 and 15040. Birds utilized originated from a research line (REL) representing the 18th generation of unselected descendants from an elite commercial broiler stock [1, 8, 35]. The REL line is maintained at the University of Arkansas Poultry Research Farm and all live animal work was performed at that facility. At hatch 600 chicks were wing-banded, and 10 μl of blood was collected via wing vein lancet puncture of chicks for DNA isolation. The birds were then challenged for six weeks using a hypobaric chamber set at 9000 ft (543 mm of Hg) above sea level [35]. Mortality data were recorded daily. All deceased birds were necropsied to determine gender and cause of death. The birds were designated ascitic/susceptible when there was excessive abdominal fluid, flaccid heart, liver lesions, and right ventricle hypertrophy (right ventricle to total ventricle; RV/TV > 0.5). At six weeks, all survivors were euthanized by cervical dislocation and necropsied for gender and ascites phenotype. Birds with normal necropsy characters were scored as resistant.
Genomic DNA isolation and purification
Genomic DNAs were isolated using a rapid isolation protocol [46] and archived at -20 oC. DNAs for next-generation sequencing were further purified by phenol-chloroform extraction, chloroform extraction, and ethanol precipitation. DNAs were quantified by Hoechst 33258 fluorescence in a GloMax (Promega Corp., Madison, WI).
WGR of genomic DNA
Duplicate pools of equal weights of 10 DNAs from each phenotype were pooled for each gender to construct 8 total pools, which were then submitted for Next Generation Sequencing library preparation. Libraries were sequenced, 2x125 bp paired-end on an Illumina HiSeq 2500, to generate approximately 66 Gb per library. Library construction and sequencing were performed by the Research Technology Support Facility at Michigan State University (East Lansing, MI).
Data analyses and bioinformatics
The adapter-trimmed FASTQ sequence reads were mapped onto the May 2018 chicken genome assembly (Galgal6) using SeqMan NGen (Lasergene Suite 16; DNAStar, Madison, WI). Specific settings in NGen were SNP filter high, diploid genome, whole genome pipeline, and Illumina adapter scan. All other parameters were the default settings. The pools were coded as resistant (R) or susceptible (S), male (M) or Female (F), and biological replicate pools as 1 or 2 (i.e., RM1, RM2, SM1, SM2, RF1, RF2, SF1, SF2). Separate templated alignments for each pool were used for SNP identification and tabulation in ArrayStar (Lasergene Suite). The SNP data were exported to Excel for further analyses. The difference in the averages of SNP frequencies of replicates for each phenotype and gender were calculated and exported to .seg files for viewing in Integrative Genomics Viewer 2.7.4 [36] by chromosome, aligned to the annotated genome from NCBI.
Exonuclease assay based genotyping
Primers and probes for exonuclease genotyping (Table 4) were designed using Primer3 (http://bioinfo.ut.ee/primer3-0.4.0/primer3/), and synthesized by Integrated DNA Technologies (IDT; Coralville, IA). Conditions for qPCR exonuclease assays were optimized for each SNP locus and then used to genotype individual DNAs from additional birds phenotyped for ascites in the hypobaric chamber. DNAs used were the same as those used previously for genotyping for CPQ [34].
Reactions (20μl) included 1x Taq buffer (50 mM Tris-Cl pH 8.3, 1 mM MgCl2, 30 μg/ml BSA), 0.2 mM MgCl2, 0.2 mM dNTP, 0.2 μM each forward and reverse primers, 0.05 μM each probe, 2.5 units Taq polymerase, and 2 μl DNA. cycling was denaturation at 90°C 3 mins, 10 cycles of 90°C 15s, 55°C 15s, 72°C 1 min, followed by another 30 cycles of 90°C 15s, 55°C 15s, 72°C 1 min with plate read. To verify qPCR genotype calls, relevant qPCR products were purified using RapidTip (Diffinity Genomics, West Chester, Pennsylvania) and quantified by Hoechst 33258 fluorescence as above. DNAs were submitted for capillary sequencing by Eurofins MWG Operon (Louisville, KY). Sequence data (.ab1 files) were aligned to the reference Jungle Fowl sequence using SeqMan Pro software (DNASTAR) for SNP scoring.
Gene expression analyses
Specific gene expression was assayed by RT-qPCR for heart, liver, brain, lung, and testis RNA previously extracted from broiler tissue samples [34]. Total RNA (500 ng) was added to a mastermix consisting of 1x First Strand Buffer (Promega Corp.), 0.5 μM dNTPs, 1 μM CT23V primer and 100 U MMLV Reverse Transcriptase (Promega Corp.). The mixtures were incubated at 50°C for 50 min and the reaction was terminated at 65°C for 5 min. The first strand cDNA was then diluted into a qPCR mixture (as described above) containing 1x EvaGreen dye (Biotium Inc., Fremont, CA). Cycling was as for exonuclease assays (above) but were followed by a high resolution melt curve from 65 to 90oC at 0.1oC steps. RT-qPCRs were run in triplicate with TATA-box binding protein (TBP) gene as the reference, and all ΔΔCt values were relative to TBP [39]. Fold change was calculated using the ΔΔCt method [47].
Statistical methods
Genotype frequencies from exonuclease assays were separately calculated for each phenotype and gender. The expected genotype counts for each phenotype were computed from the observed genotype frequencies from the entire population. Microsoft Excel chi-square test function was performed on the count versus expected to calculate P-values for allele/genotype frequencies greater than 10%. P-values were then multiplied by the number of alleles/genotypes to generate a simple Bonferroni adjusted P-value. Deviation from expected was considered statistically significant where adjusted P-values < 0.05. Gene expression differences for alleles were based on P-values from Excel t-tests (one way with unequal variance).
Availability of data and materials
Sequence data has been deposited in NCBI under BioSample accessions SAMN07312781, SAMN07327525, SAMN07327526, SAMN07327527, SAMN07327528, SAMN07327529, SAMN07327530, and SAMN07327531.
Abbreviations
- AS:
-
Ascites syndrome
- GWAS:
-
Genome wide association study
- HWE:
-
Hardy weinberg equilibrium
- Mbp:
-
Mega base pair
- NCBI:
-
National center for biotechnology information
- PGCP:
-
Plasma glutamate carboxy peptidase
- PHS:
-
Pulmonary hypertension syndrome
- qPCR:
-
Quantitative polymerase chain reaction
- QTL:
-
Quantitative trait locus
- RT-qPCR:
-
Real time quantitative polymerase chain reaction
- RV/TV:
-
Right ventricle to total ventricle
- SNP:
-
Single nucleotide polymorphism
- WGR:
-
Whole genome resequencing
References
Balog JM, Anthony NB, Cooper MA, Kidd BD, Huff GR, Huff WE, Rath NC. Ascites syndrome and related pathologies in feed restricted broilers raised in a hypobaric chamber 1. Poult Sci. 2000;79(3):318–23.
Huchzermeyer F, De Ruyck A. Pulmonary hypertension syndrome associated with ascites in broilers. Vet Rec. 1986;119(4):94.
Julian R. Rapid growth problems: ascites and skeletal deformities in broilers. Poult Sci. 1998;77(12):1773–80.
Wideman RF. Cardio-pulmonary hemodynamics and ascites in broiler chickens. Avian Biol Res. 2000;11(1):21–44.
Wideman RF. Pathophysiology of heart/lung disorders: pulmonary hypertension syndrome in broiler chickens. Worlds Poult Sci J. 2001;57(3):289–307.
Wideman RF, Rhoads DD, Erf GF, Anthony NB. Pulmonary arterial hypertension (ascites syndrome) in broilers: A review. Poult Sci. 2013;92(1):64–83.
Baghbanzadeh A, Decuypere E. Ascites syndrome in broilers: physiological and nutritional perspectives. Avian Pathol. 2008;37(2):117–26.
Balog JM, Kidd BD, Huff WE, Huff GR, Rath NC, Anthony NB. Effect of cold stress on broilers selected for resistance or susceptibility to ascites syndrome. Poult Sci. 2003;82(9):1383–7.
Chapman M, Wideman R. Pulmonary wedge pressures confirm pulmonary hypertension in broilers is initiated by an excessive pulmonary arterial (precapillary) resistance. Poult Sci. 2001;80(4):468–73.
Wideman RF Jr, Hamal KR. Idiopathic pulmonary arterial hypertension: An avian model for plexogenic arteriopathy and serotonergic vasoconstriction. J Pharmacol Toxicol Methods. 2011;63:283–95.
Wideman RF, Hamal KR, Bayona MT, Lorenzoni AG, Cross D, Khajali F, Rhoads DD, Erf GF, Anthony NB. Plexiform Lesions in the Lungs of Domestic Fowl Selected for Susceptibility to Pulmonary Arterial Hypertension: Incidence and Histology. Anat Rec (Hoboken). 2011;294(5):739–55.
Bottje WG, Wang S, Kelly FJ, Dunster C, Williams A, Mudway I. Antioxidant defenses in lung lining fluid of broilers: impact of poor ventilation conditions. Poult Sci. 1998;77(4):516–22.
Hassanzadeh M, Buyse J, Toloei T, Decuypere E. Ascites Syndrome in Broiler Chickens: A Review on the Aspect of Endogenous and Exogenous Factors Interactions. J Poult Sci. 2014;51(3):229–41.
Deaton JW, Branton SL, Simmons JD, Lott BD. The Effect of Brooding Temperature on Broiler Performance. Poult Sci. 1996;75(10):1217–20.
Acar N, Sizemore F, Leach G, Wideman R, Owen R, Barbato G. Growth of broiler chickens in response to feed restriction regimens to reduce ascites. Poult Sci. 1995;74(5):833–43.
Bottje WG, Wideman R. Potential role of free radicals in the pathogenesis of pulmonary hypertension syndrome. Avian Biol Res. 1995;6:211–31.
Bowen O, Erf G, Anthony N, Wideman R. Pulmonary hypertension triggered by lipopolysaccharide in ascites-susceptible and-resistant broilers is not amplified by aminoguanidine, a specific inhibitor of inducible nitric oxide synthase. Poult Sci. 2006;85(3):528–36.
Özkan S, Malayoğlu HB, Yalçin S, Karadaş F, Koçtürk S, Çabuk M, Oktay G, Özdemir S, Özdemir E, Ergül M. Dietary vitamin E (α-tocopherol acetate) and selenium supplementation from different sources: performance, ascites-related variables and antioxidant status in broilers reared at low and optimum temperatures. Br Poult Sci. 2007;48(5):580–93.
Lubritz DL, McPherson BN. Effect of Genotype and Cold Stress on Incidence of Ascites in Cockerels. J App Poult Res. 1994;3(2):171–8.
Lubritz DL, Smith JL, McPherson BN. Heritability of ascites and the ratio of right to total ventricle weight in broiler breeder male lines. Poult Sci. 1995;74(7):1237–41.
de Greef K, Kwakernaak C, Ducro B, Pit R, Gerritsen C. Evaluation of between-line variation for within-line selection against ascites in broilers. Poult Sci. 2001;80(1):13–21.
Deeb N, Shlosberg A, Cahaner A. Genotype-by-environment interaction with broiler genotypes differing in growth rate. 4. Association between responses to heat stress and to cold-induced ascites. Poult Sci. 2002;81(10):1454–62.
Druyan S, Ben-David A, Cahaner A. Development of ascites-resistant and ascites-susceptible broiler lines. Poult Sci. 2007;86(5):811–22.
Druyan S, Hadad Y, Cahaner A. Growth rate of ascites-resistant versus ascites-susceptible broilers in commercial and experimental lines. Poult Sci. 2008;87(5):904–11.
Druyan S, Shinder D, Shlosberg A, Cahaner A, Yahav S. Physiological parameters in broiler lines divergently selected for the incidence of ascites. Poult Sci. 2009;88(9):1984–90.
Druyan S, Shlosberg A, Cahaner A. Evaluation of growth rate, body weight, heart rate, and blood parameters as potential indicators for selection against susceptibility to the ascites syndrome in young broilers. Poult Sci. 2007;86(4):621–9.
Lahav T, Atzmon G, Blum S, Ben-Ari G, Weigend S, Cahaner A, Lavi U, Hillel J. Marker-assisted selection based on a multi-trait economic index in chicken: experimental results and simulation. Animal Genetics. 2006;37(5):482–8.
Muir WM, Wong GK-S, Zhang Y, Wang J, Groenen MAM, Crooijmans RPMA, Megens H-J, Zhang H, Okimoto R, Vereijken A, et al. Genome-wide assessment of worldwide chicken SNP genetic diversity indicates significant absence of rare alleles in commercial breeds. Proc Natl Acad Sci USA. 2008;105(45):17312–7.
Dey S, Krishna S, Anthony NB, Rhoads DD. Further investigation of a quantitative trait locus for ascites on chromosome 9 in broiler chicken lines. Poult Sci. 2017;96(4):788–97.
Krishna S, Tarrant KJ, Rhoads DD, Anthony NB. Effects of a chromosome 9 quantitative trait locus for ascites on economically important traits in broilers. Poult Sci. 2017;96(9):3039–42.
Krishnamoorthy S, Smith CD, Al-Rubaye AA, Erf GF, Wideman RF, Anthony NB, Rhoads DD. A quantitative trait locus for ascites on chromosome 9 in broiler chicken lines. Poult Sci. 2014;93(2):307–17.
Tarrant KJ, Dey S, Kinney R, Anthony NB, Rhoads DD. Multi-generational genome wide association studies identify chromosomal regions associated with ascites phenotype. Poult Sci. 2017;96(6):1544–52.
Tarrant KJ, Fulton JE, Lund A, Rhoads DD, Anthony NB. Predicting ascites incidence in a simulated altitude-challenge using single nucleotide polymorphisms identified in multi-generational genome wide association studies. Poult Sci. 2018;97(11):3801–6.
Dey S, Parveen A, Tarrant KJ, Licknack T, Kong B, Anthony NB, Rhoads DD. Whole Genome Resequencing Identifies the CPQ Gene as a Determinant of Ascites Syndrome in Broilers. PLoS One. 2018;13(1):e0189544.
Pavlidis HO, Balog JM, Stamps LK, Hughes JD Jr, Huff WE, Anthony NB. Divergent selection for ascites incidence in chickens. Poult Sci. 2007;86(12):2517–29.
Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinformatics. 2012;14(2):178–92.
Laurén J, Airaksinen MS, Saarma M. Timmusk Tõ: A novel gene family encoding leucine-rich repeat transmembrane proteins differentially expressed in the nervous system. Genomics. 2003;81(4):411–21.
Siddiqui Tabrez J, Tari Parisa K, Connor Steven A, Zhang P, Dobie Frederick A, She K, Kawabe H, Wang Yu T, Brose N, Craig Ann M. An LRRTM4-HSPG Complex Mediates Excitatory Synapse Development on Dentate Gyrus Granule Cells. Neuron. 2013;79(4):680–95.
Radonic A, Thulke S, Mackay IM, Landt O, Siegert W, Nitschea A. Guideline to reference gene selection for quantitative real-time PCR. Biochem Biophys Res Commun. 2004;313:856–62.
Hassanpour H, Bahadoran S, Farhadfar F, Chamali ZF, Nazari H, Kaewduangta W. Identification of reliable reference genes for quantitative real-time PCR in lung and heart of pulmonary hypertensive chickens. Poult Sci. 2018;97(11):4048–56.
Al-Zahrani K, Licknack T, Watson DL, Anthony NB, Rhoads DD. Further investigation of mitochondrial biogenesis and gene expression of key regulators in ascites- susceptible and ascites- resistant broiler research lines. PLoS One. 2019;14(3):e0205480.
Owen RL, Wideman JRF, Cowen BS. Changes in Pulmonary Arterial and Femoral Arterial Blood Pressure upon Acute Exposure to Hypobaric Hypoxia in Broiler Chickens. Poult Sci. 1995;74(4):708–15.
Druyan S, Cahaner A. Segregation among test-cross progeny suggests that two complementary dominant genes explain the difference between ascites-resistant and ascites-susceptible broiler lines. Poult Sci. 2007;86(11):2295–300.
de los Santos SF, Tellez G, Farnell MB, Balog JM, Anthony NB, Pavlidis HO, Donoghue AM. Hypobaric hypoxia in ascites resistant and susceptible broiler genetic lines influences gut morphology. Poult Sci. 2005;84(9):1495–8.
Ahmetov II, Egorova ES, Gabdrakhmanova LJ, Fedotovskaya ON. Genes and Athletic Performance: An Update. In: Collins M, editor. Genetics and Sports, vol. 61. Basel: Karger Publishers; 2016. p. 41–54.
Bailes S, Devers J, Kirby JD, Rhoads D. An inexpensive, simple protocol for DNA isolation from blood for high-throughput genotyping by polymerase chain reaction or restriction endonuclease digestion. Poult Sci. 2007;86:102–6.
Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25(4):402–8.
Acknowledgements
Not applicable.
Funding
This project was supported by Agriculture and Food Research Initiative Competitive Grants no. 2015-67015-22960 and 2018-67015-28244 from the USDA National Institute of Food and Agriculture from the USDA National Institute of Food and Agriculture to DR and NA. AP was supported on a Fulbright Fellowship. CJ was supported by the Arkansas INBRE program, with grants from the National Center for Research Resources - NCRR (P20RR016460) and the National Institute of General Medical Sciences - NIGMS (P20 GM103429) from the National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
AP wrote the original draft, all authors reviewed and edited the final draft, DR was responsible for the final draft, AP CJ SD and DR performed the analyses, KT produced and provided essential samples, NA and DR conceived and directed the research. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All animal procedures were approved by the University of Arkansas Institutional Animal Care and Use Committee under protocols 15039 and 15040.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Parveen, A., Jackson, C.D., Dey, S. et al. Identification and validation of quantitative trait loci for ascites syndrome in broiler chickens using whole genome resequencing. BMC Genet 21, 54 (2020). https://doi.org/10.1186/s12863-020-00859-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12863-020-00859-x