Abstract
Background
There is growing evidence that the ghrelin axis, including ghrelin (GHRL) and its receptor, the growth hormone secretagogue receptor (GHSR), play a role in cancer progression. Ghrelin gene and ghrelin receptor gene polymorphisms have been reported to have a range of effects in cancer, from increased risk, to protection from cancer, or having no association. In this study we aimed to clarify the role of ghrelin and ghrelin receptor polymorphisms in cancer by performing a meta-analysis of published case-control studies.
We conducted searches of the literature published up to January 2013 in MEDLINE using the PubMed search engine. Individual data on 8,430 cases and 14,008 controls from six case-control studies of an all Caucasian population were evaluated for three ghrelin gene (GHRL; rs696217, rs4684677, rs2075356) and one ghrelin receptor (GHSR; rs572169) polymorphism in breast cancer, esophageal cancer, colorectal cancer and non-Hodgkins lymphoma.
Results
In the overall analysis, homozygous and recessive associations indicated that the minor alleles of rs696217 and rs2075356 GHRL polymorphisms conferred reduced cancer risk (odds ratio [OR] 0.61-0.78). The risk was unchanged for breast cancer patients when analysed separately (OR 0.73-0.83). In contrast, the rs4684677 GHRL and the rs572169 GHSR polymorphisms conferred increased breast cancer risk (OR 1.97-1.98, p = 0.08 and OR 1.42-1.43, p = 0.08, respectively). All dominant and co-dominant effects showed null effects (OR 0.96-1.05), except for the rs572169 co-dominant effect, with borderline increased risk (OR 1.08, p = 0.05).
Conclusions
This study suggests that the rs696217 and rs2075356 ghrelin gene (GHRL) polymorphisms may protect carriers against breast cancer, and the rs4684677 GHRL and rs572169 GHSR polymorphisms may increase the risk among carriers. In addition, larger studies are required to confirm these findings.
Similar content being viewed by others
Background
It is appreciated that ghrelin and its receptor (members of the ghrelin axis) play a role in the development and progression of cancer [1]. Ghrelin, the endogenous ligand for the growth hormone secretagogue receptor (GHSR), has many functions, including a role in regulating growth hormone release [2] and a range of metabolic effects: regulating appetite, and influencing insulin and glucose homeostasis, energy balance and adipogenesis [3],[4]. Given the metabolic effects of ghrelin, the ghrelin axis is a promising target for interventions for obesity and diabetes mellitus type two [5].
There is growing evidence that obesity and metabolic syndrome is associated with endocrine related cancers [6] and that the ghrelin axis may play a role in cancer progression [1]. A mechanistic link has been hypothesised between obesity, ghrelin and the development of colorectal [7] and prostate cancer [8]. A number of studies have linked single nucleotide polymorphisms (SNPs) in the ghrelin (GHRL) or ghrelin receptor (GHSR) genes with cancer risk [1]. Here, we perform a meta-analysis of case-control studies that have correlated ghrelin and GHSR gene polymorphisms with cancer risk to elucidate further the association between ghrelin axis gene polymorphisms and cancer.
Methods
Data sources
Using PubMed, a literature search was performed for all association studies (available until January 2013) investigating links between cancer and the ghrelin (GHRL) and ghrelin receptor (GHSR) genes. Previous studies reporting Caucasian genotypic data with case-control designs were chosen as eligible for this meta-analysis. In the first search, we used the terms, “ghrelin”, “polymorphism” and “cancer” which yielded 11 citations, five of which were excluded. From abstracts of the remaining six, one was excluded as it described an Asian population. Full texts of the remaining five studies were obtained, all of which complied with our inclusion criteria. In the second search, we entered the terms, “GHRL” and “cancer” in PubMed yielding 13 citations. A series of exclusions reduced the number to seven, full-texts of which were retrieved. We checked the reference lists of the full-text articles from both searches to minimize the possibility of missing relevant studies. Of the seven studies, six were either duplicated by the first search, or lacked genotype data, and therefore, only one further article was suitable for inclusion. Combining outcomes from the two searches gave a total of six articles which were included in our meta-analysis [9-14].
Data extraction and power calculations
Two investigators independently extracted data and reached a consensus regarding all information. The following information was obtained from each publication: first author’s name, published year, country of origin, dominant ancestry of the study populations, state of controls, matching criteria, sample source, genotype data, number of cases and controls. We also calculated frequencies of the variant allele, deviations of controls from the Hardy-Weinberg equilibrium (HWE) and the statistical power of each study. Assuming an odds ratio (OR) of 1.5 at a genotypic risk level of α = 0.05 (two-sided), power was considered to be adequate at ≥80%.
Meta-analysis
The strength of association between the ghrelin polymorphisms and cancer risk was measured by odds ratios (ORs) with 95% confidence intervals (CIs). Pooled estimates of the OR were obtained by calculating a weighted average of OR from each study [15]. For the following genetic models using variant (var) and wild-type (wt) genotypes we estimated: (i) additive: (var-var, var-var and wt - wt) genotypes compared with the wt - wt, (ii) co-dominant: frequency of variant alleles, assuming the risk could differ across all three genotypes, (iii) recessive (var-var vs. wt-var + wt-wt) and (iv) dominant: (var-var + wt-var vs. wt-wt).
To compare effects on the same baseline, we used raw data for genotype frequencies to calculate pooled ORs, which were obtained using either the fixed effects model [16], in the absence of heterogeneity, or random effects model in the presence of heterogeneity [17]. Heterogeneity between studies was estimated using the X2-based Q test [18]. Given the low power of this test [19], significance threshold was set at P = 0.10. Heterogeneity between studies was estimated using the X2-based Q test [18] and quantified with the I2 statistic which measures degree of inconsistency among studies [20]. Sensitivity analysis, which involved omitting one study at a time and recalculating the pooled OR, was also used to test for robustness of the summary effects. Data were analyzed using Review Manager 5.3 (Copenhagen: Nordic Cochrane Centre, Cochrane Collaboration) [21] and SigmaStat 2.3 (Systat Software, San Jose, CA). Significance was set at a P-value of ≤0.05 throughout, except in heterogeneity estimation. Publication bias was not investigated because of the low sensitivity of the qualitative and quantitative tests when the number of studies is lower than ten [22].
Results
Included studies
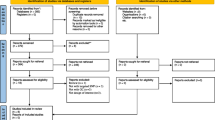
A total of six genotyping studies [10-14],[23] were included in the meta-analysis (Figure 1). The study features, which include nine ghrelin (GHRL) or ghrelin receptor (GHSR) single nucleotide polymorphisms (SNPs), the cancer type (breast, colorectal, esophageal and non-Hodgkin’s lymphoma) and study sample sizes, are outlined in Table 1. An overview of the ghrelin and ghrelin receptor SNPs examined are shown in Figure 2. Analyses of the pooled ORs revealed that five (rs495225, rs35684, rs27647, rs26802 and rs35683) of the nine SNPs investigated exhibited null effects in all genetic models (data not shown). The remaining three ghrelin SNPs (rs696217, rs4684677, rs2075356) and one GHSR SNP (rs572169) showed effects other than null, and were examined further. The features of these four SNPs (in six different studies), which included cancer type (breast, colorectal, esophageal, and non-Hodgkin’s lymphoma), ethnicity, number of cases and controls, calculated statistical power, minor allele frequency (MAF) and HWE are summarised in Table 2. The studies, that included rs696217 [10-14],[23], rs4684677 [10-13], rs2075356 [11],[12],[23] and rs572169 [11],[14],[23] had statistical power of >83%, indicating that these studies were not underpowered (Table 2). Control frequencies in two component studies [10],[14] deviated from the HWE in the rs4684677 and rs572169 polymorphisms (Table 2). Furthermore, three studies demonstrated borderline deviation from the HWE (p = 0.05-0.06) for the rs69621, rs2075356 and rs572169 polymorphisms [10],[11],[23].
Schematic diagram of the genes encoding ghrelin ( GHRL ) and the ghrelin receptor ( GHSR ). Exons are shown as boxes, introns as lines. The canonical coding exons of GHSR1a (cognate ghrelin receptor; GHSR) and ghrelin (GHRL) are shown as black boxes. Exon I and II (white boxes) are unique to the GHRL splice variant in2c-ghrelin. The SNPs examined in this study are indicated.
Quantitative effects
The overall effects (odd ratios) for the three GHRL polymorphisms (rs696217, rs4684677, rs2075356) and the rs572169 GHSR polymorphism, and the effects observed in breast cancer studies alone are shown in Table 3. Associations were observed mainly in the homozygous and recessive models and not in the dominant model, where the effects were null (OR 0.90-1.05, p = 0.19-0.92). Non-significant, decreased risks associated with the rs696217 (OR 0.61-0.63, p = 0.09-0.11) GHRL polymorphism in breast, colorectal, esophageal and colorectal cancer were not altered when analyses were confined to breast cancer studies alone (OR 0.82-0.83, p = 0.57-0.60) (Table 2). For rs696217, all of the study-specific ORs indicate reduced risk (Figure 3) and one study [11] in particular had a one-third weight contribution (33.5%) to the pooled effect (OR 0.63, p = 0.11). Similarly, a non-significant decrease in the risk of the GHRL polymorphism, rs2075356, in breast and colorectal cancer (OR 0.78, p = 0.43-0.45) was unaltered when confined to breast cancer studies (OR 0.78, p = 0.43-0.45) (Table 2). Increased risk in the GHRL SNP rs4684677 (OR 1.97-1.98, p = 0.08) associated with breast and esophageal cancers was exacerbated when confined to breast cancer (OR 2.38-2.40, p = 0.06). Figure 4 shows the contributions of study-specific ORs to the homozygous increased risk pooled effect of rs4684677 (OR 1.98, p = 0.08), mostly (60.6%) attributed to Dossus et al.[11]. On the other hand, the minimal weight contribution (5.3%) of the study by Feigelson et al.[12] is accompanied by wide confidence intervals (95% CI 0.49-169.90). The increased risk associated with the GHSR SNP rs572169 in breast and colorectal cancer (OR 1.42-1.43, p = 0.08) was also increased only when breast cancer was considered (OR 1.69-1.70, p = 0.14). While all of these effects of GHRL SNPs were obtained in zero heterogeneity (I2 = 0%), the effect of rs572169 was heterogeneous (I2 = 68-81%). Figure 5 shows heterogeneity (I2 = 68%) of the rs572169 increased risk pooled effect (OR 1.42, p = 0.08).
Forest plot of homozygous pooled effect in the rs4684677 polymorphism. Black diamond denotes the pooled OR. Blue squares indicate the OR in each study, with square sizes directly proportional to the weight contribution (%) of the study. Horizontal lines represent 95% confidence intervals. Note that case-control values for Doecke et al. [10] were non-estimable and therefore not included in the forest plot.
Increased risks were also observed in the co-dominant model of breast cancer studies for the rs4684677 GHRL SNP (OR 1.11, p = 0.23) and the GHSR rs572169 SNP, both overall and in breast cancer (OR 1.08-1.10), with borderline significance (p = 0.05), all homogeneously obtained (I2 = 0-46%).
Omitting the studies that deviated from HWE (including those at borderline level) did not materially alter the original summary effects for all four polymorphisms (data not shown). Sensitivity analysis did not alter the effects of the rs4684677 and rs2075356 GHRL polymorphisms. Removal of the Dossus et al. [11] breast cancer study generated a significant protective effect of the GHRL SNP rs696217 in the recessive model (OR 0.46, 95% CI 0.21-0.97, p = 0.04). The omission of the breast cancer study by Feigelson et al. [12] erased heterogeneity for the GHSR SNP rs572169 (from I2 = 68% to 0%), but left the pooled OR materially unchanged.
Discussion
With a combined sample size of 22,438 (8,430 cases and 14,008 controls), this meta-analysis provides evidence of overall homozygous and recessive associations, indicating a ~2-fold non-significant increase in cancer risk for the GHRL SNP rs4684677 and a ~1.4-fold non-significant increased risk for the GHSR SNP rs572169. This GHSR polymorphism showed a ~1.1-fold increased risk, with borderline significance in the co-dominant model. In contrast, the rs696217 and rs2075356 GHRL polymorphisms were both protective (22% and 38%), suggesting linkage disequilibrium (D’ = 0.90, r2 = 0.45) between the two SNPs [24]. The strength of these associations lie in the following: (i) they were obtained in total homogeneity underlying the statistical similarity of the component studies; and (ii) sensitivity analysis did not materially alter the effects underlying robustness of the pooled ORs. Interestingly, both the rs696217 (Leu72Met) and rs4684677 (Gln90Leu) GHRL SNPs have been linked with obesity [25],[26]. There is growing recognition that obesity is a risk factor for a number of cancers, including breast, endometrial, colorectal, esophageal and prostate cancer [1].
The GHRL rs2075356 (3056 T > C) and GHSR rs572169 (Gly57Gly) SNPs were found to be associated with 20% increased risk of breast cancer in a European study with 1,324 cases and 2,360 controls [11]. These findings were similar in our meta-analysis, which included an additional 2,227 cases and 3,741 controls, for rs572169 with 1.7-fold increased breast cancer risk, however, rs2075356 was 27% protective in our meta-analysis. Ghrelin and ghrelin receptor are expressed in breast cancer tissue and ghrelin could play a role in breast cancer progression [1],[27],[28]. Interestingly, the GHRL SNP rs2075356 is present within the 3′ untranslated region of the recently discovered ghrelin transcript in2c-ghrelin, which is expressed in breast tumours, but not in normal breast tissue [8]. In2c-ghrelin is predicted to encode a novel 83 amino acid preprohormone that codes for the 28 amino acid ghrelin peptide (encoded by exon 1 and 2), but not obestatin (encoded by exon 3) [8]. Depending on the cell-type, obestatin has growth promoting or suppressing functions [29], however, the function of this peptide in breast cancer remains to be determined. The in2c transcript is insulin-regulated in prostate [8] and breast cancer cell lines (data not shown). Breast and prostate tumour cells are responsive to insulin [30-32], and elevated insulin (hyperinsulinaemia) is associated with breast and prostate cancer risk [33],[34].
Strengths and weaknesses
Each of the six component studies in our meta-analysis examined multiple polymorphisms of ghrelin and its receptors. The multiplicity of calculations involved necessitated statistical adjustment to avoid false-positive findings. All six studies were adjusted for multiple testing. Three used the conservative Bonferroni correction [10],[12],[23], one used the false positive report probability [11] and the fifth used the less conservative false discovery rate [13]. The sixth study did not test for multiplicity, but compared risk SNPs with a corresponding cohort study [14].
These correction procedures, as well as the aforementioned features of the cases and controls in the component studies, reflect the overall strength of this meta-analysis. Other strengths of this study include: (i) ethnic homogeneity of the subjects given our focus on Caucasians only, resulting in minimal admixture and control for potential effects of population stratification; (ii) high sample sizes translating to robust statistical power of the component studies; (iii) statistical homogeneity in the comparisons, so that data in the included studies were similar enough to be pooled. Moreover, (iv) findings in the breast cancer subgroup were obtained in zero heterogeneity; (v) controls were either healthy or cancer-free and were matched to cases; (vi) tissue sources were blood; and (vii) all component studies were population-based which minimizes effects of selection bias, such that findings may be extrapolated to the general population. Nonetheless, limitations that need to be acknowledged are: (i) lack of representation in the various cancers (except breast cancer) disallowed further subgroup comparisons; and (ii) deviation from HWE among controls of Skibola et al.[13] and Wagner et al.[14] in the rs4684677 and rs572169 SNPs, respectively.
Conclusions
In summary, our results indicate that GHRL and GHSR SNPs may be involved in the pathophysiology of breast cancer. To our knowledge, this is the first meta-analysis to examine ghrelin polymorphisms and cancer risk. The demonstration of overall protective effects (of the rs696217 and rs2075356 SNPs) and increased susceptibility (for the rs4684677 and rs572169 SNPs) are derived from high-powered studies and are likely to increase the detection of low-penetrant effects. Further studies with larger and more well-defined sample populations are warranted to verify the role of GHRL and GHSR polymorphisms in cancer. This includes the analysis of additional metabolic, genetic and environmental contexts, which would be expected to influence the patient phenotype.
References
Chopin LK, Seim I, Walpole CM, Herington AC: The ghrelin axis-does it have an appetite for cancer progression?. Endocr Rev. 2012, 33 (6): 849-891. 10.1210/er.2011-1007.
Kojima M, Hosoda H, Date Y, Nakazato M, Matsuo H, Kangawa K: Ghrelin is a growth-hormone-releasing acylated peptide from stomach. Nature. 1999, 402 (6762): 656-660. 10.1038/45230.
Nakazato M, Murakami N, Date Y, Kojima M, Matsuo H, Kangawa K, Matsukura S: A role for ghrelin in the central regulation of feeding. Nature. 2001, 409 (6817): 194-198. 10.1038/35051587.
Wren AM, Small CJ, Abbott CR, Dhillo WS, Seal LJ, Cohen MA, Batterham RL, Taheri S, Stanley SA, Ghatei MA, Bloom SR: Ghrelin causes hyperphagia and obesity in rats. Diabetes. 2001, 50 (11): 2540-2547. 10.2337/diabetes.50.11.2540.
Seim I, Amorim L, Walpole C, Carter S, Chopin LK, Herington AC: Ghrelin gene-related peptides: multifunctional endocrine/autocrine modulators in health and disease. Clin Exp Pharmacol Physiol. 2010, 37 (1): 125-131. 10.1111/j.1440-1681.2009.05241.x.
Lorincz AM, Sukumar S: Molecular links between obesity and breast cancer. Endocr Relat Cancer. 2006, 13 (2): 279-292. 10.1677/erc.1.00729.
Waseem T, Javaid Ur R, Ahmad F, Azam M, Qureshi MA: Role of ghrelin axis in colorectal cancer: a novel association. Peptides. 2008, 29 (8): 1369-1376. 10.1016/j.peptides.2008.03.020.
Seim I, Lubik AL, Lehman M, Tomlinson N, Whiteside EJ, Herington A, Nelson C, Chopin L: Cloning of a novel insulin-regulated ghrelin transcript in prostate cancer. J Mol Endocrinol. 2013, 50: 1-14. 10.1530/JME-12-0150.
Cantor S, Drapkin R, Zhang F, Lin Y, Han J, Pamidi S, Livingston DM: The BRCA1-associated protein BACH1 is a DNA helicase targeted by clinically relevant inactivating mutations. Proc Natl Acad Sci U S A. 2004, 101 (8): 2357-2362. 10.1073/pnas.0308717101.
Doecke JD, Zhao ZZ, Stark MS, Green AC, Hayward NK, Montgomery GW, Webb PM, Whiteman DC: Single nucleotide polymorphisms in obesity-related genes and the risk of esophageal cancers. Cancer Epidemiol Biomarkers Prev. 2008, 17 (4): 1007-1012. 10.1158/1055-9965.EPI-08-0023.
Dossus L, McKay JD, Canzian F, Wilkening S, Rinaldi S, Biessy C, Olsen A, Tjonneland A, Jakobsen MU, Overvad K, Clavel-Chapelon F, Boutron-Ruault MC, Fournier A, Linseisen J, Lukanova A, Boeing H, Fisher E, Trichopoulou A, Georgila C, Trichopoulos D, Palli D, Krogh V, Tumino R, Vineis P, Quiros JR, Sala N, Martinez-Garcia C, Dorronsoro M, Chirlaque MD, Barricarte A, et al: Polymorphisms of genes coding for ghrelin and its receptor in relation to anthropometry, circulating levels of IGF-I and IGFBP-3, and breast cancer risk: a case-control study nested within the European Prospective Investigation into Cancer and Nutrition (EPIC). Carcinogenesis. 2008, 29 (7): 1360-1366. 10.1093/carcin/bgn083.
Feigelson HS, Teras LR, Diver WR, Tang W, Patel AV, Stevens VL, Calle EE, Thun MJ, Bouzyk M: Genetic variation in candidate obesity genes ADRB2, ADRB3, GHRL, HSD11B1, IRS1, IRS2, and SHC1 and risk for breast cancer in the Cancer Prevention Study II. Breast Cancer Res. 2008, 10 (4): R57-10.1186/bcr2114.
Skibola DR, Smith MT, Bracci PM, Hubbard AE, Agana L, Chi S, Holly EA: Polymorphisms in ghrelin and neuropeptide Y genes are associated with non-Hodgkin lymphoma. Cancer Epidemiol Biomarkers Prev. 2005, 14 (5): 1251-1256. 10.1158/1055-9965.EPI-04-0895.
Wagner K, Hemminki K, Grzybowska E, Klaes R, Burwinkel B, Bugert P, Schmutzler RK, Wappenschmidt B, Butkiewicz D, Pamula J, Pekala W, Forsti A: Polymorphisms in genes involved in GH1 release and their association with breast cancer risk. Carcinogenesis. 2006, 27 (9): 1867-1875. 10.1093/carcin/bgl036.
Breslow NE, Day NE: Statistical methods in cancer research. Volume II--The design and analysis of cohort studies.IARC Sci Publ 1987 82:1-406.,
Mantel N, Haenszel W: Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959, 22 (4): 719-748.
DerSimonian R, Laird N: Meta-analysis in clinical trials. Control Clin Trials. 1986, 7 (3): 177-188. 10.1016/0197-2456(86)90046-2.
Lau J, Ioannidis JP, Schmid CH: Quantitative synthesis in systematic reviews. Ann Intern Med. 1997, 127 (9): 820-826. 10.7326/0003-4819-127-9-199711010-00008.
Higgins JP, Thompson SG, Deeks JJ, Altman DG: Measuring inconsistency in meta-analyses. BMJ (Clin Res Ed. 2003, 327 (7414): 557-560. 10.1136/bmj.327.7414.557.
Higgins JP, Thompson SG: Quantifying heterogeneity in a meta-analysis. Stat Med. 2002, 21 (11): 1539-1558. 10.1002/sim.1186.
Review Manager (RevMan) [Computer program]. Version 5.3. Copenhagen: The Nordic Cochrane Centre, The Cochrane Collaboration; 2014.
Ioannidis JP, Trikalinos TA: The appropriateness of asymmetry tests for publication bias in meta-analyses: a large survey. CMAJ. 2007, 176 (8): 1091-1096. 10.1503/cmaj.060410.
Campa D, Pardini B, Naccarati A, Vodickova L, Novotny J, Steinke V, Rahner N, Holinski-Feder E, Morak M, Schackert HK, Gorgens H, Kotting J, Betz B, Kloor M, Engel C, Buttner R, Propping P, Forsti A, Hemminki K, Barale R, Vodicka P, Canzian F: Polymorphisms of genes coding for ghrelin and its receptor in relation to colorectal cancer risk: a two-step gene-wide case-control study. BMC Gastroenterol. 2010, 10: 112-10.1186/1471-230X-10-112.
Ando T, Komaki G, Naruo T, Okabe K, Takii M, Kawai K, Konjiki F, Takei M, Oka T, Takeuchi K, Masuda A, Ozaki N, Suematsu H, Denda K, Kurokawa N, Itakura K, Yamaguchi C, Kono M, Suzuki T, Nakai Y, Nishizono-Maher A, Koide M, Murakami K, Nagamine K, Tomita Y, Ookuma K, Tomita K, Tonai E, Ooshima A, Ishikawa T, et al: Possible role of preproghrelin gene polymorphisms in susceptibility to bulimia nervosa. Am J Med Genet B Neuropsychiatr Genet. 2006, 141B (8): 929-934. 10.1002/ajmg.b.30387.
Gueorguiev M, Lecoeur C, Meyre D, Benzinou M, Mein CA, Hinney A, Vatin V, Weill J, Heude B, Hebebrand J, Grossman AB, Korbonits M, Froguel P: Association studies on ghrelin and ghrelin receptor gene polymorphisms with obesity. Obesity (Silver Spring Md). 2009, 17 (4): 745-754. 10.1038/oby.2008.589.
Ukkola O, Ravussin E, Jacobson P, Perusse L, Rankinen T, Tschop M, Heiman ML, Leon AS, Rao DC, Skinner JS, Wilmore JH, Sjostrom L, Bouchard C: Role of ghrelin polymorphisms in obesity based on three different studies. Obes Res. 2002, 10 (8): 782-791. 10.1038/oby.2002.106.
Gahete MD, Cordoba-Chacon J, Hergueta-Redondo M, Martinez-Fuentes AJ, Kineman RD, Moreno-Bueno G, Luque RM, Castano JP: A novel human ghrelin variant (In1-ghrelin) and ghrelin-O-acyltransferase are overexpressed in breast cancer: potential pathophysiological relevance. PLoS One. 2011, 6 (8): e23302-10.1371/journal.pone.0023302.
Jeffery PL, Murray RE, Yeh AH, McNamara JF, Duncan RP, Francis GD, Herington AC, Chopin LK: Expression and function of the ghrelin axis, including a novel preproghrelin isoform, in human breast cancer tissues and cell lines. Endocr Relat Cancer. 2005, 12 (4): 839-850. 10.1677/erc.1.00984.
Seim I, Walpole C, Amorim L, Josh P, Herington A, Chopin L: The expanding roles of the ghrelin-gene derived peptide obestatin in health and disease. Mol Cell Endocrinol. 2011, 340 (1): 111-7. 10.1016/j.mce.2011.03.018.
Lann D, LeRoith D: The role of endocrine insulin-like growth factor-I and insulin in breast cancer. J Mammary Gland Biol Neoplasia. 2008, 13 (4): 371-379. 10.1007/s10911-008-9100-x.
Lubik AA, Gunter JH, Hendy SC, Locke JA, Adomat HH, Thompson V, Herington A, Gleave ME, Pollak M, Nelson CC: Insulin increases de novo steroidogenesis in prostate cancer cells. Cancer Res. 2011, 71 (17): 5754-5764. 10.1158/0008-5472.CAN-10-2470.
Pollak M: Targeting insulin and insulin-like growth factor signalling in oncology. Curr Opin Pharmacol. 2008, 8 (4): 384-392. 10.1016/j.coph.2008.07.004.
Suissa S, Azoulay L, Dell-Aniello S, Evans M, Vora J, Pollak M: Long-term effects of insulin glargine on the risk of breast cancer. Diabetologia. 2011, 54 (9): 2254-2262. 10.1007/s00125-011-2190-9.
Grossmann M, Wittert G: Androgens, diabetes and prostate cancer. Endocr Relat Cancer. 2012, 19 (5): F47-62. 10.1530/ERC-12-0067.
Acknowledgements
We thank Ofelia Francisco-Pabalan. NP was supported by the Saint Louis University special multigrant. We acknowledge the National Breast Cancer Foundation, Australia, for pilot project funding for part of this work (to LKC).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
None of the authors reports any competing interests relative to the work presented in this manuscript.
Authors’ contributions
Conceived and designed the experiments: NP. Analyzed the data: NP, HJ, IS, LKC. Wrote the paper: NP, HJ, IS, and LKC. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Pabalan, N.A., Seim, I., Jarjanazi, H. et al. Associations between ghrelin and ghrelin receptor polymorphisms and cancer in Caucasian populations: a meta-analysis. BMC Genet 15, 118 (2014). https://doi.org/10.1186/s12863-014-0118-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12863-014-0118-3