Abstract
Background
The orogeny of the eastern Mediterranean region has substantially affected ecological speciation patterns, particularly of mountain-dwelling species. Mountain vipers of the genus Montivipera are among the paramount examples of Mediterranean neo-endemism, with restricted ranges in the mountains of Anatolia, the Levant, Caucasus, Alborz, and Zagros. Here we explore the phylogenetic and ecological diversification of Montivipera to reconstruct its ecological niche evolution and biogeographic history. Using 177 sequences of three mitochondrial genes, a dated molecular phylogeny of mountain vipers was reconstructed. Based on 320 occurrence points within the entire range of the genus and six climatic variables, ecological niches were modelled and used to infer ancestral niche occupancy. In addition, the biogeographic history and ancestral states of the species were reconstructed across climate gradients.
Results
Dated phylogenetic reconstruction revealed that the ancestor of mountain vipers split into two major clades at around 12.18 Mya followed by multiple vicariance events due to rapid orogeny. Montivipera colonised coastal regions from a mountain-dwelling ancestor. We detected a highly complex ecological niche evolution of mountain vipers to temperature seasonality, a variable that also showed a strong phylogenetic signal and high contribution in niche occupation.
Conclusion
Raising mountain belts in the Eastern Mediterranean region and subsequent remarkable changes in temperature seasonality have led to the formation of important centres of diversification and endemism in this biodiversity hotspot. High rates of niche conservatism, low genetic diversity, and segregation of ranges into the endemic distribution negatively influenced the adaptive capacity of mountain vipers. We suggest that these species should be considered as evolutionary significant units and priority species for conservation in Mediterranean mountain ecosystems.
Similar content being viewed by others
Background
The recent decline in biodiversity necessitates a deeper understanding of current species distribution patterns and their evolution through time. This knowledge is particularly important in ecology, evolution, and conservation planning [1, 2]. In this regard, limited resources for nature conservation need to be concentrated on species having unique evolutionary histories [3, 4]. One of the main prerequisites for maintaining a species in the face of global change, and specifically climate change, is having sufficient genetic diversity to support environmental tolerances and occupy different ranges of ecological conditions (i.e., niche evolution), which is referred to as evolutionary flexibility or adaptive capacity [5, 6]. Species that have slowly evolved their ecological niches in the past, i.e., represent high degrees of niche conservatism [7], are more sensitive to demographic changes and population declines [6]. Significant efforts have been made recently to identify and model characteristics that allow species to adapt to environmental changes [e.g. 8, 9]. However, for many species, detailed demographic information and local adaptation rates are unavailable. A practical tool in this regard is to combine comparative phylogenetic methods and ecological niche modelling (ENM) to shed light on differential rates of niche evolution among sister taxa [6, 10, 11].
Ecological divergence has been emphasised as one of the main drivers of speciation [12, 13]. During ecological divergence, natural populations are influenced by a variety of biotic and abiotic factors, such as inter- and intra-species interactions (e.g., predation, competition) and fluctuations in climate and food resources [14, 15]. This may result in separate evolutionary responses and the formation of distinct climatic tolerances (i.e., niche divergence) [12, 16, 17]. In contrast, preservation of similar ecological conditions over time, in line with allopatric speciation, limits adaptation to local conditions and confines genetic differentiation between vicariant populations (i. e., niche conservatism) [18,19,20].
Mountains are one of the main cradles of diversification, as half of the world’s biodiversity hotspots are located in mountainous landscapes [21, 22]. A tremendous variety of microclimates and diverse local conditions in mountain ranges facilitates local adaptation and the formation of distinct genetic structures during ecological speciation [23, 24]. However, fragmentation and isolation in populations of mountain-dwelling species lead to limited gene flow, resulting in allopatric and peripatric speciation during orogeny [25, 26] that do not require niche adaptation as a motivator. Such processes play an important role, particularly in mountains that undergo rapid orogeny, where newly emerging extreme environments provide room for generating regional diversification pulses [22, 24].
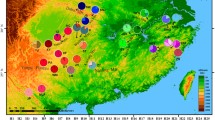
Our research focuses on one such mountain system, the Iran-Anatoly-Caucasus mountain chains, a long and newly developed rugged landscape that stretches from the Taurus Mountains in the south of Turkey, elongates toward the Anatolian Diagonal to the Caucasus and ends at the Alborz and Zagros Mountains in the north and southwest of Iran (Fig. 1). In general, the interaction of tectonic, topographic, and climatic physiognomies has shaped a heterogeneous assemblage of biodiversity in this region [26, 27]. This area embraces two biodiversity hotspots, the Caucasus and Irano-Anatoly, characterised by high degrees of endemism [28]. Moreover, severe ecological conditions have resulted in a repetitive pattern of expansion, contraction, and isolation in the Irano-Anatolian and Caucasus regions, and accordingly, the emergence of a substantial degree of recent speciation events, i.e. neo-endemism. Consequently, this region includes one of the world’s richest herpetofauna and preserves the highest diversity of true vipers within the Palearctic biome [29]. One of the striking models of neo-endemism in the Irano-Anatoly and Caucasus regions are mountain vipers of the genus Montivipera, which have experienced considerable speciation in mountainous landscapes [26, 30]. Their global distribution includes isolated populations of two species complexes, the Xanthina and the Raddei complex. The Xanthina complex includes the species M. xanthina, M. wagneri, and M. bornmuelleri, and a clade of two closely-related sister species M. albizona and M. bulgardaghica, although M. albizona has also been considered as a subspecies of M. bulgardaghica based on the low genetic divergence between them [26]. The Raddei complex contains the species M. raddei, M. latifii, M. albicornuta and, M. kuhrangica. While in the Xanthina complex M. xanthina ranges from the semi-elevated uplands of southern Turkey to low-elevated plains and islands of the Mediterranean Sea, other species of both complexes disperse in mountain-top isolated populations (Figs. 1, 2). Despite a wealth of studies on the morphology and phylogeny of vipers in general, and mountain vipers in particular, their biogeographic history remains to be more carefully analysed [26, 30, 31].
Geographic distribution (top), dated maximum clade credibility tree (bottom, reconstructed in BEAST based on multi-locus mitochondrial data), and ancestral range estimates (inferred with BIOGEOBEARS using a DIVA-like model) of the genus Montivipera. Numbers at each node indicate mean age estimates, and blue bars the 95% highest posterior density (HPD) confidence intervals. Boxes at tips show geographic areas including each species, colour-coded for the eight areas illustrated as polygons on the map. Pie charts indicate the estimated probability of each area. Dots, triangles, and stars next to the phylogenetic tree is the species-specific indication of the occurrence points and coloured polygons on the map are the approximate range of the biogeographic areas
Given the complex dynamics and idiosyncratic paleontological history of the mountain belts of the Irano-Anatoly and Caucasus regions and the taxonomic controversy over mountain vipers, the main question to be answered is how past changes in environmental conditions have driven the divergence among these species. Specifically, we aimed to unveil that whether the formation of the eastern Mediterranean mountain belt and the consequent climate changes is associated with cladogenic niche divergence. We also intended to explore if and where this orogeny has forced closely related taxa to preserve ecological adaptations after vicariant speciation, i.e., niche conservatism. We further aimed to examine patterns of phylogenetic niche conservatism in range-restricted Montivipera species allowing us to identify those species with the highest vulnerability to environmental change. To test these hypotheses, we reconstructed a dated phylogenetic tree and combined evolutionary analyses with ecological niche modelling to explore patterns of niche evolution among sister taxa of the mountain vipers.
Results
Phylogenetic relationships and divergence dating
The dated phylogenetic tree of mountain vipers (Fig. 1) allowed the identification of two Montivipera groups, i.e., the Xanthina and Raddei complexes, estimated to have diverged around 12.18 Mya (95% HPD 9.9–14.46). The Raddei complex consists of four species, M. raddei, M. albicornuta, M. latifii, and M. kuhrangica, showing a strongly supported monophyly that has split around 1.97 Mya (HDP = 0.79–3.15). The species inside this complex have diverged between 1.38 Mya (95% HDP = 0.64–2.12) and 1.01 Mya (95% HDP = 0.62–1.39). A strong cladogenic divergence is visible in the Xanthina complex, with the complex having split into the Xanthina and Bornmuelleri clades around 6.65 Mya (95% HDP = 5–8.3). A comparably early split in the Xanthina clade was the divergence of the coastal groups (M. xanthina Lycia, M. xanthina Aegean, and M. xanthina Greece) from their mountain-dwelling sister group (M. xanthina Taurus) at 4.49 Mya (95% HDP = 3.47–5.51). In the Bornmuelleri clade, consisting of M. bornmuelleri, M. wagneri, M. albizona and M. bulgardaghica, the divergence from the most recent common ancestor (MRCA) was estimated at 3.16 Mya (95% HDP = 2.18–4.14), 2.54 Mya (95% HDP = 1.73–3.35), and 1.62 Mya (95% HDP 1.42–1.82), subsequently. The complete unpruned phylogenetic tree of the species is shown in the Additional file 1: Fig. S1.
ENM and history of niche evolution
The distribution models based on six bioclimatic variables resulted in spatially well-defined potential ranges for each of the mountain viper species (Fig. 2). AUC and TSS values obtained for the distribution models among the eight studied species ranged from 0.88 and 0.89 in M. xanthina to 0.95 and 0.94 in M. bornmuelleri, respectively (Additional file 1: Table S2). The contribution of climatic variables to explain their distribution varied among species. Yet, temperature seasonality, temperature of the warmest month, precipitation seasonality, and precipitation of the driest month revealed the highest levels of importance for almost all species (Additional file 1: Table S2). As a general rule, all Montivipera species prefer areas with relatively cold climate conditions and with relatively high temperature seasonality, except for M. bornmuelleri and coastal lineages of M. xanthina, which show strong preferences for comparably warmer climates having low temperature seasonality and high precipitation seasonality.
The ancestral niche occupancy profiles (Fig. 3) provided evidence of both phylogenetic niche conservatism and niche divergence. The most noticeable phylogenetic niche conservatism was seen for M. latifii and M. kuhrangica with respect to almost all of the six climatic variables, and this is surprising considering the large geographic distance between their ranges. This pattern was also found for M. albizona and M. bulgardaghica, however, they are geographically in close proximity. Within the Raddei complex, M. latifii, M. kuhrangica, and M. albicornuta showed similar environmental affinities, while M. raddei revealed evidence of divergent evolution, particularly under extreme warm and dry climatic conditions. The most obvious phylogenetic niche divergence was seen for M. bornmuelleri and coastal lineages of M. xanthina (M. xanthina Lycia, M. xanthina Aegean, M. xanthina Greece), which have evolved in different directions compared to their close sister taxa. Notwithstanding, they showed between-group convergent evolution, particularly with respect to the precipitation variables.
Inferred history of the evolution of environmental tolerances of six climatic variables within the mountain viper species based on a BEAST chronogram. Internal nodes denote the mean of climatic tolerances as estimated for the most recent common ancestor of the related extant taxa. Vertical bars and point marks show the 80% central density of environmental tolerance for each extant taxon and the related mean values, respectively. Lines and points are coloured according to clades defined in Fig. 1
Reconstructing biogeographic history
Ancestral range estimation analyses revealed that based on the likelihood-ratio test comparing six biogeographic models, the DIVALIKE model is supported best (See Additional file 1: Table S3 for complete biogeographic comparison outputs). The resulting parameters of the DIVALIKE model included the anagenetic dispersal rate (d) of 0 and a high cladogenetic dispersal rate (j) of 0.407. Reconstruction of the biogeographic history of mountain vipers using a BEAST chronogram in BIOGEOBEARS is illustrated in Fig. 1, showing the area or combined areas with the highest probability for each node. The ancestral range estimations showed that the most probable ancestral area for extant species of the Xanthina complex is highly likely split by a vicariance event between three areas: the Taurus Mountains (gray polygon in Fig. 1), the Levant (orange polygon in Fig. 1), and the Anatolian Diagonal (green polygon in Fig. 1). A striking biogeographic history was revealed in the M. xanthina clade, where coastal lineages (M. xanthina Lycia, M. xanthina Aegean, and M. xanthina Greece) dispersed toward Mediterranean coastal habitats in southwest and western Turkey from a mountain-dwelling ancestor. Reconstructions within the Bornmuelleri clade indicated two subsequent dispersal events from the ancestor to the Levant, i.e. M. bornmuelleri, and to the northeastern Turkey, i.e. M. wagneri, followed by a vicariance in Anatolian Diagonal between M. bulgardaghica and M. albizona. Supporting our results, Stümpel et al. [26] estimated that the geographic location of the ancestor of the Xanthina complex, including the Bornmuelleri clade, occurred in Anatolian Taurus Mountains in southern Turkey. The estimated ancestral areas for the Raddei basal node seemed to have diverged rapidly by repeated vicariance events between three areas of the Caucasus—Kurdistan, the Alborz Mountains, and the Zagros Mountains.
Ancestral state estimation
We found phylogenetic signals under both Pagel’s λ (P < 0.01) and Blomberg’s K (P < 0.05) for four of the six climatic variables: temperature seasonality, maximum temperature of the warmest month, minimum temperature of the coldest month, and precipitation of the driest month (Table 1). Based on the AICc statistic, the Brownian motion model showed the best fit of trait evolution for these bioclimatic variables, and precipitation of the wettest month and precipitation seasonality followed the white noise evolutionary model (Additional file 1: Table S3). Maximum likelihood ancestral state estimation (Fig. 4) based on the BM model indicated a mix of phylogenetic niche conservatism and divergent trait evolution (here climatic variables) in mountain vipers. Similar to the niche evolution chronogram, the ancestral state phenogram also confirmed that coastal lineages of M. xanthina (M. xanthina Lycia, M. xanthina Aegean, M. xanthina Greece) have highly likely diverged from a mountain-dwelling population. Relatively firm phylogenetic conservatism was seen in species of the Raddei complex. However, in this group, M. latifii in comparison to its ancestor has evolved toward extreme environmental conditions with a colder climate and higher temperature seasonality. Similarly, in the Bornmuelleri clade, M. bornmuelleri and M. wagneri have evolved divergently toward opposite climatic conditions compared to their most recent common ancestor.
Discussion
The taxonomic status of mountain vipers of the genus Montivipera has long been the subject of debate among many herpetologists and taxonomists. All species in this group experienced a complex taxonomic history. For example, Mertens et al. [32] resolved M. bornmuelleri as Vipera lebetina bornmuelleri, a subspecies of the later Macrovipera genus. Sigg et al. [33] suggested that this species was Vipera xanthina when the Montivipera genus had not yet been introduced, and many species of this genus along Macrovipera and Daboia were considered to be monophyletic groups under the genus Vipera. Nilson and Andrén [34] first suggested Montivipera and Macrovipera as subgenera of Vipera, and Joger [35] later resurrected them as different genera. The present research, due to the use of a complete genetic dataset of mountain vipers, can be considered as the most comprehensive study of the phylogeny and biogeography of the genus Montivipera. Since our phylogenetic inference is based on only mitochondrial data we acknowledge that it might raise levels of uncertainty in species tree reconstruction as reliance on mitochondrial data can have ramifications for both evolutionary inference and taxonomy [36]. Indeed, male-biased dispersal, i.e. gene-flow, might violate inferences of mitochondrial markers especially in continuous species [37] or even in snakes [36, 38]. Although mountain vipers are extremely range-limited with low movement abilities, any interpretation on their taxonomic delimitation using only mitochondrial data should be taken cautiously.
The results of this study highlight the role of mountain ranges in producing and maintaining diversity in the eastern Mediterranean region. Particularly during times of orogeny, mountains facilitate geographic isolation and formation of allopatric speciation [25], a pattern that has been reported for a number of species groups [21, 22, 24, 39]. Based on the inferred phylogeny, the three monophyletic clades Xanthina, Bornmuelleri, and Raddei were verified. The first split in the ancestor of mountain vipers occurred around 12.18 Mya, which coincides with the Middle-to-Late Miocene. Similarly, Stümpel et al. [26] estimated the divergence time between the Xanthina and Raddei complex around 12.3 Mya. This period overlaps with widespread tectonic changes in the Near and Middle East, which include the uplift of the Irano-Anatolian mountains and disconnection between the Tethys Sea and Indian Ocean [40]. With continued tectonic changes in the region and the gradual disappearance of the Tethys Sea and formation of the Para-Tethys Sea in the Late Miocene, two clades, Xanthina in the west and Bornmuelleri in the central east of Anatolia, were separated. The increase of elevation to 1.5–2 km a.s.l in this period [41] likely stimulated major changes in climatic conditions and repetitive altitudinal shifts in biotic communities of the region, and highly likely, led to changes in the ecological niches and tolerances of Montivipera species. Generally, our estimated divergence times in the Montivipera genus are consistent with those of previous studies, for example see Stümpel et al. [26] and Behrooz et al. [30].
A considerable divergence in mountain vipers is the split of coastal vipers from their mountain-dwelling ancestor around 4.6 Mya, confirmed by niche evolution and ancestral range estimations. A possible reason for this divergence could be the absence of Macrovipera lebetina in the Mediterranean coastal ranges [31], a dominant competitor of Montivipera that occupies lower-elevated habitats in co-existing ranges. Overall, in the Bornmuelleri and Raddei clades, all the divergence events have occurred during Pliocene and Pleistocene climatic oscillations. Avise et al. [42] reported 57% of speciation in herpetofauna as a consequence of Pleistocene glaciations. Likewise, Veith et al. [43] and Plötner et al. [44] argued that the highest rates of speciation in the amphibians of Anatolia coincides with this time era. As suggested by BIOGEOBEARS range estimations, a progressive dispersal-vicariance provoked by the repeated fragmentation-reconnection during climatic cycles in this time period derived vicariance in different populations of mountain vipers and the foundation of allopatric speciation. This expedited phylogenetic niche conservatism, especially in M. latifii, M. kuhrangica, M. xanthina Taurus, M. albizona, and M. bulgardaghica, as supported by niche evolution and ancestral state estimation, emphasises the classic model of niche conservatism: conservation of niches after allopatric speciation in sister taxa [7, 20]. Here, allopatric speciation spatially overlaps with the floral glaciation refuges in the Taurus, Levant, and Anatolian Diagonal documented by Médail and Diadema [45].
Overall, the results of this research reveal a complex mixture of niche evolution in mountain vipers in terms of phylogenetic niche conservatism and divergence. The extreme divergent evolution in the climatic tolerance of mountain vipers was seen in the species of the Bornmuelleri clade. While M. wagneri and M. latifii have convergently evolved toward areas with the highest temperature seasonality and negative extremes of temperature, M. bornmuelleri has adapted to humid ranges with the lowest temperature seasonality. Consistently, temperature seasonality showed the strongest phylogenetic signal and was ranked among the most important variables contributing to the current distribution of mountain vipers (Additional file 1: Table S2). This consistency is one line of evidence that the Mediterranean ranges of Irano-Anatolia, along with Irano-Turanian, are characterised by their continentality and high temperature seasonality [46]. A similar pattern has also been revealed by divergent climatic tolerance in lizards of the genus Timon occupying opposite sides of the Mediterranean [47]. It should be naoted that the inherent low sample size of the species occurrence points might increase uncertainty in the predicted ENMs niche evolution analyses of the species. Pooling data of closely related taxa into one set or simply removing those with low sample sizes were the options we could consider in this study. The former might end with the inflation in the predicted niche of the species [48] which in turn might generalise distinct processes that mountain viper have been evolved through. Interestingly, those species with the lowest distribution area and the least number of occurrence points, e.g. M. kuhrangica, M. latifii, and M. bornmuelleri, are among the few mountain vipers that are classified as true species from a taxonomic viewpoint. As a consequence, we chose not to remove them from the analyses as they each show a noteworthy status in the phylogenetic and niche modelling analyses. For the ENM analysis, we chose MaxEnt as this method has been reported the most capable in producing useful results with small sample sizes, even less than 25 occurrences [48].
We considered the rate of niche evolution as the difference between the occupied niche of sister taxa and the projected niche of their most recent ancestor (y axes of Fig. 3) given the estimated divergence time (x axes of Fig. 3). Our results indicate that species of the Raddei complex have experienced slower rates of niche evolution, and given PNO and ancestral state reconstruction, have adapted to similar gradients of environmental space. Small population sizes, restricted distributions, and low genetic and haplotype diversity [30, 49] in the species of this complex are highly threatening and could negatively influence their adaptive capacities. Species with slower evolutionary changes in their niches compared to those experiencing rapid niche evolution are more vulnerable to demographic decline and population fluctuation [6, 50]. Particularly, niche conservatism in mountain vipers leads to lower resistance and adaptive capacity toward environmental changes [6]. Conversely, species with higher genetic diversity often have had rapid evolutionary rates in the past, and presently have broader environmental tolerance and niche breath, thus occupying wider geographic ranges [51, 52].
Generally, our results demonstrate that severe environmental changes in the past due to rapid orogeny and striking topographic heterogeneity in the mountain belts of Irano-Anatolia and the Caucasus have forced mountain vipers to experience a variety of niche evolution and diversification patterns. Due to their tendency towards niche conservatism and their low genetic diversity, these species are being forced to move their ranges or become extinct as the environment changes. Altitudinal range shift in mountain vipers has been demonstrated as a response to climatic changes [49]. This results in greater isolation and limited gene flow, intensifying extinction risk in these sky-island taxa. Population decline, isolated distributions, low genetic diversity, and low adaptive capacity create challenges for the conservation of mountain vipers. According to Fraser and Bernatchez [53], mountain-dwelling species with isolated distribution and limited gene flow are high-priority evolutionary distinct conservation units. The concept of evolutionary significant units (ESUs) and/or cryptic species can be considered as a consolidation of evolutionary biology, taxonomy, and conservation planning. While identifying evolutionary distinct taxa and their geographic distribution patterns is the cornerstone of biogeography and speciation [2], focus on these species as evolutionary ESUs and the existence of threatened taxa within cryptic complexes reflect the necessity for conservation considerations [3]. In this regard, focus on ecological diversification along with the use of ENMs facilitate understanding speciation process as well as taxonomic delimitations [12, 13, 54]. Here by using mountain vipers as striking models of neo-endemism in mountains of Eastern Mediterranean we bring together biogeographic, taxonomic, and conservation objectives into one set. Due to the unique biogeographic history and limited adaptive capacity we suggest that all the isolated populations of mountain vipers thus could be considered as ESUs requiring immediate conservation attention. Establishing conservation zones and adaptive conservation strategies such as assisted migration, and reducing threats such as overgrazing, habitat destruction, and over-collection for anti-venom production, are among the most important potential conservation strategies.
Conclusion
Mountain vipers of the genus Montivipera are interesting examples of ecological species on the plateaus of Iran, Anatolia, and the Caucasus. On the one hand, their entire global reach is limited to this area, on the other hand, a high rate of neo-endemism has occurred in this genus. The combination of information obtained from phylogenetic reconstruction and ENM allowed unveiling the niche evolution of the species during diversification processes. Our results demonstrate that mountain vipers have been isolated from their common ancestors through common dispersal—vicariance processes, especially during the Pliocene–Pleistocene glacial fluctuations. The orogenic system of the region has additionally forced the genus to experience a high and complex diversity of ecological niche evolution of the species. In general, the species of Bornmuelleri clade have experienced the highest rate of evolutionary divergence toward climatic gradients while in the Raddei clade the species have had slow and convergence evolution through the climatic niche conditions. In conclusion, our study reveals limited evolutionary flexibility, and thus, adaptive capacity in mountain vipers especially for those distributed across mountains of Alborz and Zagros in Iran.
Materials and methods
Data collection
Species occurrence points for ENM analysis were recorded during tissue sampling of direct field surveys conducted by three authors of this study (M.A., R.B., and F.G., n = 105), and also obtained from other herpetologists’ field surveys (n = 62), existing databases (i.e. HerpNet, n = 10), and scientific publications (n = 143). Particularly, the study by Mebert et al. [55] has noticeably provided improved distribution data of the Montivipera of central-eastern Anatolia. Overall, we obtained 320 presence points covering the entire distribution of mountain vipers’ (Fig. 1) and ranging from 12 occurrences for M. kuhrangica to 51 localities for M. albizona.
In order to reconstruct a dated molecular tree, we downloaded 177 mitochondrial sequences from NCBI GenBank generated by previous studies. Specifically, our dataset comprised 82 sequences of cytochrome b (Cyt-b, 1061 bp) and ND4 (640 bp) generated by Behrooz et al. [30] for the Raddei complex. Additionally, 95 genetic sequences of three mitochondrial genes (Cyt-b, COX1, and ND5) produced by Stümpel et al. [26] for both Xanthina and Raddei complexes were downloaded. Details of the number of sequences and presence points are shown in Additional file 1: Table S1.
Phylogenetic analysis and estimation of divergence times
As congruence plays an important role in crucial phylogenetic analyses based on multi-locus data [56], we tested congruence among four mitochondrial genes using Shimodaira–Hasegawa (SH) test. Results showed congruence among Cytb, COX1, and ND5, and accordingly a dataset matrix with 2488 bp (Cytb 1061 bp, COX1 824 bp, and ND5 602 bp) was considered for conducting the phylogenetic analysis. All sequences were aligned using ClustalW, implemented in MEGA v.6 [57]. We then concatenated the genetic sequences to one multi-locus dataset ranging from five sequences for M. bornmuelleri to 53 sequences for M. albicornuta (see Additional file 1). Best models of sequence evolution based on a partitioning scheme were chosen using a “greedy” algorithm and the Bayesian Information Criterion (BIC) in PartitionFinder 1.1.1 [58].
Divergence dates among 12 main lineages of Montivipera sp, discovered also in previous studies (Stümpel et al. [26], were calculated using BEAST 1.8.0 [59]. Molecular clock was carried out with two calibration points recommended by Stümpel et al. [26] including (i) the divergence between Macrovipera and Montivipera at ~ 15.5 Mya using a normal distribution with SD of 0.5, and (ii) the split between species of M. xanthina and M. raddei complexes using a normal distribution with a mean of 12.6 Mya and a standard deviation of 1.2 Mya. Accordingly, additional sequences of Cytb for two species of Macrovipera, including Levantine viper Macrovopera lebetina and Razi’s viper Macrovipera razii, were obtained from GenBank and incorporated to our dataset. Two independent runs were performed at 200 million generations and sampled every 10,000 generations, and omitting 25% as burn-in. Tracer 1.5 [60] was used for evaluating acceptable levels of MCMC chain mixing, and for checking convergence and effective sample sizes for all parameters. LOGCOMBINER V. 1.8.2 was applied to combine trees and log files. Finally, the maximum clade credibility tree with mean ages was summarised in TREEANNOTATOR v1.8.0 with a PP limit of 0.95.
Ecological niche evolution
We used the ancestral niche occupancy reconstruction approach proposed by Evans et al. [61] to reconstruct patterns of niche evolution within mountain vipers. This method integrates each species’ potential distribution with the dated, ultrametric tree to explore environmental niche evolution by constructing predicted niche occupancy (PNO) profiles [61]. We limited ancestral niche reconstructions to a single terminal per species by pruning replicate branches off the Montivipera BEAST tree using the drop.tip function in the R package ape [62].
Species’ potential distribution was modelled by using the maximum entropy (MaxEnt) method [63] and six climatic variables representing climate extremes and seasonal variability in the region, including maximum temperature of the warmest month (°C), minimum temperature of the coldest month (°C), temperature seasonality, precipitation of the driest month (mm), precipitation of the wettest month (mm), and precipitation seasonality. By reflecting adaptation to extreme climatic conditions, these variables represent important environmental constraints on species distributions, niche evolution, and adaptability [64]. Also, this selection avoids collinearity problems as the pairwise correlation fulfils |r|< 0.7. Data on climatic variables with a spatial resolution of 30 arc-second were obtained from the WorldClim database [65]. We used MaxEnt model as this method has proven to obtain more reliable results with small sample sizes even lower than 25 occurrences [48]. Additionally, we adopted two methods to reduce the uncertainty caused by the inherent low sample size of mountain vipers’ presence data. First, to cope with the negative impacts of spatial autocorrelation (SAC), we set the minimum distance between species presence to 1 km. Second, for each species we repeated MaxEnt modelling 10 times based on a bootstrap procedure. The predictive performance of models was evaluated based on the area under the curve (AUC) of receiver operating characteristic (ROC) plots. Because of the drawbacks of AUC to measure the predictive performance of distribution models [66], we also calculated the true skill statistic (TSS) as a measure of the model’s classification accuracy. We considered the minimum suitability at the presence points as a suitability threshold to calculate TSS.
To reconstruct ancestral niche occupancy profiles, we combined each species’ potential distribution at a given grid cell in the study area with the corresponding scores of the six original climatic variables to create a vector of probabilities per binned climatic values (here we calculated it for 100 intervals). To generate profiles of predicted niche occupancy (PNO), the map of each climatic variable was converted to a histogram of 100 equal-interval bins, and the total MaxEnt raw probabilities in each bin were calculated for each species. By using this method, instead of extracting climatic data from species localities to reconstruct ancestral climatic tolerances, we linked habitat suitability with bins of climatic variables [61]. We then reconstructed the climatic tolerances of ancestral nodes using the PNO profiles under the assumption of Brownian motion evolution. To do so, the PNO profiles were resampled 1000 times, and the ancestral character reconstructions were performed using the generalised least squares method [for more detail see 62]. The ancestral niche reconstructions were computed using the package phyloclim [67].
Ancestral range estimation
We inferred ancestral ranges and the biogeographic history of mountain vipers based on the pruned divergence time tree and three models of biogeographical range expansion, including Dispersal-Extinction-Cladogenesis (DEC), Dispersal-Vicariance (DIVA-like), and Bayesian inference (BAYAREA-like) models, all using the BioGeoBEARS package [68] in the R environment. Using these models enabled us to explore different possibilities of dispersal, vicariance, and extinction. We also incorporated a founder-event parameter (+ J) in the analysis allowing for cladogenic dispersal outside of the parental areas, i.e., jump speciation [69]. We divided mountain vipers into eight biogeographic areas: (1) Taurus Mountains, (2) Mediterranean coasts and islands, (3) the Levant, (4) Anatolian Diagonal, (5) Northeast Anatolia, (6) South Caucasus—Kurdistan, (7) Alborz Mountains, and (8) South Zagros Mountains. These biogeographic areas were selected given the topology of mountain vipers in their phylogenetic time tree as well as the physiography of the selected mountain patches in the region. Specifically, these biogeographic areas are geographically isolated except for the Anatolian Diagonal and Northeast Anatolia, for which the reason for considering them as different groups was that we additionally intended to reveal the species-specific biogeographic history of mountain vipers. To facilitate clear estimations, we allowed inferred ancestral ranges to occupy up to two areas, with relative probabilities of dispersal and the possibility to disperse in four time-slices (0–0.40, 0.40–4, 4–8, and 8–11.5 Mya) considering divergence times within the complex. Likelihood estimates of three biogeographic models and their modified J types were compared using the Akaike Information Criterion (AIC) to identify the best-fit model.
Ancestral state estimation
We illustrated the ancestral state estimations of six climatic variables to assess phylogenetic niche conservatism (PNC) in mountain vipers [7]. First, phylogenetic signals were evaluated to examine whether similarity in lineage traits (here, the climatic variables) is correlated with phylogenetic relationships. As input data, we averaged each of the six climatic variables extracted from each of the species presence points. We tested for phylogenetic signals in the six climatic variables based on Blomberg’s K [70] and Pagel’s λ [71] using the phylosig function of package phytools [72]. We then tested for PNC under three models of trait evolution: Brownian motion (BM), Ornstein–Uhlenbeck (OU), and white noise (WN) using the fitContinuous function of package geiger [73]. Selection of the WN model indicates that traits are evolving independently without phylogenetic signal; BM assumes that the correlation structure among trait values is proportional to the extent of shared ancestry for pairs of species, and OU implies stabilising selection over time. The best model was ranked based on the AICc. Finally, a maximum likelihood framework was used to estimate the ancestral state of internal nodes under the selected evolutionary model, and the inferred states were interpolated along the edges of each branch based on Felsenstein’s equation [74] using the contMap function of package phytools [75].
Availability of data and materials
The XML file used to reconstruct phylogenetic analysis, the R code, and the input data for the niche evolution and the ancestral estimation analysis are available at: https://github.com/mahmadi-iut/Montivipera-niche-evolution.git
References
Moritz C, Potter S. The importance of an evolutionary perspective in conservation policy planning. Mol Ecol. 2013;22(24):5969–71.
Richardson DM, Whittaker RJ. Conservation biogeography—foundations, concepts and challenges. Divers Distrib. 2010;16(3):313–20.
Collen B, Turvey ST, Waterman C, Meredith HM, Kuhn TS, Baillie JE, et al. Investing in evolutionary history: implementing a phylogenetic approach for mammal conservation. Phil Trans R Soc Lond B Biol Sci. 2011;366(1578):2611–22.
Faith DP. Quantifying biodiversity: a phylogenetic perspective. Conserv Biol. 2002;16(1):248–52.
Giennap P, Teplitsky C, Alho JS, Mills JA, Merila J. Climate change and evolution: disentangling environmental and genetic responses. Mol Ecol. 2008;17(1):167–78.
Lavergne S, Evans ME, Burfield IJ, Jiguet F, Thuiller W. Are species’ responses to global change predicted by past niche evolution? Phil Trans R Soc Lond B Biol Sci. 2013;368(1610):20120091.
Wiens JJ, Ackerly DD, Allen AP, Anacker BL, Buckley LB, Cornell HV, et al. Niche conservatism as an emerging principle in ecology and conservation biology. Ecol Lett. 2010;13(10):1310–24.
Kirkpatrick M, Peischl S. Evolutionary rescue by beneficial mutations in environments that change in space and time. Phil Trans R Soc Lond B Biol Sci. 2013;368(1610):20120082.
Schiffers K, Bourne EC, Lavergne S, Thuiller W, Travis JM. Limited evolutionary rescue of locally adapted populations facing climate change. Phil Trans R Soc Lond B Biol Sci. 2013;368(1610):20120083.
Salamin N, Wüest RO, Lavergne S, Thuiller W, Pearman PB. Assessing rapid evolution in a changing environment. Trends Ecol Evol. 2010;25(12):692–8.
Ahmadi M, Naderi M, Kaboli M, Nazarizadeh M, Karami M, Beitollahi SM. Evolutionary applications of phylogenetically-informed ecological niche modelling (ENM) to explore cryptic diversification over cryptic refugia. Mol Phylogenet Evol. 2018;127:712–22.
McCormack JE, Zellmer AJ, Knowles LL. Does niche divergence accompany allopatric divergence in Aphelocoma jays as predicted under ecological speciation?: insights from tests with niche models. Evolution. 2010;64(5):1231–44.
Schluter D. Evidence for ecological speciation and its alternative. Science. 2009;323(5915):737–41.
Coyne JA, Orr HA. Speciation. MA: Sinauer Associates Sunderland; 2004.
Schluter D, Conte GL. Genetics and ecological speciation. Proc Natl Acad Sci. 2009;106(Supplement 1):9955–62.
Wooten J, Gibbs H. Niche divergence and lineage diversification among closely related Sistrurus rattlesnakes. J Evol Biol. 2012;25(2):317–28.
Ahmadzadeh F, Flecks M, Carretero MA, Böhme W, Ilgaz C, Engler JO, et al. Rapid lizard radiation lacking niche conservatism: ecological diversification within a complex landscape. J Biogeogr. 2013;40(9):1807–18.
Peterson AT. Ecological niche conservatism: a time-structured review of evidence. J Biogeogr. 2011;38(5):817–27.
Pyron RA, Costa GC, Patten MA, Burbrink FT. Phylogenetic niche conservatism and the evolutionary basis of ecological speciation. Biol Rev. 2015;90(4):1248–62.
Wiens JJ, Graham CH. Niche conservatism: integrating evolution, ecology, and conservation biology. Annu Rev Ecol Evol Syst. 2005;36:519–39.
Fjeldså J, Bowie RC, Rahbek C. The role of mountain ranges in the diversification of birds. Annu Rev Ecol Evol Syst. 2012;43:249–65.
Garcia-Porta J, Simó-Riudalbas M, Robinson M, Carranza S. Diversification in arid mountains: biogeography and cryptic diversity of Pristurus rupestris rupestris in Arabia. J Biogeogr. 2017;44(8):1694–704.
Antonelli A, Kissling WD, Flantua SGA, Bermúdez MA, Mulch A, Muellner-Riehl AN, et al. Geological and climatic influences on mountain biodiversity. Nat Geosci. 2018;11(10):718–25.
Eronen JT, Janis CM, Chamberlain CP, Mulch A. Mountain uplift explains differences in Palaeogene patterns of mammalian evolution and extinction between North America and Europe. Proc R Soc B Biol Sci. 1809;2015(282):20150136.
Antonelli A, Nylander JA, Persson C, Sanmartín I. Tracing the impact of the Andean uplift on Neotropical plant evolution. Proc Natl Acad Sci. 2009;106(24):9749–54.
Stümpel N, Rajabizadeh M, Avcı A, Wüster W, PJoger U. Phylogeny and diversification of mountain vipers (Montivipera, Nilson et al., 2001) triggered by multiple Plio-Pleistocene refugia and high-mountain topography in the Near and Middle East. Mol Phylogenet Evol. 2001;2016(101):336–51.
Steinbauer MJ, Irl SD, Beierkuhnlein C. Elevation-driven ecological isolation promotes diversification on Mediterranean islands. Acta Oecologica. 2013;47:52–6.
Mittermeier RA, Turner WR, Larsen FW, Brooks TM, Gascon C. Global biodiversity conservation: the critical role of hotspots. In: Biodiversity hotspots. Springer, Berlin; 2011. p. 3–22
Sindaco R, Venchi A, Carpaneto GM, Bologna MA. The reptiles of Anatolia: a checklist and zoogeographical analysis. Biogeographia. 2000. https://doi.org/10.21426/B6110017
Behrooz R, Kaboli M, Arnal V, Nazarizadeh M, Asadi A, Salmanian A, et al. Conservation below the species level: suitable evolutionarily significant units among Mountain Vipers (the Montivipera raddei complex) in Iran. J Hered. 2018;109(4):416–25.
Stümpel N, Joger U. Recent advances in phylogeny and taxonomy of Near and Middle Eastern Vipers—an update. ZooKeys. 2009;31:179.
Mertens R, Darevsky I, Klemmer K. Vipera latifii, eine neue Giftschlange aus dem Iran. Senck Biol. 1967;48:161–8.
Sigg H, Baran IB, Schätti B. Rediscovery of the Bolkar viper: morphological variation and systematic implications on the “Vipera xanthina complex.” Amphibia-Reptilia. 1991;12(3):305–27.
Nilson G, Andrén C. The species concept in the Vipera xanthina complex: reflecting evolutionary history or hiding biological diversity? Amphibia-Reptilia. 1992;13(4):421–4.
Joger U. Montivipera Nilson, Tuniyev, Andrén, Orlov, Joger, and Herrmann (1999) In: Joger U, Stümpel N (Eds) (2005) Handbuch der Reptilien und Amphibien Europas, Schlangen (Serpentes) III, Aula-Verlag. 2005
Folt B, Bauder J, Spear S, Stevenson D, Hoffman M, Oaks JR, et al. Taxonomic and conservation implications of population genetic admixture, mito-nuclear discordance, and male-biased dispersal of a large endangered snake, Drymarchon couperi. PLoS ONE. 2019;14(3): e024439.
Ashrafzadeh M-R, Khosravi R, Ahmadi M, Kaboli M. Landscape heterogeneity and ecological niche isolation shape the distribution of spatial genetic variation in Iranian brown bears, Ursus arctos (Carnivora: Ursidae). Mamm Biol. 2018;93(1):64–75.
Marshall TL, Chambers EA, Matz MV, Hillis DM. How mitonuclear discordance and geographic variation have confounded species boundaries in a widely studied snake. Mol Phylogenet Evol. 2021. https://doi.org/10.1016/j.ympev.2021.107194.
Toussaint EF, Hall R, Monaghan MT, Sagata K, Ibalim S, Shaverdo HV, et al. The towering orogeny of New Guinea as a trigger for arthropod megadiversity. Nat Commun. 2014;5(1):1–10.
Popov SV, Rögl F, Rozanov AY, Steininger FF, Shcherba IG, Kovac M. Lithological-Paleogeographic maps of Paratethys-10 maps Late Eocene to Pliocene. 2004. p. 1–46.
Şapaş A, Boztepe-Güney A. Shear wave splitting in the Isparta Angle, southwestern Turkey: anisotropic complexity in the mantle. J Earth Syst Sci. 2009;118(1):71–80.
Avise JC, Walker D, Johns GC. Speciation durations and Pleistocene effects on vertebrate phylogeography. Proc R Soc Lond B. 1998;265(1407):1707–12.
Veith M, Schmidtler J, Kosuch J, Baran I, Seitz A. Palaeoclimatic changes explain Anatolian mountain frog evolution: a test for alternating vicariance and dispersal events. Mol Ecol. 2003;12(1):185–99.
Plötner J, Uzzell T, Beerli P, Akın Ç, Bilgin CC, Haefeli C, et al. Genetic divergence and evolution of reproductive isolation in eastern Mediterranean water frogs. In: Evolution in action. Springer, Berlin; 2010. p. 373–403.
Médail F, Diadema K. Glacial refugia influence plant diversity patterns in the Mediterranean Basin. J Biogeogr. 2009;36(7):1333–45.
Djamali M, Baumel A, Brewer S, Jackson ST, Kadereit JW, López-Vinyallonga S, et al. Ecological implications of Cousinia Cass. (Asteraceae) persistence through the last two glacial–interglacial cycles in the continental Middle East for the Irano-Turanian flora. Rev Palaeobotany Palynol. 2012;172:10–20.
Ahmadzadeh F, Flecks M, Carretero MA, Böhme W, Ihlow F, Kapli P, et al. Separate histories in both sides of the Mediterranean: phylogeny and niche evolution of ocellated lizards. J Biogeogr. 2016;43(6):1242–53.
Hernandez PA, Graham CH, Master LL, Albert DL. The effect of sample size and species characteristics on performance of different species distribution modeling methods. Ecography. 2006;29(5):773–85.
Ahmadi M, Hemami M-R, Kaboli M, Malekian M, Zimmermann NE. Extinction risks of a Mediterranean neo-endemism complex of mountain vipers triggered by climate change. Sci Rep. 2019;9(1):1–12.
Jezkova T, Wiens JJ. Rates of change in climatic niches in plant and animal populations are much slower than projected climate change. Proc R Soc B Biol Sci. 1843;2016(283):20162104.
Gonzalez A, Ronce O, Ferriere R, Hochberg ME. Evolutionary rescue: an emerging focus at the intersection between ecology and evolution. Phil Trans R Soc B. 2013. https://doi.org/10.1098/rstb.2012.0404.
Sexton JP, Montiel J, Shay JE, Stephens MR, Slatyer RA. Evolution of ecological niche breadth. Annu Rev Ecol Evol Syst. 2017;48:183–206.
Fraser DJ, Bernatchez L. Adaptive evolutionary conservation: towards a unified concept for defining conservation units. Mol Ecol. 2001;10(12):2741–52.
Ahmadzadeh F, Flecks M, Carretero MA, Mozaffari O, Böhme W, Harris DJ, et al. Cryptic speciation patterns in Iranian rock lizards uncovered by integrative taxonomy. PLoS ONE. 2013;8(12): e80563.
Mebert K, Göçmen B, İğci N, Kariş M, Oğuz MA, Yildiz MZ, et al. Mountain vipers in central-eastern Turkey: huge range extensions for four taxa reshape decades of misleading perspectives. Herpetol Conserv Biol. 2020;15(1):169–87.
Leigh JW, Lapointe F-J, Lopez P, Bapteste E. Evaluating phylogenetic congruence in the post-genomic era. Genome Biol Evol. 2011;3:571–87.
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–9.
Lanfear R, Calcott B, Ho SY, Guindon S. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 2012;29(6):1695–701.
Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29(8):1969–73.
Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst Biol. 2018;67(5):901–4.
Evans ME, Smith SA, Flynn RS, Donoghue MJ. Climate, niche evolution, and diversification of the “bird-cage” evening primroses (Oenothera, Sections Anogra and Kleinia). Am Nat. 2009;173(2):225–40.
Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20(2):289–90.
Phillips SJ, Anderson RP, Schapire RE. Maximum entropy modeling of species geographic distributions. Ecol Model. 2006;190(3–4):231–59.
Saladin B, Thuiller W, Graham CH, Lavergne S, Maiorano L, Salamin N, et al. Environment and evolutionary history shape phylogenetic turnover in European tetrapods. Nat Commun. 2019;10(1):249.
Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. Very high resolution interpolated climate surfaces for global land areas. Int J Climatol. 2005;25(15):1965–78.
Lobo JM, Jiménez-Valverde A, Real R. AUC: a misleading measure of the performance of predictive distribution models. Glob Ecol Biogeogr. 2008;17(2):145–51.
Heibl C, Calenge C. phyloclim: integrating phylogenetics and climatic niche modelling. http://cran.r-project.org/web/packages/phyloclim/index.html. 2013.
Matzke NJ. Probabilistic historical biogeography: new models for founder-event speciation, imperfect detection, and fossils allow improved accuracy and model-testing. Front Biogeogr. 2013. https://doi.org/10.21425/F5FBG19694.
Matzke NJ. Model selection in historical biogeography reveals that founder-event speciation is a crucial process in island clades. Syst Biol. 2014;63(6):951–70.
Blomberg SP, Garland T, Ives AR. Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution. 2003;57(4):717–45.
Pagel M. Inferring the historical patterns of biological evolution. Nature. 1999;401(6756):877–84.
Revell LJ. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol Evol. 2012;3(2):217–23.
Harmon LJ, Weir JT, Brock CD, Glor RE, Challenger W. GEIGER: investigating evolutionary radiations. Bioinformatics. 2008;24(1):129–31.
Felsenstein J. Phylogenies and the comparative method. Am Nat. 1985;125(1):1–15.
Revell LJ. Two new graphical methods for mapping trait evolution on phylogenies. Methods Ecol Evol. 2013;4(8):754–9.
Acknowledgements
We would like to thank the Department of Environment of Iran and the Isfahan University of Technology for their technical support. Our special thanks go to the Iranian team that helped us during field sampling: Kamran Almasieh, Mohammadreza Masoud, Saeid Khaki, Jalil Imani Harsini, and kind environmental guards and local people from Alborz and Zagros Mountains.
Funding
No funding was obtained for this study.
Author information
Authors and Affiliations
Contributions
M.A., M-R.H. and M.K. conceived the ideas; M.A., M.K. and P.G. collected the data; M.A. and M.N. analysed the data; and M.A., M-R.H., M.K., M.M., P.G., J.A. and N.E.Z. led the writing. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All field survey was included only recording occurrence points of the species. The sampling was conducted under a scientific permit and was approved by the Department of Environment (DoE) of Iran under the permission (94/1431).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1:
Table S1. Number of occurrence points and mitochondrial sequences used to reconstruct phylogenetic time tree and perform ecological niche modelling of mountain vipers. Table S2. Average percent contribution of six climatic variables, area under the receiver operating characteristic curve (AUC) and true statistics skill (TSS) of the ecological niche models performed for mountain vipers of the genus Montivipera in the eastern Mediterranean region. Table S3. Results of assessing the best evolutionary models of bioclimatic variables affecting Montivipera based on AICc statistics, using the BEAST chronogram and the fitContinuous function of geiger package. Bold values show the best model. AICc: Akaike Information Criterion for small sample-sized data, Ll: Log-likelihood. Table S4. List of samples, geographical origin, id and accession numbers for genes of the genus Montivipera and out-groups used in the phylogeographic and molecular dating analyses. Fig. S1. The Bayesian phylogeny tree reconstructed from the 177 sequences of mountain vipers (Montivipera). The tree was reconstructed based on a partitioned analysis with three Mitochondrial genes (CYTB, COX1, ND5). Values above and below the branches are the posterior probabilities and mean age estimates, respectively. Colors of the branches corresponds to the geographic origin of the species as shown in Fig. 1.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Ahmadi, M., Hemami, MR., Kaboli, M. et al. The legacy of Eastern Mediterranean mountain uplifts: rapid disparity of phylogenetic niche conservatism and divergence in mountain vipers. BMC Ecol Evo 21, 130 (2021). https://doi.org/10.1186/s12862-021-01863-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12862-021-01863-0