Abstract
Background
Skippers (Family: Hesperiidae) are a large group of butterflies with ca. 4000 species under 567 genera. The lack of a time-calibrated higher-level phylogeny of the group has precluded understanding of its evolutionary past. We here use a 10-gene dataset to reconstruct the most comprehensive time-calibrated phylogeny of the group, and explore factors that affected the diversification of these butterflies.
Results
Ancestral state reconstructions show that the early hesperiid lineages utilized dicots as larval hostplants. The ability to feed on monocots evolved once at the K-Pg boundary (ca. 65 million years ago (Mya)), and allowed monocot-feeders to diversify much faster on average than dicot-feeders. The increased diversification rate of the monocot-feeding clade is specifically attributed to rate shifts in two of its descendant lineages. The first rate shift, a four-fold increase compared to background rates, happened ca. 50 Mya, soon after the Paleocene-Eocene thermal maximum, in a lineage of the subfamily Hesperiinae that mostly fed on forest monocots. The second rate shift happened ca. 40 Mya in a grass-feeding lineage of Hesperiinae when open-habitat grasslands appeared in the Neotropics owing to gradual cooling of the atmospheric temperature.
Conclusions
The evolution of monocot feeding strongly influenced diversification of skippers. We hypothesize that although monocot feeding was an intrinsic trait that allowed exploration of novel niches, the lack of extensive availability of monocots comprised an extrinsic limitation for niche exploration. The shifts in diversification rate coincided with paleoclimatic events during which grasses and forest monocots were diversified.
Similar content being viewed by others
Background
The remarkable diversity of life is often attributed to either the gradual accumulation of species over long periods of time [1, 2] or to dramatic changes in diversification rates across lineages and time [3, 4]. However, paleontological evidence [3] and phylogenetic comparisons [4] across the tree of life predict the latter scenario as the major cause, where the fluctuations in speciation and extinction rates explain the historical pattern of lineage accumulation. A major cause for these varying rates is rapid diversification promoted by ecological opportunity (EO) [5,6,7,8]. EO is generated when a macroevolutionary process affects a lineage in two ways: (i) the lineage evolves the ability to utilize resources otherwise unavailable (by evolutionary innovation; intrinsic factor), and (ii) when new resources become available to the lineage (by colonization or antagonistic extinction or in situ availability of new resource; extrinsic factors) [5,6,7,8,9].
Many comparative studies have shown how rapid diversification is associated with evolutionary innovations ([7,8,9]; but see [10, 11]). Of particular note is the evolution of herbivory, i.e., shifts from carnivory to herbivory, which has repeatedly elevated diversification rates [12, 13], and this is especially true in the case of insects [13]. Indeed, herbivorous insects comprise about half of all terrestrial eukaryotic species [13], exemplifying the importance of insect-plant interactions in generating the diversity of life [14,15,16]. Among butterflies, major shifts in hostplant use have led to bursts in diversification rates [17, 18]. For instance, Satyrini butterflies (Subfamily Satyrinae: Tribe Satyrini), which specialise on grasses and include ca. 2200 species, diversified simultaneously with the expansion of grasslands [19]. Therefore, feeding on grasses appears to be closely associated with increased diversification rate in Satyrini.
Paleontological records and comparative phylogenetic analyses indicate that diversification of a taxon is often associated with the extinction of others [20,21,22]. For instance, nymphalid butterflies (Family Nymphalidae) radiated immediately after the K-Pg extinction (~65 million years ago (Mya)) [23]. Alternatively, in situ availability of new resources can elevate diversification rates of the lineage (see [7, 8]). For instance, paleoclimatic events trigger the appearance and diversification of several groups of animals and plants [24,25,26,27], which in turn influence diversification of other taxa. Phytophagous insects, for example, diversified following the evolution of flowering plants ([22, 28]; but see [29, 30]). Furthermore, colonization of a novel geographic area, newly formed islands for instance, are often associated with increased diversification [31,32,33].
Thus, evidence supporting the role of EO in accelerating diversification largely comes from analyses of distinct macroevolutionary processes that represent either intrinsic or extrinsic factors. However, theory predicts that both availability of resources due to extrinsic factors and the intrinsic ability to utilize them are necessary to generate EO [7,8,9]. For instance, a lineage capable of utilizing a new but scarce resource may undergo rapid diversification when those resources become abundant in situ. Although intrinsic and extrinsic factors of EO are often presented independently, their role in generating EO are not mutually exclusive; and, as the aforementioned predictions suggest, the factors promoting EO may appear one after another over a phylogeny, but EO is generated only when all the factors become available to the lineage. However, this possibility is not explored in detail for any group of organisms.
We here investigate the pattern of appearance of intrinsic and extrinsic factors of EO and their effect on diversification in a highly speciose but hitherto ignored group of butterflies - the skippers (Family Hesperiidae). The skippers comprise ca. 4000 species distributed among ca. 567 genera [34]. About 50% of skippers feed on monocots during larval stages. We compiled a 10-gene dataset of 7726 bp (base pair) from 290 genera representing all the known major clades. Based on estimated times of divergence, we checked whether skippers have experienced rapid shifts in diversification rates, indicative of adaptive radiation(s). To test the hypothesis that the rate shifts have occurred in response to ecological opportunity, we specifically investigate whether (i) rate shifts are associated with feeding preferences of the lineages, (ii) the K-Pg extinction had an impact on diversification rate, (iii) the diversification pattern reflects the biogeographic distribution of lineages, and (iv) paleoclimate has influenced diversification rate.
Methods
Phylogenetic and molecular dating analyses
We used a concatenated dataset of nine nuclear genes and one mitochondrial gene for the divergence time estimation. Our dataset was built upon the previous work [34], to which we added 34 specimens, resulting in a dataset of 290 genera that accounts for nearly 60% of known skipper genera. Many skipper genera have highly restricted distributions and comprise very few species, making them logistically very challenging to sample. Therefore, despite the massive field effort of several collaborators worldwide spanning more than a decade, we were unable to achieve a more comprehensive sampling of the taxa. However, we believe we have included representatives of all the major clades.
A previous study [34] with 270 skipper samples showed that there are two equally plausible topologies of hesperiid relationships. Hence, we first investigated whether the existence of the contrasting topologies (see [34]) has any effect on the age estimates of its major clades. We used 105 branch-optimized trees obtained from independent Maximum Likelihood (ML) analysis of the previously published dataset [34] with gene partitions. We estimated ultrametric trees using the PATHd8 method [35], which uses the mean path length algorithm with correction for a molecular clock, implemented in the program PATHd8 v1.0 [35]; and assigned an arbitrary time unit of one to the crown age of the ingroup to estimate relative times of divergence for nodes. We then calculated the distribution of relative age estimates of the deeper clades from these ultrametric trees.
We used the software BEAST v2.4.2 [36] on the CIPRES Science Gateway [37] to simultaneously estimate phylogenetic relationships and times of divergences for our current dataset. Sequences of two species from Hedylidae, which is the sister family to Hesperiidae [38], were acquired from Genbank and treated as outgroups. We estimated the most appropriate partitioning scheme for the data matrix using TIGER v1.02 (Tree Independent Generation of Evolutionary Rates) [39] and the nucleotide substitution models for the partitions by PartitionFinder v1.1.1 [40]. We assumed a relaxed clock model that allowed branch lengths to vary as per an uncorrelated lognormal distribution, and assigned a Birth-Death Process as the tree prior. We assigned an exponential distribution of mean 10.0 as the prior for the hyperparameter ‘ucldMean’. Priors of all other parameters were kept at their default values.
We used the previously estimated age ranges (81–114 Mya) [38] to calibrate the split between Hesperiidae and Hedylidae (uniform distribution), along with a recently described fossil [41] to constraint the minimum stem age of Hesperiinae (25 Mya; log-normal distribution). Posteriors of the parameters were estimated from two independent runs of 50 million generations each. We checked the convergence of independent runs from the distribution of their log-likelihood scores using the program Tracer v1.6 [42]. To confirm proper mixing of samples from every generation, we checked the Effective Sample Size of all parameters after discarding the initial 20% of trees as burnin. The tree files, after discarding burnin, were combined using LogCombiner v2.4.5 and the parameter values were annotated to the Maximum Clade Credibility (MCC) tree using TreeAnnotater v2.4.5 (a part of BEAST v2.4.5 package; [36]).
Diversification analysis
For subsequent analyses, we retained only one randomly chosen representative species for each genus if multiple species for that genus were available. We estimated gamma (γ) statistic [43] from the MCC tree using the R (v3.2.3; [44]) package Laser v2.4.1 [45]. The gamma statistic indicates whether internal nodes are closer to the root (γ < 0) or to the tips (γ > 0) of the tree than expected under a constant rate model (γ = 0). We accounted for incomplete taxon sampling by adjusting the critical value for gamma using the Monte Carlo Constant Rates (MCCR) test [43].
We generated a Lineage Through Time (LTT) plot from the MCC tree. To evaluate the pattern of lineage accumulation in the empirical LTT plot, we generated 1000 trees under the birth-death model with 290 taxa using the R package TreeSim v2.2 [46]. These trees were then used to construct a mean LTT curve with 95% confidence interval and compared with the empirical LTT curve. We evaluated the fit of the lineage accumulation on the MCC tree to different models of diversification [47] using hierarchical likelihood ratio test and Akaike Information Criteria (AIC) in the R package Laser v2.4.1 [45].
We calculated the diversification rates of higher level clades following the method-of-moments estimator for stem-group ages [48] and checked using Phylogenetic Generalized Least Squares (PGLS) [49] in the R package APE v3.5 [50] whether clade age or diversification rate predicts species richness. PGLS accounts for phylogenetic non-independence of clades while modeling regression between the parameters.
We employed the program BAMM v2.5.0 [51] to model the diversification rate shifts. This is a Bayesian approach that describes the number and locations of rate shifts as posterior distributions. We specified the sampling probability as the proportion of representative species in our dataset out of total known species in each tribe or subfamily. To check for the prior sensitivity of BAMM to the number of rate shifts detected, we performed multiple BAMM analyses with varying priors (1, 5 and 10) for expected rate shifts. We ran the analyses for 5 million generations each and sampled every 5000 generations. Since priors had little effect on the results, we used one as the rate shift prior during the final analysis, performed with four independent chains of 20 million generations each and sampled every 20,000 generations. After removing the first 20% of generations as burnin, the number of rate shifts and rate shift configurations were estimated using the R package BAMMtools v2.1.0 [52]. We note that although BAMM is very popular, there has been recent debate about the reliability of the method. In particular, the likelihood function and the prior used in BAMM have been argued to be flawed [53], but [54] later argued that these problems do not apply to most datasets. Rather than relying solely on BAMM, we have adopted multiple modelling approaches and we believe our results are robust to the potential flaws of BAMM (see Discussion for more detail).
In another approach, we estimated trait-specific diversification rates considering hostplants as binary characters - monocots or dicots. About 16% of the taxa in our dataset feed on magnoliids or both monocot and dicot; we assigned them as data unavailable. The character states for each tip in the phylogeny were compiled from multiple sources (Additional file 1: Appendix S1). We applied the Binary State Speciation and Extinction model (BiSSE) [55] in the R package diversitree v0.9.7 [56]. BiSSE calculates speciation, extinction and transition rates for each assigned character state. We accounted for the incomplete sampling in our dataset by providing the total proportion of missing taxa in our dataset. We used a uniform prior probability and 10,000 MCMC steps to estimate the posterior probability distribution of each parameter.
Nevertheless, the presence of unmeasured factors could potentially influence the diversification pattern estimated for the states of the observed trait over a phylogeny, and hence can lead to erroneous inference when using SSE (State dependent Speciation and Extinction) models [57, 58]. Thus, we also tested whether shifts in diversification rates correlate with the shift in feeding habit or any unmeasuerd factors, by applying Hidden State Speciation and Extinction model (HiSSE) [58].
Character mapping
We mapped the known biogeographic distribution of the sampled taxa onto the phylogeny using the online tool Interactive Tree Of Life (iTOL) v3 [59]. For each missing genus, we randomly assigned the distribution to a sampled genus from the same tribe or subfamily. The geographic range of genera was compiled from various sources (Additional file 1: Appendix S1) and was divided into six biogeographic regions – Neotropical, Nearctic, Palearctic, Afrotropical, Oriental and Australian.
We also annotated the phylogeny with the larval hostplant data of sampled genera using iTOL v3 [59] and accounted for the hostplants of missing genera in the manner as was done for geographic ranges above. Based on information about larval hostplant use, we divided the genera into three broad categories – dicots, monocots and magnoliids. Additionally, the monocots were subdivided into Poales, Arecales, Zingiberales and Asparagales.
Results
Although a previous study [34] indicated two equally likely tree topologies for higher relationships in skipper butterflies, after scaling those topologies in relative times with the crown age of skippers assigned to one unit time, we observed that the relative times of divergences across the contrasting topologies were similar for all major clades, except Euschemoninae, Eudaminae and two clades of Pyrginae (Additional file 1: Figure S1). To assess the influence of topological uncertainties on diversification analyses, we performed all diversification analyses additionally on 100 randomly selected posterior trees from the dating analysis, which showed no such influence.
Diversification rates and pattern
The MCC tree indicates that Hesperiidae began diversifying in the late Cretaceous ca. 82 Mya, when Coeliadinae diverged from the rest of the family. Based on the Monte Carlo Constant Rates (MCCR) test, internal nodes were significantly closer to the root than expected, indicative of a decrease in net diversification rate over time (gamma = −13.88, critical gamma = −12.72 at p = 0.004). The LTT plot deviated significantly from a simulated curve generated under constant diversification rate with incomplete taxon sampling (Fig. 1d), indicating heterogeneity in diversification rate through time. Moreover, a model of diversification specifying change in rate best fits the lineage accumulation on the MCC tree (Additional file 1: Table S1).
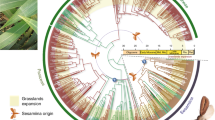
Diversification rates across the hesperiid phylogeny and the comparison of rates with paleoclimatic events. A more detailed figure with names of genera is shown in Additional file 2. a The hesperiid time tree mapped with hostplant data and geographic distributions. The names at nodes represent subfamilies. The terminal branches are colored based on the broad category of hostplant use. Monocot feeding taxa are additionally shown with the known categories of plants (black Circle: Poales; Outward triangle: Arecales; Star: Zingiberales; Inward triangle: Asparagales) they feed upon. Coloured circles represent known distributions of the taxa (a filled circle indicates presence in the area). The arrow indicates the shift from dicot to monocot feeding. The red stars at nodes indicate the points of diversification rate shifts (numbered as shift 1 and 2) from the BAMM analysis. b Posterior distributions from the BiSSE analysis for net diversification rates of monocot- and dicot-feeding hesperiids. Lines below each distribution are 95% confidence intervals. c Net diversification rate from the BAMM analysis for the dicot- (blue) and monocot-(red) feeding lineages. d The LTT curve (in red) of the MCC tree superimposed on the LTT curves (in grey) from 1000 trees simulated under constant diversification rate for 290 taxa. e The change in paleoclimatic temperature (as in [93]) (in grey) plotted using the R package RPADNA v1.2 [94]. This climatic plot is superimposed with the mean speciation rate (along with the posterior distributions) for the whole hesperiid phylogeny (in dark grey) and the speciation rates of the lineages those experienced rate shifts (shift 1 and 2 as in (a))
The pattern of variation in diversification rates was also reflected in the BAMM analysis accounting for extant species richness. There was a slow and continuous increase in the net diversification rate up to 50 Mya (Fig. 1e; black curve), after which the slope of the curve increased steadily indicating an increase in the rate. After ~40 Mya, the diversification rate gradually declined.
This disparity in diversification rates across the phylogeny explains the species richness of higher-level clades (PGLS: r = 0.87; p < 0.0001). The trait-independent analysis in BAMM estimated two major shifts in diversification rate over the phylogeny (Fig. 1a) (basal rate: 0.10, rate at 1 st shift = 0.48 and 2nd shift = 0.24), both within the subfamily Hesperiinae. We note that while the probability for two rate shifts on the phylogeny is 0.83, the point estimates for those shifts have varying probabilities, i.e. 0.9 for 1st shift and 0.6 for 2nd shift, as evident from credibility set of rate shifts (Additional file 1: Figure S2).
Influence of the K-Pg event
The LTT plot shows that the diversification rate slowed down around K-Pg boundary (~ 65 Mya), suggesting either slower speciation rate or lineage extinctions (Fig. 1d). The rate of lineage accumulation remained low up to ca. 50 Mya, after which the rate suddenly increased.
Hostplant use and Biogeography
Mapping of the larval hostplant data onto the phylogeny (Fig. 1a) indicated that the early hesperiid lineages fed on dicots. Around ca. 65 Mya, one lineage shifted to monocot feeding and later this lineage gave rise to three subfamilies including the most speciose subfamily Hesperiinae.
The posterior distribution from the BAMM analysis indicates that the net diversification rate of the monocot-feeding clade was higher than that of the dicot-feeding clades (Fig. 1c). The differential diversification rates across the monocot- and dicot-feeding clades were corroborated in the BiSSE analysis (Fig. 1b), where the higher diversification rate of the monocot-feeding lineages corresponded to a higher speciation rate although extinction rates did not differ between dicot- and monocot-feeding groups (Additional file 1: Figure S3). Topological uncertainties had no major influence on these estimations, as the AIC value from the MCC tree analysis is close to the average of the AIC values from 100 randomly selected posterior trees under a full model analysis (Additional file 1: Figure S4). However, a comparison of the SSE models (Additional file 1: Table S2) indicated that the model with hidden (unmeasured) states of the trait best fits the diversification pattern.
We did not find any strong associations between diversification rate shifts and biogeographic patterns (Fig. 1a). However, the mapping illustrated that the first rate shift happened in a lineage distributed across the Oriental and Afrotropical regions. The second rate shift occurred in a Neotropical lineage.
Discussion
We present the most comprehensive phylogenetic hypothesis of hesperiid butterflies, including ca. 300 taxa. Based on this phylogeny, we infer the macroevolutionary history of the group in relation to historical events and coevolutionary interactions with their hostplants.
Skippers started evolving ca. 90 Mya, when dicots were the dominant plants [60], and early lineages of the family diversified on these plants. Evolution of monocot feeding ca. 65 Mya provided the intrinsic ability to utilize otherwise unavailable ecological resources. This novel interaction with the environment resulted a higher net diversification rate of the monocot-feeding group (Fig. 1b, c). The robustness of this correlation (monocot feeding ~ diversification) to fundamentally different approaches of analyses (BAMM, and BiSSE; see Fig. 1b, c) lends further support to the higher diversification of monocot-feeding group. Although the monocot-feeding lineage represented <20% of the lineages at the K-Pg boundary (~65 Mya), it includes ca. 50% of extant species.
However, model comparisons indicated a possible influence of unknown factors on the estimated diversification pattern suggesting that the association between monocot-feeding and diversification rate must be influenced by certain unmeasured factors. Interestingly, we found that the change to monocot feeding did not increase diversification immediately – the rate shift, a ca. four-fold increase compared to the background rate, occurred ca. 50 Mya (Fig. 1a, e) within the subfamily Hesperiinae. Although the intrinsic ability to utilize monocots provided new capability for niche exploration, the low availability of monocots probably imposed an extrinsic limitation that did not allow high rates of diversification initially. The earliest monocot-feeding lineages specialized on grasses (Fig. 1a); the limited availability of open grassland habitats may have also imposed a limitation on diversification. The rate shift occurred soon after the Paleocene–Eocene Thermal Maximum (PETM) ca. 55–56 Mya, a period when the earth’s temperature increased dramatically as a result of addition of carbon to the oceans and the atmosphere [61, 62].
The PETM followed less-severe warming periods up to early Eocene (~ 50 Mya) mainly due to shuffling of carbon between atmosphere and ocean [63]. These warm periods are associated with diversification of forest dwelling monocots and also experienced compositional changes in insect fauna, as evidenced by fossil records [64, 65]. Eocene fossils indicate elevated levels of insect herbivory (leaf damage frequency) and greater insect herbivore diversity per hostplant (leaf damage type) [28, 64]. Thus, the rapid radiation of skippers ca. 50 Mya coincides with the historical diversification pattern of herbivorous insects in the Eocene, although the selective forces behind these radiations are not known [65].
The increase in diversification beginning ca. 50 Mya happened in lineages distributed in the Oriental and Afrotropical regions. This was followed by a downward shift in diversification rate, during the late Eocene to Oligocene, possibly due to saturation of ecological niches in these biogeographic areas. The diversification rate again increased (about two-fold compared to the background rate) ca. 40 Mya in a Neotropical grass-feeding (Poales) lineage (Fig. 1a, e). This shift coincides with the appearance of open-habitat grasses in the Neotropics when the atmospheric temperature gradually decreased [66,67,68]. Therefore, the emergence of open-habitat grasslands appears to provide the extrinsic factor of EO for the diversification of skipper butterflies. Although grasses became dominant in Neotropics during the Miocene (~ 20 Mya) [68], this increased availability of grasses, interestingly, had little impact on hesperiids.
Our study suggests that the extrinsic availability of resources can modulate the potential of intrinsic ability in generating EO for rapid diversification. The diversification pattern in Satyrinae butterflies [19] is consistent with this hypothesis. In Satyrinae, the ability to feed on grasses appeared early in the lineage; however, rapid diversification occurred in one lineage – the tribe Satyrini – when grasses became abundant during Oligocene (~ 33–26 Mya) [19].
Although grass feeding has been thought to be an important evolutionary innovation that is closely tied to rapid radiations of most herbivorous insects [69], the mechanistic basis of increased speciation rates due to grass feeding is unclear. Diffuse coevolution [70,71,72] between insects and their hostplants, which has been widely shown to increase insect speciation rates [13, 73, 74], has been attributed to the pressure on herbivorous insects to specialize. As first postulated by Ehrlich and Raven [14], plants evolve chemical defenses against herbivores [75], which forces herbivores to specialize [76], leading to a coevolutionary arms race [15, 16]. Episodes of generalization and specialization can then lead to increased speciation rates [77, 78]. However, grass-feeding insects tend to be broad generalists, with rare examples of narrow specialists (see [79, 80]). This is likely because most grasses are expected to rely predominantly on physical defenses such as silicifaction [81,82,83] (also see [84, 85]). Although chemical defenses in the form of secondary metabolites [86, 87] and fungal-mediated toxins [88, 89] have been reported in grasses (also see [90, 91]), the extent to which these chemicals confer defense against herbivory are still obscure and limited to few taxa.
Yet, satyrines and skippers, the major butterfly groups that feed on grasses, both appear to have radiated rapidly as a result of grass feeding. We opine that this is likely not because of the classical insect-hostplant coevolutionary arms race postulated by Ehlrich and Raven [14]. Rather, the opportunity to invade grasslands (savannahs) could have been the key. The dominance of grasslands over forest communities may have increased rates of allopatric speciation.
We did not find evidence for the effect of the K-Pg boundary on the diversification of skipper butterflies, unlike in nymphalid butterflies that radiated explosively immediately after this event [23]. However, the evolution of monocot feeding coincided with the K-Pg event. We hypothesize that the catastrophic events during this period led to the extinction of competing herbivores on monocots, and thus allowed the inclusion of monocots in the hostplant repertoire of skippers.
Potential methodological concerns
BiSSE
Although hugely popular, BiSSE is known to perform poorly when the number of tips is low, and when the tip-ratio bias (ratio of numbers of tips with different character states) is high [92]. Davis et al. [92] concluded that BiSSE results should be inferred with extreme caution when the phylogeny included fewer than 300 terminals and/or when fewer than 10% of species are of one character state. Although tip-ratio bias is unlikely to be a problem in our dataset, we acknowledge that the number of tips is a concern. However, the main result from BiSSE, i.e., increased diversification of monocot feeders compared to that of dicot feeders, is also corroborated by BAMM, a fundamentally different modelling approach. Therefore we believe that the result is robust.
BAMM
Moore et al. [53] argued that BAMM is strongly affected by the priors specified, and that the estimates of diversification rate parameters are unreliable (but see [54]). To check the effect of priors on the BAMM results, we performed multiple analyses with varying priors for rate shifts, but found that priors had no effect on the number or positions of rate shifts detected.
Therefore, we have confidence in the main results presented here.
Conclusions
We have shown that the ability of skipper butterflies to feed on monocots evolved at the K-Pg boundary and provided a novel intrinsic EO that eventually allowed monocot-feeders to have a higher diversification rate compared to dicot-feeders. The diversification rate (a four-fold increase compared to background rates) itself shifted ca. 50 Mya, soon after the PETM, in a lineage of the subfamily Hesperiinae. We attribute this delayed increase in diversification to the limited availability of monocots, which comprised an extrinsic limitation for niche exploration. There was another increase in diversification rate (a ca. two-fold increase) in a Neotropical Hesperiinae lineage ca. 40 Mya, coinciding with the appearance of grassland communities in the region. We suggest that grass feeding allowed rapid radiations in the two major groups of grass-feeding butterflies - skippers and satyrines - not through the classical insect-hostplant coevolutionary arms race, but by enhancing the chances of allopatric speciation.
References
McPeek MA, Brown JM. Clade age and not diversification rate explains species richness among animal taxa. Am Nat. 2007;169:E97–106.
Hedges SB, Marin J, Suleski M, Paymer M, Kumar S. Tree of life reveals clock-like speciation and diversification. Mol Biol Evol. 2015;32:835–45.
Condamine FL, Clapham ME, Kergoat GJ. Global patterns of insect diversification: towards a reconciliation of fossil and molecular evidence? Sci Rep. 2016;6:19208.
Scholl JP, Wiens JJ. Diversification rates and species richness across the tree of life. Proc Royal Soc Biol Sci. 2016;283:20161334.
Simpson GG. Tempo and mode in evolution. New York: Columbia University Press; 1949.
Simpson GG. The major features of evolution. New York: Columbia University Press; 1953.
Yoder JB, Clancey E, DesRoches S, Eastman JM, Gentry L, Godsoe W, et al. Ecological opportunity and the origin of adaptive radiations. J Evol Biol. 2010;23:1581–96.
Stroud JT, Losos JB. Ecological opportunity and adaptive radiation. Annu Rev Ecol Evol Syst. 2016;47:507–32.
Wellborn GA, Langerhans RB. Ecological opportunity and the adaptive diversification of lineages. Ecology and Evolution. 2015;5:176–95.
McGee MD, Borstein SR, Neches RY, Buescher HH, Seehausen O, Wainwright PC. A pharyngeal jaw evolutionary innovation facilitated extinction in Lake Victoria cichlids. Science. 2015;350(6264):1077–9.
Wainwright PC, Price SA. The impact of organismal innovation on functional and ecological diversification. Integr Comp Biol. 2016;56(3):479–88.
Price S, Hopkins S, Smith K, Roth V. Tempo of trophic evolution and its impact on mammalian diversification. Proc Natl Acad Sci. 2012;109:7008–12.
Wiens J, Lapoint R, Whiteman N. Herbivory increases diversification across insect clades. Nat Commun. 2015;6:8370.
Ehrlich PR, Raven PH. Butterflies and plants: a study in coevolution. Evolution. 1964;18:586–608.
Mitter C, Farrell B, Futuyma D. Phylogenetic studies of insect-plant interactions: Insights into the genesis of diversity. Trends Ecol Evol. 1991;6:290–3.
Vermeij GJ. The evolutionary interaction among species: selection, escalation, and coevolution. Annu Rev Ecol Syst. 1994;25:219–36.
Fordyce JA. Host shifts and evolutionary radiations of butterflies. Proc R Soc Lond B Biol Sci. 2010;277:3735–43.
Hardy NB, Otto SP. Specialization and generalization in the diversification of phytophagous insects: tests of the musical chairs and oscillation hypotheses. Proc R Soc Lond B Biol Sci. 2014;281:20132960.
Peña C, Wahlberg N. Prehistorical climate change increased diversification of a group of butterflies. Biol Lett. 2008;4:274–8.
Niklas KJ, Tiffney BH, Knoll AH. Patterns in vascular land plant diversification. Nature. 1983;303:614–6.
Penny D, Phillips MJ. The rise of birds and mammals: are microevolutionary processes sufficient for macroevolution? Trends Ecol Evol. 2004;19:516–22.
Labandeira CC, Johnson KR, Wilf P. Impact of the terminal Cretaceous event on plant-insect associations. Proc Natl Acad Sci. 2002;99:2061–6.
Wahlberg N, Leneveu J, Kodandaramaiah U, Peña C, Nylin S, Freitas AV, et al. Nymphalid butterflies diversify following near demise at the Cretaceous/Tertiary boundary. Proc R Soc Lond B Biol Sci. 2009;276:4294–302.
Heimhofer U, Hochuli PA, Burla S, Dinis JML, Weissert H. Timing of early Cretaceous angiosperm diversification and possible links to major paleoenvironmental change. Geology. 2005;33:141–4.
Scheibner C, Speijer RP, Marzouk AM. Turnover of larger foraminifera during the Paleocene-Eocene Thermal Maximum and paleoclimatic control on the evolution of platform ecosystems. Geology. 2005;33:493–6.
Vieites DR, Min M-S, Wake DB. Rapid diversification and dispersal during periods of global warming by plethodontid salamanders. Proc Natl Acad Sci. 2007;104:19903–7.
Groves J, Yue W. Foraminiferal diversification during the late Paleozoic ice age. Paleobiology. 2009;40:367–92.
Currano E, Labandeira C, Wilf P. Fossil insect folivory tracks paleotemperature for six million years. Ecol Monogr. 2010;80:547–67.
Labandeira CC, Currano ED. The fossil record of plant-insect dynamics. Annu Rev Earth Planet Sci. 2013;41:287–311.
Labandeira CC, Sepkoski JJJ. Insect diversity in the fossil record. Science. 1993;261:310–5.
Gillespie R. Community assembly through adaptive radiation in Hawaiian spiders. Science. 2004;303:356–9.
Baldwin BG. Adaptive radiation of shrubby tarweeds (Deinandra) in the California Islands parallels diversification of the Hawaiian silversword alliance (Compositae–Madiinae). Am J Bot. 2007;94:237–48.
Givnish TJ, Millam KC, Mast AR, Paterson TB, Theim TJ, Hipp AL, et al. Origin, adaptive radiation and diversification of the Hawaiian lobeliads (Asterales: Campanulaceae). Proc R Soc B Biol Sci. 2009;276:407–16.
Sahoo RK, Warren AD, Wahlberg N, Brower AV, Lukhtanov VA, Kodandaramaiah U. Ten genes and two topologies: an exploration of higher relationships in skipper butterflies (Hesperiidae). PeerJ. 2016;4:e2653.
Britton T, Anderson CL, Jacquet D, Lundqvist S, Bremer K. Estimating divergence times in large phylogenetic trees. Syst Biol. 2007;56:741–52.
Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu C-H, Xie D, et al. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 2014;10:e1003537.
Miller M, Pfeiffer W, Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees in Proceedings of the Gateway Computing Environments Workshop (GCE). New Orleans, LA. 2010;1–8. doi:10.1109/GCE.2010.5676129.
Heikkilä M, Kaila L, Mutanen M, Peña C, Wahlberg N. Cretaceous origin and repeated tertiary diversification of the redefined butterflies. Proc R Soc Lond B Biol Sci. 2012;279:1093–9.
Cummins CA, McInerney JO. A method for inferring the rate of evolution of homologous characters that can potentially improve phylogenetic inference, resolve deep divergence and correct systematic biases. Syst Biol. 2011;60:833–44.
Lanfear R, Calcott B, Ho S, Guindon S. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 2012;29:1695–701.
De Jong R. Reconstructing a 55-million-year-old butterfly (Lepidoptera: Hesperiidae). Eur J Entomol. 2016;113:423–8.
Rambaut A. Suchard MA, Xie D. Tracer: Drummond AJ; 2014.
Pybus O, Harvey P. Testing macro-evolutionary models using incomplete molecular phylogenies. Proc Royal Soc B Biological Sci. 2000;267:2267–72.
R Core team. R: a language and environment for statistical computing. 2015.
Rabosky D. LASER: a maximum likelihood toolkit for detecting temporal shifts in diversification rates from molecular phylogenies. Evol Bioinforma. 2006;2:273–6.
Stadler T. Simulating trees with a fixed number of extant species. Syst Biol. 2011;60:676–84.
Rabosky DL, Lovette IJ. Explosive evolutionary radiations: decreasing speciation or increasing extinction thrrough time? Evolution. 2008;62:1866–75.
Magallón S, Sanderson MJ. Absolute diversification rates in angiosperm clades. Evolution. 2001;55:1762–80.
Martins EP, Hansen TF. Phylogenies and the comparative method: a general approach to incorporating phylogenetic information into the analysis of interspecific data. Am Nat. 1997;149:646–67.
Popescu A-A, Huber K, Paradis E. ape 3.0: new tools for distance-based phylogenetics and evolutionary analysis in R. Bioinformatics. 2012;28:1536–7.
Rabosky DL. Automatic detection of key innovations, rate shifts, and diversity-dependence on phylogenetic trees. PLoS One. 2014;9:e89543.
Rabosky D, Grundler M, Anderson C, Title P, Shi J, Brown J, et al. BAMMtools: an R package for the analysis of evolutionary dynamics on phylogenetic trees. Methods Ecol Evol. 2014;5(7):701–7.
Moore BR, Höhna S, May MR, Rannala B, Huelsenbeck JP. Critically evaluating the theory and performance of Bayesian analysis of macroevolutionary mixtures. Proc Natl Acad Sci. 2016;113:9569–74.
Rabosky DL, Mitchell JS, Chang J. Is BAMM flawed? Theoretical and practical concerns in the analysis of multi-rate diversification models. Syst Biol. 2017;66(4):477–98.
Maddison WP, Midford PE, Otto SP. Estimating a binary character’s effect on speciation and extinction. Syst Biol. 2007;56:701–10.
FitzJohn RG. Diversitree: comparative phylogenetic analyses of diversification in R. Methods Ecol Evol. 2012;3:1084–92.
Rabosky DL, Goldberg EE. Model inadequacy and mistaken inferences of trait-dependent speciation. Syst Biol. 2015;64(2):340–55.
Beaulieu JM, O’Meara BC. Detecting hidden diversification shifts in models of trait-dependent speciation and extinction. Syst Biol. 2016;65(4):583–601.
Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–5.
Willis K, McElwain J. The evolution of plants. Oxford: Oxford University Press; 2014.
McInerney FA, Wing SL. The Paleocene-Eocene Thermal Maximum: a perturbation of carbon cycle, climate, and biosphere with implications for the future. Earth Planet Sci. 2011;39:489–516.
Schaller MF, Fung MK, Wright JD, Katz ME, Kent DV. Impact ejecta at the Paleocene-Eocene boundary. Science. 2016;354:225–9.
Sexton P, Norris R, Wilson P, Pälike H, Westerhold T, Röhl U, et al. Eocene global warming events driven by ventilation of oceanic dissolved organic carbon. Nature. 2011;471:349–52.
Wilf P, Labandeira CC. Response of plant-insect associations to Paleocene-Eocene warming. Science. 1999;284:2153–6.
Wilf P, Labandeira CC, Johnson KR, Cúneo RN. Richness of plant–insect associations in Eocene Patagonia: a legacy for South American biodiversity. Proc Natl Acad Sci. 2005;102:8944–8.
Edwards EJ, Osborne CP, Stromberg CA, Smith SA, C4 Grasses Consortium. The origins of C4 grasslands: integrating evolutionary and ecosystem science. Science. 2010;328:587–91.
Strömberg CA. Evolution of grasses and grassland ecosystems. Annu Rev Earth Planet Sci. 2011;39:517–44.
Strömberg CA, Dunn RE, Madden RH, Kohn MJ, Carlini AA. Decoupling the spread of grasslands from the evolution of grazer-type herbivores in South America. Nat Commun. 2013;4:1478.
van Bergen E, Barlow HS, Brattström O, Griffiths H, Kodandaramaiah U, Osborne CP, Brakefield PM. The stable isotope ecology of mycalesine butterflies: implications for plant-insect co-evolution. Funct Ecol. 2016;30:1936–46.
Janzen DH. When is it Coevolution? Evolution. 1980;34:611–2.
Fox LR. Diffuse coevolution within complex communities. Ecology. 1988;69:906–7.
Strauss S, Sahli H, Conner J. Toward a more trait-centered approach to diffuse (co)evolution. New Phytol. 2005;165:81–90.
Mitter C, Farrell B, Wiegmann B. The phylogenetic study of adaptive zones: has phytophagy promoted insect diversification? Am Nat. 1988;132:107–28.
Winkler IS, Mitter C. The phylogenetic dimension of insect-plant interactions: a summary of recent evidence. Berkeley: Univ. California Press; 2008.
Becerra JX, Noge K, Venable LD. Macroevolutionary chemical escalation in an ancient plant–herbivore arms race. Proc Natl Acad Sci. 2009;106:18062–6.
Karban R, Agrawal AA. Herbivore offense. Annu Rev Ecol Syst. 2002;33:641–64.
Janz N, Nylin S, Wahlberg N. Diversity begets diversity: host expansions and the diversification of plant-feeding insects. BMC Evol Biol. 2006;6:1–10.
Hamm C, Fordyce J. Patterns of host plant utilization and diversification in the brush-footed butterflies. Evolution. 2015;69:589–601.
Denno RF, Roderick GK. Population biology of planthoppers. Annu Rev Entomol. 1990;35:489–520.
Tscharntke T, Greiler HJ. Insect communities, grasses, and grasslands. Annu Rev Entomol. 1995;40:535–58.
Keeping MG, Meyer JH. Silicon-mediated resistance of sugarcane to Eldana saccharina Walker (Lepidoptera: Pyralidae): effects of silicon source and cultivar. J Appl Entomol. 2006;130:410–20.
Massey FP, Ennos AR, Hartley SE. Silica in grasses as a defence against insect herbivores: contrasting effects on folivores and a phloem feeder. J Anim Ecol. 2006;75:595–603.
Massey FP, Hartley SE. Experimental demonstration of the antiherbivore effects of silica in grasses: impacts on foliage digestibility and vole growth rates. Proc R Soc Lond B Biol Sci. 2006;273:2299–304.
Reynolds OL, Keeping MG, Meyer JH. Silicon-augmented resistance of plants to herbivorous insects: a review. Ann Appl Biol. 2009;155:171–86.
Reynolds OL, Padula MP, Zeng R, Gurr GM. Silicon: potential to promote direct and indirect effects on plant defense against arthropod pests in agriculture. Front Plant Sci. 2016;7:744.
Vicari M, Bazely DR. Do grasses fight back? The case for antiherbivore defenses. Trends Ecol Evol. 1993;8(4):137–41.
Cheeke PR. Endogenous toxins and mycotoxins in forage grasses and their effects on livestock. J Anim Sci. 1995;73(3):909–18.
Clay K. Fungal endophytes of grasses. Annu Rev Ecol Syst. 1990;21:275–97.
Powell RG, Petroski RJ. Alkaloid toxins in endophyte infected grasses. Nat Toxins. 1992;1(3):163–70.
Huitu O, Forbes KM, Helander M, Julkunen-Tiitto R, Lambin X, Saikkonen K, Stuart P, Sulkama S, Hartley S. Silicon, endophytes and secondary metabolites as grass defenses against mammalian herbivores. Front Plant Sci. 2014;5:478.
Moore BD, Johnson SN. Get tough, get toxic, or get a bodyguard: Identifying candidate traits conferring belowground resistance to herbivores in grasses. Front Plant Sci. 2017;7:1925.
Davis M, Midford P, Maddison W. Exploring power and parameter estimation of the BiSSE method for analyzing species diversification. BMC Evol Biol. 2013;13:1–11.
Zachos J, Pagani M, Sloan L, Thomas E, Billups K. Trends, rhythms, and aberrations in global climate 65 Ma to present. Science. 2001;292:686–93.
Morlon H, Lewitus E, Condamine F, Manceau M, Clavel J, Drury J. RPANDA: an R package for macroevolutionary analyses on phylogenetic trees. Methods Ecol Evol. 2016;7:589–97.
Acknowledgements
We thank Niklas Wahlberg for his suggestions to improve the analyses, and for comments on a previous manuscript version.
Funding
The project was funded by the Department of Science and Technology
(DST-RFBR-P-155) and INSPIRE Faculty Award (DST/INSPIRE/04/2013/000476) to Ullasa Kodandaramaiah. Ranjit Kumar Sahoo was supported by a research fellowship from Council of Scientific and Industrial Research, India.
Availability of data and materials
The sequences generated in the study are deposited into Genbank (Accession # MF555197 - MF555484). The time tree generated and analysed during this study is presented in Additional file 2.
Author information
Authors and Affiliations
Contributions
The study was conceptualized by UK and ADW. RKS did the molecular work and analyses with input from UK. Specimen collection was done by ADW and SCC. All authors participated in the manuscript drafting, which was led by RKS. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
“Hostplant change and paleoclimatic events explain diversification shifts in skipper butterflies (Family: Hesperiidae).” Figure S1. Comparison of ultrametric trees of contrasting topologies calibrated using one time unit as the crown age. Table S1. Fitting of models of diversification to the lineage accumulation pattern in MCC tree. Figure S2. 95% credibility shift configurations with the configuration frequencies summarized with the posteriors from the BAMM analysis. Figure S3. Posterior distributions of speciation and extinction rates of dicot- and monocot-feeding lineages from the BiSSE analysis. Figure S4. Graphs showing the relative positions of AIC values from the BiSSE analysis of the MCC tree on the distribution of the AIC values from 100 random posterior trees from the dating analysis. Table S2. Exploring the effect of hidden states on the diversification rate estimation. Appendix S1. Data References. (PDF 510 kb)
Additional file 2:
The hesperiid time tree. (PDF 4563 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Sahoo, R.K., Warren, A.D., Collins, S.C. et al. Hostplant change and paleoclimatic events explain diversification shifts in skipper butterflies (Family: Hesperiidae). BMC Evol Biol 17, 174 (2017). https://doi.org/10.1186/s12862-017-1016-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12862-017-1016-x