Abstract
Analyzing the interactions of circular RNAs (circRNAs) is a crucial step in understanding their functional impacts. While there are numerous visualization tools available for investigating circRNA interaction networks, these tools are typically limited to known circRNAs from specific databases. Moreover, these existing tools usually require complex installation procedures which can be time-consuming and challenging for users. There is a lack of a user-friendly web application that facilitates interactive exploration and visualization of circRNA interaction networks. CircNetVis is an interactive online web application to enhance the analysis of human/mouse circRNA interactions. The tool allows three different input formats of circRNAs including circRNA IDs from CircBase, circRNA coordinates (chromosome, start position, end position), and circRNA sequences in the FASTA format. It integrates multiple interaction networks for visualization and investigation of the interplay between circRNA, microRNAs, mRNAs and RNA binding proteins. CircNetVis also enables users to interactively explore the interactions of unknown circRNAs which are not reported from previous databases. The tool can generate interactive plots and allows users to save results as output files for offline usage. CircNetVis is implemented as a web application using R-shiny and freely available for academic use at https://www.meb.ki.se/shiny/truvu/CircNetVis/.
Similar content being viewed by others
Introduction
Circular RNAs (circRNAs) are a distinct class of single-stranded non-coding RNA molecules that are covalently linked in a closed-loop structure. CircRNAs exhibit diverse biological functions within cells through their interactions with other molecules [1]. For example, a circRNA can function as a competing endogenous RNA (ceRNA) by acting as a sponge to bind to microRNAs (miRNAs) and regulate their function [2]. Since miRNAs can play a role in post-transcriptional expression of target genes, the circRNA is able to indirectly regulate the gene expression [1, 3]. Additionally, circRNAs can affect gene expression by interacting with RNA binding proteins (RBPs) and modulating mRNA stability [4]. Some recent studies have demonstrated that circRNAs not only serve as molecular markers but also participate in cancer proliferation and invasion by regulating miRNAs in various types of cancer. This regulation can occur through circRNA-miRNA-mRNA regulatory network [5, 6] or circRNA-RBP interaction network [7].
A number of tools have been developed to support the discovery of biological functions of circRNAs through interaction network analysis, for example, circlncRNAnet [8], RAID [9], circPlant [10], CircMiMi [11], Circr [12]. However, most of the existing tools primarily focus on circRNAs provided by specific circRNA databases such as circBase [13], which limits their applicability for studying novel circRNAs. Of note, the circRNA databases are mainly constructed using data from previous studies and may not encompass all potential circRNAs in newly generated data. Furthermore, these tools often require experienced users to download, configure and install the software, which can be time-consuming and challenging. Finally, most of the tools lack an interactive interface that would facilitate the exploration and analysis of circRNA interaction networks.
Therefore, we have developed CircNetVis, an online web application designed to facilitate the visualization and analysis of circRNA interaction networks. CircNetVis enables users to visualize and export the results of different types of interactions including circRNA-miRNA, miRNA-mRNA, circRNA-RBP from one or multiple (unknown) circRNAs of interest. For circRNA-miRNA, three commonly-used interaction predictors including TargetScan [14], RNAhybrid [15] and miRanda [16] are utilized. CircNetVis facilitates the investigation of interaction network by allowing users to input miRNAs and genes of interest and apply different filters for the interaction predictors. CircNetVis is implemented using R-shiny with external tools running in a Linux environment, and available at https://www.meb.ki.se/shiny/truvu/CircNetVis/. The source codes and data resources to run the web application are provided in its GitHub page at https://github.com/nghiavtr/CircNetVis.
Implementation
CircNetVis utilizes R-shiny [17] to build the web interface (frontend) with all calculations (backend) running on a Linux environment. The current version of this application supports only circRNAs detected in the human genome reference hg19 and Mus musculus (mouse) genome reference mm9. The details of methodologies, tools and their parameters are provided in the web application. A brief comparison of CircNetVis with existing tools are provided in Table 1.
Backend description
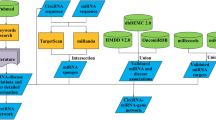
The overview of the backend of the CircNetVis web application is presented in Additional file 1: Figure S1. In the backend of CircNetVis, the following key public databases are collected: circBase [13] for annotated circRNAs, miRbase [18] for miRNAs, CircInteractome database [19] for circRNA-RBP interactions, TargetScan database [14] version 72 for miRNA-mRNA interactions. To identify circRNA-miRNA interactions, three widely used prediction tools are utilized including miRanda [16] version v3.3a, RNAhybrid [15] version 2.1.2, and TargetScan [14] version 72. These tools get input from the pseudo-sequences of circRNAs which are generated using Circall-simulator [20]. The further detailed generation of pseudo-sequences of circRNAs are provided in the Additional file 1. For annotated circRNAs from circBase, a miRNA-mRNA interaction database is built in advance to speed up the processing. For novel circRNAs not found in circBase, the prediction tools will be performed on-the-fly during the use of CircNetVis.
Frontend description
In the frontend, CircNetVis takes input from a single or multiple circRNAs of interest, Fig. 1A. The input circRNAs can be provided in three different types of format. First, users can use directly the circRNA.ID from circBase, for example, hsa_circ_0001946. Second, the circRNA information can be provided as the format of normID in Circall [20] (of note, circRNA can be predicted from other circRNA prediction tools, however for the purpose of consistency, the circRNA IDs need to be re-formatted as the normID). A normID includes a chromosome name (chr), a start position and an end position (in the one-based coordinate system of the genome reference) concatenated together as chr__start__end, for example X__139865340__139866824. Finally, CircNetVis also allows the input from the sequences of the circRNAs in the standard FASTA format.
The exploration of circRNA-miRNA interactions can be controlled by the filters specific to each of the three interaction predictors in the setting control panel, Fig. 1A. Users can use one or combine multiple predictors to obtain confident interactions. The default thresholds for the filters are often used in previous studies of circRNA-miRNA-mRNA networks [5, 6]. These values are easily adjustable by users. In addition, users can limit the results by inputting the list of miRNAs and genes of interest. These miRNAs and genes can be derived from specific analyses, such as differential expression analysis of a research project. This function enhances the flexibility of CircNetVis for application in different projects.
The circRNA-miRNA-mRNA interaction network is visualized using a flexible interactive network visualization R-package, visNetwork version 2.1.0 [21]. visNetwork allows users to move nodes, change edge position, and interact with the network easily. The data of the circRNA-miRNA and miRNA-mRNA interactions are reported in tables for further investigation or exporting to files.
In addition, CircNetVis provides a separate circRNA-RBP interaction network for the circRNAs of interest collected from the CircInteractome database which was previously built using CLIP-seq data [19], Fig. 1B. CircNetVis also utilizes the gene set enrichment analysis of Reactome [22] to suggest biological pathways relevant to the genes in the circRNA-miRNA-mRNA network, Fig. 1C.
Application
To illustrate how CircNetVis works, in Fig. 1, we present a case study of hsa_circ_0001946, a circRNA of CDR1as gene, which has been extensively studied in the literature [23].
The obtained circRNA-miRNA network reports eight miRNAs passing the default filters of the three circRNA-miRNA prediction tools. Among those, the interaction of hsa-miR-7-5p with hsa_circ_0001946 is well validated in human [23]. This interaction, which is highlighted in the table at the bottom of Fig. 1A, shows strong signals with a high number of binding sites of 8mer1a, 7merm8, 7mer1a and 6mer reported by TargetScan, a high miRanda score (Max score = 175) and a low RNAhybrid mfe (minimum of free energy) of -32.1. Multiple mRNAs may interact with this miRNA, as illustrated in the figure. Among these interactions, the association between hsa-miR-7-5p and paired box gene 6 (PAX6) has been investigated in the literature [24]. Two other top interactions of hsa_circ_0001946 with miRNAs (ranked by Max_Score of miRanda) are also previously validated, including hsa-miR-1270 [25] and hsa-miR-671-5p [26]. Regarding circRNA-RBP, CircNetVis provides a visualization of the corresponding interactions from CircInteractome database, Fig. 1B. AGO2 is one of RBPs densely bounding around the circRNA of CDR1as with 43 sites. This circRNA-RBP interaction is also reported in a study of hsa_circ_0001946 [23]. Taking all together, CircNetVis is not only able to recall well the interactions reported in previous studies but also provide a chance to explore other interactions from the networks.
Conclusion
We have developed CircNetVis, an online web application for interactive visualization and investigation of interaction networks of circRNA. The interplays between circRNAs, miRNAs, mRNAs and RBPs are important to explore molecular functions of circRNAs and their signaling pathways.
Availability of data and materials
CircNetVis is implemented using R-shiny with external tools running in a Linux environment, and available at https://www.meb.ki.se/shiny/truvu/CircNetVis/. The source codes and data resources to run the web application are provided in its GitHub page at https://github.com/nghiavtr/CircNetVis.
References
Kristensen LS, Jakobsen T, Hager H, Kjems J. The emerging roles of circRNAs in cancer and oncology. Nat Rev Clin Oncol. 2022;19:188–206.
Zhong Y, Du Y, Yang X, Mo Y, Fan C, Xiong F, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17:79.
Chen L-L. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol. 2020;21:475–90.
Zhou W-Y, Cai Z-R, Liu J, Wang D-S, Ju H-Q, Xu R-H. Circular RNA: metabolism, functions and interactions with proteins. Mol Cancer. 2020;19:1–19.
Liang Z-Z, Guo C, Zou M-M, Meng P, Zhang T-T. circRNA-miRNA-mRNA regulatory network in human lung cancer: an update. Cancer Cell Int. 2020;20:173.
Guan Y, Ma J, Song W. Identification of circRNA–miRNA–mRNA regulatory network in gastric cancer by analysis of microarray data. Cancer Cell Int. 2019;19:183.
Wang Z, Lei X, Wu F-X. Identifying cancer-specific circRNA–RBP binding sites based on deep learning. Molecules. 2019;24.
Wu S-M, Liu H, Huang P-J, Chang IY-F, Lee C-C, Yang C-Y, et al. circlncRNAnet: an integrated web-based resource for mapping functional networks of long or circular forms of noncoding RNAs. GigaScience. 2018;7.
Yi Y, Zhao Y, Li C, Zhang L, Huang H, Li Y, et al. RAID v2.0: an updated resource of RNA-associated interactions across organisms. Nucleic Acids Res. 2017;45:D115–8.
Zhang P, Liu Y, Chen H, Meng X, Xue J, Chen K, et al. CircPlant: an integrated tool for CircRNA detection and functional prediction in plants. Genomics Proteomics Bioinform. 2020. https://doi.org/10.1016/j.gpb.2020.10.001.
Chiang T-W, Mai T-L, Chuang T-J. CircMiMi: a stand-alone software for constructing circular RNA-microRNA-mRNA interactions across species. BMC Bioinform. 2022;23:164.
Dori M, Caroli J, Forcato M. Circr, a computational tool to identify miRNA: circRNA associations. Front Bioinform. 2022;2.
Glažar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20:1666–70.
Agarwal V, Bell GW, Nam J-W, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:e05005.
Rehmsmeier M, Steffen P, Höchsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–17.
Miranda KC, Huynh T, Tay Y, Ang Y-S, Tam W-L, Thomson AM, et al. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–17.
Chang W, Cheng J, Allaire JJ, Sievert C, Schloerke B, Xie Y et al. Shiny: web application framework for R. 2023.
Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47:D155–62.
Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13:34–42.
Nguyen D, Trac QT, Nguyen T-H, Nguyen H-N, Ohad N, Pawitan Y, et al. Circall: fast and accurate methodology for discovery of circular RNAs from paired-end RNA-sequencing data. BMC Bioinform. 2021.
Almende BV, Thieurmel B, Robert T. visNetwork: network visualization using “vis.js” Library. 2021.
Jassal B, Matthews L, Viteri G, Gong C, Lorente P, Fabregat A, et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2020;48:D498-503.
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–8.
Needhamsen M, White RB, Giles KM, Dunlop SA, Thomas MG. Regulation of human PAX6 expression by miR-7. Evol Bioinform Online. 2014;10:107–13.
Su Y, Lv X, Yin W, Zhou L, Hu Y, Zhou A, et al. CircRNA Cdr1as functions as a competitive endogenous RNA to promote hepatocellular carcinoma progression. Aging. 2019;11:8183–203.
Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30:4414–22.
Acknowledgements
The computations were enabled by re-sources provided by the National Academic Infrastructure for Supercomputing in Sweden (NAISS) at UPPMAX partially funded by the Swedish Research Council through grant Agreement No. 2018-05973.
Funding
Open access funding provided by Karolinska Institute. This work was partially supported by funding from the Swedish Research Council (VR) and CancerFonden.
Author information
Authors and Affiliations
Contributions
TNV conceived and oversaw the study. TN and TNV developed the tool and performed the analysis. TN prepared the first draft with input from TNV and HN. HN involved in discussion and contributed to manuscript writing. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors have declared no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1.
Supplementary documents and figures.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Nguyen, TH., Nguyen, HN. & Vu, T.N. CircNetVis: an interactive web application for visualizing interaction networks of circular RNAs. BMC Bioinformatics 25, 31 (2024). https://doi.org/10.1186/s12859-024-05646-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12859-024-05646-4