Abstract
The study’s goal was to look into the protective properties of quercetin (QU) in natural form and QU nanoparticles-loaded chitosan nanoparticles (QU-CHSNPs) against cardiotoxicity. The ionotropic gelation approach was adopted to form QU-CHSNPs. The characterizations were performed using advanced techniques. In vitro, the release profile of QU was studied. Cardiotoxicity was induced by doxorubicin (DOX) and protected via concurrent administration of QU and QU-CHSNPs. The heart's preventive effects of QU and QU-CHSNPs were manifested by a decrease in elevated serum activities of cardiac enzymes, as well as an improvement in the heart's antioxidant defence system and histological changes. The findings substantiated QU-CHSNPs' structure with an entrapment efficiency of 92.56%. The mean of the zeta size distribution was 150 nm, the real average particle size was 50 nm, and the zeta potential value was − 27.9 mV, exhibiting low physical stability. The percent of the free QU-cumulative release was about 70% after 12 h, and QU-CHSNPs showed a 49% continued release with a pattern of sustained release, reaching 98% after 48 h. And as such, QU and QU-CHSNPs restrained the induced cardiotoxicity of DOX in male Wistar rats, with the QU-CHSNPs being more efficient.
Similar content being viewed by others

Background
Cardiotoxicity is caused by anticancer drug classes (Iqubal et al. 2018). Doxorubicin is a potent anthracycline chemotherapeutic agent for solid and hematological tumors such as leukemia, soft-tissue sarcomas, osteosarcomas, breast cancer, and lymphomas (Cappetta, et al. 2017). Because of its toxicity on various tissues, including the renal and cardiac, it is used in controlled doses in certain types of cancer (Pugazhendhi et al. 2018). According to the study of Zhao et al. 2018 (Zhao et al. 2018), the main factors of DOX toxicity are lipid peroxidation (LPO), oxidative stress, autophagy, DNA damage, and mitochondrial injury. The essential reason for DOX-induced cardiac toxicity is the production of reactive oxygen species (ROS), in addition to mitochondrial dysfunction (Pereira, et al. 2011). Since cardiomyocytes have relatively low levels of antioxidant enzymes such as superoxide dismutase (SOD), catalase (CAT), glutathione peroxidase (GPx), Glutathione-S-transferase, and glucose-6-phosphatedehydrogenase (Hashish et al. 2021; do Nascimento, T.C.,, et al. 2020; Doroshow et al. 1980), they are more vulnerable to free radical-mediated damage induced by DOX. The implication of SOD in cardiovascular diseases has been described. As an example, inhibition of SOD2 expression induced both mitochondrial oxidative stress and cardiomyocytes hypertrophy (Dubois-Deruy et al. 2017).
Polyphenolic compounds have both curative and preventive properties for many diseases, including heart disease, cancer, diabetes, and hypertension (Hussain et al. 2020; Lovegrove et al. 2017; Maaliki et al. 2019). Quercetin is a polyphenolic agent (flavonoid class) and is enriched in onions, plums, peppers, mangos, and berries (Khan 2021). It has anti-inflammatory, anti-cancer, anti-viral, and anti-allergic activity (Ferenczyova et al. 2020). Additionally, QU greedily scavenges superoxide anion, lipid proxy radicals, and singlet oxygen (Nathiya et al. 2014). Further, it inhibits the activation of extracellular signal-regulated kinase and mitogen-activated protein kinase in ROS-mediated cardiomyopathy (Hashish et al. 2021). Also, QU was a robust agent versus cardiac ischemia–reperfusion injury, diabetic cardiomyopathy, and cardiotoxicity, indicating cardioprotective effects (Ertuğ et al. 2013). The cardioprotective effects of QU are linked to a variety of signaling pathways in the heart, including oxidative stress reduction, apoptosis inhibition, and inflammatory protein regulation (Castillo et al. 2018). However, problems like low oral bioavailability and poor aqueous solubility make quercetin an unreliable candidate for therapeutic purposes. Additionally, the rapid gastrointestinal digestion of quercetin is also a major barrier to its clinical translation (Ferenczyova et al. 2020). Hence, to overcome these disadvantages, quercetin-based nanoformulations have been considered in recent times. Nanoformulations of quercetin have shown promising results in their uptake by the epithelial system as well as enhanced delivery to the target site.
In pharmaceutical management, CHSNPs are widely accepted as carriers for hydrophobic drugs to advance their therapeutic potency and diminish their toxic effects. The reason is their biocompatibility, bioactivity, biodegradability, and polycationicity. Further, the CHSNPs exhibit drug targeting delivery, solubilize various hydrophobic drugs, increase bioavailability and blood circulation, enhance encapsulation efficiency, and sustain drug release (Li et al. 2016). To overcome the limitations of QU's natural free form and to achieve sustained release, the current study aimed to design a nanoformula of QU nano that was loaded on CHSNPs and to study their protective effect against DOX-induced cardiotoxicity in male Wister rats.
Materials and methods
Materials
Extra-pure QU, CHS medium-molecular weight MW (75 kDa), TPP, acetic acid, dimethyl sulfoxide (DMSO), and ethanol were purchased from Sigma (Sigma-Aldrich Co., St. Louis, Missouri, USA). "DOX hydrochloride (50 mg in a 25 ml injection vial) was purchased from Hikma Pharmaceutical, Bader City, Cairo." All used chemicals and solvents were of analytical grade and high purity. Experiments were carried out with deionized water.
Synthesis of chitosan and quercetin nanoparticles
According to the ionic gelation of chitosan with TPP anions (Zhao et al. 2011), the CHSNPs were formed. In brief, a solution incorporating 50 ml of 1% acetic acid, 500 mg of 85% deacetylated chitosan, and 1 mg of TPP per mL was stirred at 1400 rpm for 30 min to form CHSNPs. The collected pellet was adopted for characterization. Regarding quercetin nanoparticles preparation, the procedures for the nanoparticipation technique were performed by adding ethanol to water at a volume ratio of 1:35 and a fixed flow rate of 8 and 10 ml/min under magnetic stirring (1000 rpm) (Kakran et al. 2015).
Chitosan nanoparticles loaded with quercetin
The procedures followed the Xu et al. 2007 method with modifications (Fan et al. 2012). 1 ml of a solution containing dissolved 0.5 g QU nanoparticles in ethanol was added dropwise to the CHS solution, which contained 1 g of CHS dissolved in 50 ml of glacial acetic acid (1%), under constant stirring for 60 min. Next, at a rate of 1 mL/min, a solution of 0.2 g of sodium TPP in 20 mL of deionized water was added with further stirring for 60 min. Later, the entire mixture was centrifuged at 15,000 rpm for 30 min. A formula of loaded, cross-linked QU-CHSNPs was obtained, washed with distilled water three times, and dried under vacuum at 40 ◦C to be characterized.
Characterizations techniques
The XRD system was employed to characterize the crystallinity of the prepared items. Scherrer’s equation determined the crystallite size:
where D is the crystallite size, λ is the wavelength, β is the corrected full width at half the maximum of the diffraction peak, and θ is the Bragg’s diffraction angle. The Fourier transformation infrared (FTIR) spectra of QU nano, CHS, CHSNPs, and QU-CHSNPs were reported in the transmission mode to determine the functional groups.
The surface morphology and the particle size of the prepared CHSNPs and QU-CHSNPs were investigated by a high-resolution transmission electron microscope (HR-TEM) and a field emission scanning electron microscope. The particle size distribution of the dispersed QU and QU-QU-CHSNPs was measured by the dynamic light scattering (DLS) method using the Stokes–Einstein equation:
where D0 is the diffusion coefficient, kB is the Boltzmann constant, T is the absolute temperature, and η is the viscosity of the solvent water dispersant with a refractive index of 1.330 (Xu et al. 2007). The change in the surface charge (zeta potential) reflecting the colloidal stability was determined by electrophoretic laser Doppler velocimetry.
In vitro quercetin entrapment efficiency and release study
The entrapment efficiency and the release percentage of QU were calculated using the UV–Vis spectrometer at the absorption peak of QU (λ max) at 370–415 nm. A modified dialysis bag method was used to assess the release of QU from QU-CHSNPs (Luque-Alcaraz 2012; Tamilselvan and Raghavan 2015). The entrapment efficiency of QU on CHSNPs was calculated as follows:
(Rashedi et al. 2019). The concentration of the released QU was calculated as a percentage of the initial amount of the incorporated QU. All measurements were performed in triplicate.
In vivo study
Experimental animal
In this study, male Wistar rats weighing 140–160 g served as the experimental animals. They were purchased from the Pharmacy Faculty's Animal House at Nahda University in Beni-Suef, Egypt. They were kept under tight care and monitoring for 14 days prior to the beginning of the study to rule out any concurrent infections. Animals were kept in cages with six rats per cage throughout the experiment. The animals were kept in standard polypropylene cages at room temperature (22 °C), humidity (55 5%), and lighting (12-h light/dark cycles). At all times, water and a typical, balanced diet were made available to the rats. All animal procedures have been approved by the experimental animal ethics committee of the Faculty of Science at Beni-Suef University in Egypt and are in compliance with the guidelines for the humane care and use of laboratory animals published by the Canadian Council on Animal Care (CCAC) in 1993. (BSU/FS/2019/2 is the ethics approval number.) Every effort was made to lessen the number of animals used and their suffering.
Experimental design
In the current study, 30 rats were used. These rats were divided into five (n = 6) groups as follows: Group 1 (Normal control): Normal, healthy rats were given an equivalent volume of 0.9% NaCl (2 ml/kg, three days per week) by oral gavage for six weeks. Group 2 (doxorubicin-administered control): the rats were given doxorubicin intraperitoneally at a dose of 1.25 mg/kg/48 h for six consecutive doses over two weeks to induce cardiotoxicity (I.P. injection). Group 3 (the DOX-administered group treated with QU natural): on the same day that DOX administration began, QU natural form that was suspended in a 0.9% NaCl saline solution (10 mg/kg, three times per week, orally) began as well. After DOX was stopped, QU natural was administered concurrently for an additional four weeks (a 6-week total period). Group 4 (the DOX-administered group treated with QU-CHSNPs): On the same day that DOX began, QU-CHSNPs (10 mg/kg, three times per week) began and continued for an additional four weeks after DOX was stopped (a 6-week total period). Group 5 (normal group): Normal healthy rats were orally treated with QU-CHSNPs (10 mg/kg b.w.t./day) by oral gavage for 6 weeks (Raj et al. 2015; Matook, et al. 2012).
Serum and tissue sampling
At the end of the 6-week period, six rats from each group were slaughtered under mild diethyl ether anesthesia. Jugular vein blood was drawn from each rat and placed in a centrifuge tube, where it was allowed to coagulate for 45 min at room temperature. The serum samples were separated by centrifugation at 3000 rpm for 10 min at 30 °C, and they were then frozen and kept at − 20 °C for biochemical analysis, interleukins, and cytokine parameter measurements. Each rat's heart was quickly removed and infused with sterile normal saline. Part of the cardiac tissue was immediately removed, weighed, and stored in RNA lysate for gene expression investigation. For a 24-h period, one part of the heart was fixed in 10% neutral buffered formalin before being moved to 70% alcohol for histopathological analysis. To determine some oxidative stress and antioxidant parameters, 0.5 g was stored at − 20 °C and homogenized in 5 ml of 0.9% sterilized sodium chloride (10% w/v) using a Teflon homogenizer (Glass-Col, Terre Haute, USA). Part of the cardiac tissue was immediately excised and fixed in 7.2% glutaraldehyde for examination under an electron microscope. A portion of the cardiac tissue was immediately removed and frozen for DNA fragmentation gel electrophoresis.
Instruments
The instruments used were a glass column homogenizer (USA), A Bruker (Vertex 70 FTIR-FT Raman) spectrometer, high-resolution transmission electron microscope (HRTEM, JEM 1400, software version DM_3, Japan), field emission scanning electron microscopy (FESEM, JEOL JSM-5900, Japan), XRD, (PANalytical Empyrean, 202964, Sweden), light microscope, probe sonicator, cooling centrifuge, Lyophilizer, zeta sizer instrument (Malvern, UK), shaking water bath (Precision Scientific Group, USA), deep freezer -80 C (forma Scientific, USA), deep freezer – 20 C (forma Scientific, USA), Ohaus Pioneer digital balance (PA114), centrifuge, Hospitex diagnostic, UV–VIS spectrophotometer (CARY100, Germany), vortex mixer (VM-300), Eppendorf centrifuge (model 5804R), pH meter (HI 110, USA), and electro-thermal incubator (Model DNP, USA).
Biochemical analyses
Determination of serum cardiac enzymes
Creatine kinase-MB (CK-MB) activity in serum was measured by a kinetic method using kits developed by Spectrum Diagnostics, Egypt, based on the method of Ibrahim et al. (2009). Using reagent kits purchased from Bio System Company (Spain), aspartate aminotransferase (AST) activity in serum was determined by a kinetic method in accordance with the method of Friedman and Young (1997). The kinetic method described in Burtis et al. (2005) was used to measure the lactate dehydrogenase (LDH) activity in serum using reagent kits obtained from humans (Germany). The Rat Cardiac Troponin-I ELISA Kit with Catalog Number CSB-E08594r from CUSABIO was used to measure serum cardiac Troponin-I levels. This assay is used to determine serum levels of rat cardiac troponin (c Tn-I). The quantitative sandwich enzyme immunoassay method is used in this assay.
Determination of serum lipid profile
Using a reagent kit obtained from Spinreact Co. (Spain), the amount of cholesterol in the serum was measured using the method of Witt and Trendelenburg (1982). A reagent kit purchased from Spinreact Company (Spain) was used to evaluate the serum triglyceride concentration (TG) using the method described in Young (2001). The method of Friedewald et al. (1972) was used to assess the serum HDL-c concentration using reagent kits purchased from Spinreact Co. (Spain). The concentration of LDL-c in the blood was calculated as follows: total cholesterol—(TG/5)—HDL-c = LDL-c concentration (mg/dL). The serum VLDL-c concentration was calculated as follows: VLDL-c concentration (mg/dL) = TG /5.
Estimation of activity and concentration of antioxidant markers and oxidative stress
The activities of myocardial superoxide dismutase (SOD), glutathione S-transferase (GST) (Tietz et al. 1995), glutathione peroxidase (GPx) (Habig et al. 1974), glutathione (GSH) (Paglia and Valentine 1967), lipid peroxidation (malondialdehyde MDA) (Beutler et al. 1963), and nitric oxide (NO) (Ohkawa et al. 1979) in cardiac tissue homogenate were measured using a commercial kit (Bio Diagnostic, Egypt) and based on a previously described colorimetric method.
Measurement of serum pro-inflammatory interleukin-1beta (IL-1β), tumor necrosis factor-alpha (TNF-α) and anti-inflammatory interleukin-10 (IL-10)
Rat interleukin-10 (IL-10) ELISA Kit, Cat. No. MBS764911, from My BioSource, depending on sandwich enzyme-linked immunosorbent assay technology, was used to assess IL-10 activity. Also, rat Interleukin-1B activity was measured using the rat Interleukin-1B (IL-1B) ELISA Kit User Manual, Catalog # MBS825017. The CUSABIO CSB-E11987 rat (TNF-α) ELISA Kit was used to measure TNF α-activity. The concentrations of IL-10, TNF-α, and IL-1B were determined using ELISA assay kits (Assay Pro, USA).
Measurement of serum apoptotic marker caspase-3 (Casp-3)
Using the Rat Casp-3 ELISA Kit, Cat. No. CSB-E08857r from CUSABIO, casp-3 activity in serum was determined. The quantitative sandwich enzyme immunoassay method is used in this assay.
Detection of NrF2, PPAR-γ and Annexin-V molecular gene expression by quantitative real-time polymerase chain reaction (QRT-PCR)
The quantitative real-time polymerase chain reaction (QRT-PCR) technique was used to measure the levels of nuclear factor erythroid 2-related factor 2 (Nrf2), peroxisome proliferator-activated receptor gamma (PPAR-γ), and Annexin-V in heart tissue. In brief, the Qiagen tissue extraction kit (Qiagen, USA) was used to extract RNA from the heart tissue of all rats. Total RNA (0.5–2 ug) was used for cDNA transformation using a high-capacity cDNA reverse transcription kit according to the manufacturer's instructions (Fermentas, USA). As a reference, the β-actin gene was used. An Applied Biosystem with software version 3.1 (Step OneTM, USA) was used for the QRT-PCR amplification and analysis. The QRT-PCR assay using the primer sets was calibrated to work best at the annealing temperature. SYBR Green was used to monitor the synthesis of double-stranded DNA. The amplifications were completed by using 40 cycles of denaturation at 95 °C for 15 min, annealing at 72 °C for one minute, and extension at 60 °C for one minute. The Step One Plus real-time thermal cycler (Applied Biosystems, Life Technology, USA) was used for analysis and amplification. The following primers were used: Nrf2: forward: 5′-TTGTAGATGACCATGAGTCGC-3′, reverse: 5′-TGTCCTGCTGTATGCTGCTT-3′. Annexin-V: 5′-GTATCCACAGCCACCTTCACAGTC-3′ (forward) and 5′-TCCAAADAAACAGGAGAGAAAACAG-3′. PPAR-γ Forward: 5′-GGACGCTGAAGAAGAGACCTG-3′, Reverse 5′- CCGGGTCCTGTCTGAGTATG-3′. Forward primer GCA CCA CAC CTT CTA CAA TG and reverse primer TGC TTG CTG ATC CAC ATC TG were used for the -actin primers.
DNA fragmentation assay by agarose gel electrophoresis
Apoptosis is linked to the detection of traditional DNA laddering in cardiac extracts. The lysis solution (1% SDS, 10 mM Tris HCl, 50 mM EDTA, pH 7.4) was used to homogenize 1 g of frozen hearts using an Ultra-Turrax T25 homogenizer from Janke and Kunkel IKA-Laboratory. Samples were treated with RNAsa (1 mg/ml) from Sigma Chemical Corporation for two hours at 37 °C and proteinase K (1 mg/ml) for 45 min at 48 °C. DNA was extracted using phenol, chloroform, and isoamyl alcohol (25:24:1) (Sigma Chemical Corporation) and precipitated for 12 h at 20 °C with isopropanol (v/v) and 0.5 M NaCl. Centrifugation at 15000 g for 20 min was used to recover the DNA, and the pellet was then washed with 70% ethanol, dried, and resuspended in Tris–EDTA buffer (10 mM Tris, 50 mM EDTA), which has a pH of 7.4. Ethidium bromide (0.5 ug/ml) was added to samples (250 g DNA) for analysis on a 2% agarose gel.
Transmission electron microscopy (TEM) examination of heart
The samples were fixed for two hours at room temperature in 3 percent glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.0), washed in the same solution, and then post-fixed for two hours in 1 percent osmium tetroxide. The samples were dehydrated for 15 min in each ethyl alcohol dilution, ranging from 10 to 90%, and then for 30 min in absolute ethanol. In a graded series of tests, samples were penetrated with epoxy resin and acetone until finally reaching pure resin. Formvar-coated copper grids were used to collect ultrathin sections. The sections were then dyed twice with uranyl acetate, followed by lead citrate. Stained sections were examined using a JEOL JEM 1010 TEM at 70 kV et al.-Azhar University's Regional Center for Mycology and Biotechnology (RCMB) (Montogomery and Dymock 1961).
Histopathological examination of heart
The heart was removed immediately after the rats were slaughtered. It was washed with saline, fixed in 10 percent neutral buffered formalin for 24 h, dehydrated with serial dilutions of ethyl alcohol solutions (70%, 95%, and absolute ethanol), immersed in paraffin, sectioned with a microtome at 4–5 m thickness, and stained with hematoxylin and eosin for histopathological examination under the light electric microscope.
Statistical analysis
The one-way ANOVA statistical analysis was used to compare the data between groups, and then the Tukey–Kramer post hoc analysis procedure was used. The results were expressed as a mean ± standard error. For all the data, the statistical significance level is P < 0.05. To evaluate all of the data, the Statistical Package for Social/Science (SPSS) version 22 software was used on Windows 10.
Results and discussion
Qualitative analysis of XRD
The XRD spectra for QU nano, CHS, CHSNPs, and QU-CHSNPs are shown in Fig. 1. For CHS, the XRD spectra had a low diffraction signal intensity, which reflected its low level of crystallinity. With a d-spacing of 4.43, the highest peak at 19.9° represents a relative intensity of 100%. According to Eq. 1, the calculated crystallite size of the pure CHS was 128.878 Å with a micro-strain of 1.72%. It is important to note that as the width of the peak increases, the crystallite size decreases.
The XRD spectrum of CHSNPs had a disordered arrangement of CHS chains, showing a wide diffuse hump peak at around 30°, which is a typical fingerprint of semi-crystalline chitosan (Abdel-Aziz et al. 2017). Our findings were proportionate with those of a former study (Abdel-Moneim et al. 2020), which showed that TPP counter-ions induce cross-linking among CHS chains and thus the construction of an opaque network that vanishes in the diffraction peaks of CHS, which form a single floppy peak. The broad peak indicated a decrease in crystallite size, reflecting a reduction in periodicity, i.e., the long-range order of atoms, ions, or molecules in the particles, and consequently, there was low ordering of the hkl planes, which decreased crystallinity. The low degree of crystalline perfection may also be attributed to the high number of defects in the nucleation and the growth rate of the crystals as a result of using polymers like chitosan (Stachurski 2011).
According to the chemical structure of QU (Scheme 1), the previously published XRD spectrum for QU confirmed its crystallinity state (Stachurski 2011). In our study, the nanoscale crystallinity of QU was determined by high-resolution XRD to enhance our understanding of its chemical action. The obtained XRD spectrum of QU nano had similar peaks to Ritchie et al. (2013); they were in slightly different positions. The XRD spectrum of QU nano had peaks at 8.9°, 9.7°, 10.7°, 12.4°, 13.1°, 13.6°, 16.6°, 23.1°, 24.8°, 26.1°, 27.4°, 28.1°, 28.5°, 38.6°, and 41.9°, respectively. There was a high basal diffraction peak at the diffraction angle of 13.6° with 100% relative intensity, a d-spacing of 6.77 Å, a crystallite size of 485.6 Å, and a micro-strain of 0.69%. This peak was coordinated with prior research (Abd El-Rahmanand and Suhailah 2014). These distinct diffraction peaks indicate that the crystallinity of QU nano did not change, in agreement with previous research (Zhao et al. 2012).
The same Fig. 1 paraded the crystal form of QU-CHSNPs with a trick peak shifting to the left. The position of the diffracted peak can shift in response to several different factors, including substitution doping, temperature, and stress. Here, the shifting could be because of differences in interaction angles caused by changes in the QU's arrangement structure plans during the loading process. Another explanation for peak shifting was the changes in the interatomic distances of the QU-CHSNPs, which resulted from the entrapped QU within the CHSNPs. We propose that a chemical bonding occurred between the CHS's amino group NH2 and the QU's CO carbonyl and OH hydroxyl functional groups based on the chemical structures of CHS and QU (Scheme 1). The diffracted peak of QU disappeared, enforcing this suggestion. The shifting may also be because of the adsorption of QU on the surface of CHSNPs. Overall, the QU-CHSNPs spectrum showed languid diffracted signal changes in shape, position, and the relative integrated intensity between 2 theta angles, which is in agreement with Zhang et al. (Muller 2004). These alterations show that there was QU loading on the CHSNPs.
FTIR spectra analysis
In Fig. 2, compared to the standard QU (Stachurski 2011), the FTIR spectrum of QU nano presented a variation of sharp, intense, and faint peaks as well as essential practical groups that corresponded to the stretching vibrations of O–H, = C–H, –CH2, C = O, C–O, and C = C. While the spectrum exhibited peaks similar to those previously reported, there were shifts in position and intensity. The spectra revealed a specific strong stretching peak of the carbonyl group C = O at 1660 cm−1, a weak peak referred to as the = C–H stretching vibration at 2937 cm−1, and a peak that indicates C = C aromatic stretching at 1511 cm−1. There was a distinct peak at 3283 cm−1 that coincides with the stretching vibration of the hydroxyl group OH, and another at 880 cm−1 that corresponds to a benzene ring. The obtained results reflect the molecular structure of QU.
The FTIR spectra for CHS and CHSNPs are also presented in Fig. 2. Considering the chemical structure of CHS, the broad peak at 3437 cm−1 is attributed to the stretching vibration of the OH group. A polysaccharide peak of CHS, which is in-plane N–H bending vibration, appeared at 1637 cm−1. The C–O stretching vibration of the primary alcoholic group’s evidence is at 1385.34 cm−1, while a bending vibration peak of C–N is presented at 1076 cm−1. Regarding the CHSNPs spectrum, the two spectra appeared to be similar and showed various characteristic peaks with a faint diversity in the wideness and non-considerable shifting of the peak position. The broad peak ranged from 3353.09–3172.46 cm−1, which corresponded to overlapping among the stretching vibrations of the O–H and N–H groups. The peaks at 2919.18 cm−1 were associated with aliphatic sp3 C–H stretching, those at 1636.91 cm−1 with in-plane N–H bending vibrations, and those at 1405.34 cm−1 with C–O stretching vibrations. The peak at 1020 cm−1, which appears in the FTIR spectrum for CHSNPs, shows a characteristic P = O stretching vibration from the phosphate groups of the TPP. A previous study (Zhang et al. 2006) reported similar results for the formation of CHSNPs treated with TPP.
The pattern of intramolecular hydrogen bonds may explain the broad divergence between CHS and CHSNPs. Furthermore, the observed hypochromic shifting in peak positions was caused by interactions between the CHS's NH3+ groups and the TPP’s phosphate groups. For this reason, the FTIR spectra were employed to verify TPP and CHSNPs. The decline in amide I band intensity for CHSNPs (1636 cm−1) when compared to CHS (1637 cm−1) enforced this interaction. The –CH2 wagging peak at 1405 cm−1 was another substantial one for CHSNPs.
The spectrum of QU-CHSNPs revealed significant variations in the peak patterns, intensities, and positions. For example, the broad band extended from 3500–3000 cm−1 due to the intermolecular hydrogen bonding, as well as the disappearance of some peaks. These variations reflected a type of interaction between CHSNPs and QU, confirming the loading process. These results were in accordance with those of previous research (Lustriane et al. 2018).
Surface morphology TEM and SEM study
Employing high-resolution transmission electron microscopy (HRTEM) to study the shape and particle size, the HRTEM images of CHSNPs and QU-CHSNPs are shown in Fig. 3. An optimized spherical shape of the prepared CHSNPs with an average particle size of approximately 50 nm is shown in Fig. 3 (A), while an HRTEM with a low field of view revealing stack clusters of the prepared QU-CHSNPs is shown in Fig. 3 (B). The images display two different signal intensities, dark and gray, which might be due to a variation in the attenuation of the incident electron beam on the QU-CHSNPs. This attenuation is based on the variations in the electron densities of the QU and CHSNPs, confirming the loading process. In the HRTEM images, the appearance of darker regions inside the particles indicates the presence of lipophilic QU in the matrix of the CHSNPs, and grey regions may indicate soluble CHS. The surface microstructures show that the sample was coated with gold to avoid charges and promote the signals needed for surface analysis with the SEM (Figure 3). The CHSNPs had a non-homogeneous, rough, and irregular surface texture (Fig. 3 C); this irregularity could be due to the complex's formation scheme, which includes incorporating two aqueous phases, one containing polymer CHS and the other containing polyanion TPP. In contrast, the SEM image of QU-CHSNPs had a smooth surface pattern with morphology like a pineapple (Fig. 3 (D)).
A HRTEM images of the spherical shape of the prepared CHSNPs with an average particle size of approximately 50 nm, B stacked clusters of the prepared QU-CHSNPs, C SEM image of CHSNPs with a non-homogeneous, rough, and irregular surface texture; D SEM image of QU-CHSNPs with a smooth surface pattern and pineapple morphology
Zeta sizer and potential measurements
The hydrodynamic size distribution of the QU nano and QU-CHSNPs was determined by the intensity distribution method via the dynamic light scattering (DLS) technique, as shown in Fig. 4(A). The sizes of the intensity-distribution and cumulant fit were comparable, and the Z average, also called the cumulant mean or log mean, of the hydrodynamic diameter size distribution of QU nano was 150 nm. The polydispersity index PDI value (0.992) predicted a wide width of the distribution peak, indicating a heterogeneous size distribution. The intercept value of 1.09 indicated an excellent signal-to-noise ratio. For the QU-CHSNPs, the reported hydrodynamic size, PDI, and intercept were 329.4 nm, 0.541, and 0.983, respectively (Fig. 4). The hydrodynamic diameter size distribution of QU-CHSNPs was slightly bigger than that identified using the HRTEM.
A QU nano and QU-CHSNP hydrodynamic size and size distribution, the polydispersity index of QU nano indicated a narrow width and a homogeneous size distribution, while that of QU-CHSNPs displayed a wide width and a heterogeneous size distribution. B 48-h in vitro release profile of QU-CHSNPs versus free quercetin nano at pH 6.8 and 37 °C. The results are averages (n = 3). Because of the slow diffusion of QU from the CHSNPs matrix, free QU had a lower cumulative drug percent than QU-CHSNPs, confirming a pattern of sustained release
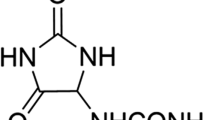
However, this discrepancy is reasonable as the distribution number (from electron microscopy) is expected to be smaller than the distribution intensity (from DLS). In other words, the particle size determined using the DLS represents its hydrodynamic diameter, whereas that obtained using HRTEM represents its real diameter. The surface charge of the QU-CHSNPs formula was estimated by measuring the zeta potential. In total, a zeta potential value of greater than 30 mV is thought to be a standard value that will give ample repulsion force to limit particle aggregation. The repulsion among particles indicated that the low zeta potential value was -27.9 ± 6.8 mV, which indicates low physical stability. QU is a hydrophobic polyphenol with a pentahydroxy flavone and five hydroxy groups at the 3, 3', 4', 5, and 7-positions (Scheme 1). The negative charge of the formula was due to the hydroxy group, indicating the successful entrapment of QU by cross-linked CHSNPs.
Entrapment efficiency and release profiles
According to Eq. 3, the average entrapment efficiency for the QU in CHSNPs was 92.56% 7.72%. The release profiles for the free QU and the QU from the CHSNPs in the PBS solution at a constant temperature of 37 °C and a pH of 6.8 for 48 h are shown in Fig. 4 (B). A surge release of 47.2% was observed from free QU within the first 4 h, whereas QU-CHSNPs showed a slaw burst release of 29.21%. Free QU was released faster than QU-CHSNPs, which could be attributed to the existence of QU in the CHS’s network. The initial rapid release of the loaded QU may be due to the rapid dispersion of the QU present on the surface of the CHSNPs, while at a later stage, QU may also be constantly released from the core of the CHS matrix because of CHS hydration and swelling. The drug was therefore released more gradually, and the rate of release was influenced not by polymer erosion but by drug diffusion through the amorphous territory of the polymer matrix. The free QU cumulative release in a saturation state after 12 h was approximately 70%, while that of QU-CHSNPs was 49%, and it showed a pattern of sustained release, reaching 98% after 48 h. This was explained previously (Sarkar et al. 2013), as CHS was found to be a biodegradable polymer. However, its biodegradation is substantially slower than that of other degradable polymers. In addition, the deterioration of CHS was particularly limited in an aqueous medium because of its poor crystallinity and hydrophobicity.
Therefore, the only possible mechanism of QU release was diffusion, and not the degradation of the CHS polymer. Furthermore, it is worth noting that the diffraction pattern of the QU-CHSNPs showed that some of the QU crystal peaks had disappeared, suggesting the amorphous pattern of QU in the matrix of the CHSNPs polymer or that it was dispersed in the amorphous region of the CHSNPs. As a result of the slow diffusion of QU from the CHSNPs matrix, the cumulative drug percentages of free QU were lower than those of QU-CHSNPs.
Biological study
Doxorubicin (brand name: Adriamycin) is a chemotherapy drug made from Streptomyces peucetius. It is used together with other chemotherapy agents against malignant neoplasms, including breast cancer, lung cancer, leukemia, Hodgkin’s disease, Kaposi’s sarcoma, acute lymphoblastic leukemia, lymphomas, and several metastatic tumors. DOX intercalates with DNA and inhibits macromolecular biosynthesis by limiting DNA double helix formation and blocking the replication process (Ha et al. 1997). However, the long-term use of DOX produces severe effects, which limit its clinical applications (Tacar et al. 2013). Cardiotoxicity is one of the extremely serious effects of DOX, as it can lead to dilated cardiomyopathy, which results in congestive heart failure. The DOX aggregation dose controls the cardiomyopathy rate. Oxidative stress, downregulation of genes for contractile proteins, and p53-mediated apoptosis are the avenues of cardiomyopathy promoted by DOX. (Songbo et al. 2019).
Oxidative stress
There is abundant evidence that oxidative stress plays an essential role in the pathophysiology of DOX-induced cardiotoxicity. In cardiomyocytes, DOX can lose one electron through the metabolic efficiency of Nicotinamide Adenine Dinucleotide/Phosphate-Cytochrome P450 reductase. The DOX reduction forms semiquinone free radicals, which produce radicals of hydroxyl (·OH), hydrogen peroxide (H2O2), and proxynitrite (ONOO) (Al-Harthi et al. 2014). These species trigger proteins and LPO via damaging the macromolecular cellular components of the cell membrane, inducing oxidative stress, and starting cell apoptosis (Ahmed 2013). Antioxidants regulate the cellular oxidative stress that is induced by an inequality in the cellular production of ROS and reactive nitrogen species (RNS) (Ayla, et al. 2011). The heart lacks antioxidant enzymes; this results in extensive destruction of the cardiac cellular mitochondrial membranes, nucleic acids, and endoplasmic reticulum (Mahmoud et al. 2020a).
Our investigation showed a marked elevation in cardiac nitric oxide (NO) levels in DOX-rats when compared with control rats (Fig. 5; (p < 0.001), in agreement with previous research (Kelleni et al. 2015). The capability of DOX to intercede with the induction of NOS expression and NO release in the heart is the reason for the ascent in NO levels. Further, DOX causes a surge in e-NOS transcription and protein activity in cardiac endothelial cells by H2O2 and calcium influx triggering, leading to the synthesis of NO (Saad et al. 2004). A recent study gives evidence of the upregulation of iNOS genes and protein expression in DOX-induced cardiomyopathy. The concomitant overabundance of NO and ROS yields excessive levels of peroxynitrite and RNS, which may invade and damage vital cellular biomolecules (Goetz et al. 1999).
Effect of QU and QU-CHSNPs on heart NO, MDA, GSH, GPX, GST, and SOD in normal control and DOX-administered rats. The data are presented as mean SE. The number of samples in each group is six. P < 0.001 values for F-probability values that share the same superscript symbol are not significantly different. Glutathione content (GSH), glutathione peroxidase (GPx), glutathione-S-transferase (GST), superoxide dismutase (SOD), DOX (doxorubicin), QU (quercetin), QU-CHS (QU-loaded CHSNPs), DOX (doxorubicin), QU-CHS (QU-loaded CHSNPs), a, b, c, and d show similarity and non-similarity between groups. Groups that have the same superscript symbol(s) are not significantly different; groups that have different superscript symbols are significantly different
In our study, we investigated the protective effects of QU and QU-CHSNPs on DOX-induced cardiotoxicity and the underlying mechanisms of this protection (Ibrahim et al. 2011). As displayed in Fig. 5, QU decreased NO levels with an even greater decrease in QU-CHSNPs. This may be because QU leads to the scavenging of free radicals, which decreases their interactions with nitric oxide and thus reduces the amount of damage (Chen et al. 2019). The results suggest that QU provides cardiac protection against DOX by decreasing oxidative stress and damage. It is worth mentioning that the administration of QU-CHSNPs to normal rats showed a non-significant change when compared to the control, indicating safety.
Oxidative stress provides deleterious effects either by triggering LPO or by operating as a second messenger for primary free radicals that start LPO (Baghel et al. 2012; Nandakumar et al. 2011). MDA levels were measured in the present study as an indicator for LPO (Jaganjac et al. 2013). In Fig. 5, DOX administration caused a significant increase in MDA levels compared to the control (p < 0.001), consistent with preceding research that employed an analogous drug regimen (Korkina and Afanas’ 1996; Kim et al. 2017; Elberry et al. 2010). The initial targets of DOX-mediated free radical damage are cellular membranes that are rich in lipids susceptible to peroxidation. This radical damage produces stable and toxic aldehydes, such as MDA. These aldehydes can diffuse within the cell or even cross the plasma membrane and attack macromolecular targets far from where they were generated, thus acting as "second cytotoxic messengers." The treatment of DOX-injected rats with QU showed a decrease in MDA levels, while QU-CHSNPs showed a high pattern of decrease (Fig. 5). The existing research supports the potency of the antioxidant activity of QU versus DOX-induced oxidative stress in vivo, as evidenced by the inhibition of ROS generation and the reduction in MDA LPO activity, which corresponds with the findings (Balachandar et al. 2003).
Furthermore, our results confirmed that DOX decreased the antioxidant content and performance of non-enzymatic (GSH), enzymatic GPx, SOD, and glutathione-S-transferase (GST), significantly, as shown in Fig. 5 (p < 0.001). These results are in accord with preceding reviews (Dong et al. 2014; Hozayen et al. 2014; QuanJun et al. 2017), which supported the competence of DOX to provide ROS and overthrow the antioxidant defence policy. Our findings linked a decrease in cardiac GSH content to increased responses of GSH metabolizing enzymes. The first enzyme is GPx, which catalyzes the disintegration of H2O2 and organic peroxides through its four selenium co-factors, using GSH as a reducing agent. The data in Fig. 5 revealed a decline in GPx in the DOX-rats, and this was because of the mopping up of GPx by the free radicals generated by DOX (Mahmoud et al. 2020b). The second enzyme is GST, which employs GSH in the conjugation of DOX toxic metabolites (Omóbòwálé et al. 2018). GST detoxifies xenobiotics, drugs, and carcinogens and supports cells against redox cycling and oxidative stress. The heart possesses low levels of GST, and an overwhelming generation of free radicals by DOX may result in low GST levels higher than natural levels.
The administration of QU and QU-CHSNPs enhanced the activities of the antioxidants GSH, GPx, SOD, and GST against DOX, in agreement with previous research (Ibrahim et al. 2010). Thus, QU scavenges for superoxide radicals and reduces myocardial damage. Indeed, evidence indicates that QU has cardioprotective properties because of its antioxidant activity. QU is an excellent metal chelator that chelates transition metals such as iron, which can initiate the formation of ROS. Both free radical scavenging and chelating effects are involved in its cardio-treatment and protective effects (Raj et al. 2015).
Lipids
Lipids play a role in the complications of cardiovascular disease. DOX interferes with the lipid metabolism, changing the lipid profile. In Fig. 6, the levels of T. cholesterol, TG, LDL-c, and VLDL-c were found to increase in the DOX-rats, consistent with previous research (p < 0.001) (Arafa et al. 2005; Chennuru and Saleem 2013). The increase in cholesterol concentration could be because of a reduction in HDL levels, as HDLs transport cholesterol from tissues to the liver for catabolism. In this context, we observed decreased levels of HDLs in the DOX-rats. We attribute the elevation in TGs to the low activity of lipoprotein lipase. These changes in lipid levels may be due to enhanced lipid biosynthesis by cardiac cyclic adenosine monophosphate (Mubarak et al. 2018). Lipid-lowering drugs preserve the myocardium from DOX-induced toxicity (Sakthivel et al. 2013).
Displays effect of QU and QU-CHSNPs on the serum lipid profile of normal control and DOX-administered rats. Data are expressed as mean ± SE. Numbers of samples in each group is six. F-probability: P < 0.001 values that share the same superscript symbol, are not significantly different. Low- density lipoprotein-cholesterol (LDL-c), very-low-density lipoprotein-cholesterol (VLDL-c), High density lipoprotein-cholesterol (HDL-c), DOX (doxorubicin), QU (quercetin), QU-CHS (QU-loaded CHSNPs), a, b, c, and d show similarity and non-similarity between groups. Groups that have the same superscript symbol(s) are not significantly different; groups that have different superscript symbols are significantly different
The administration of QU and QU-CHSNPs decreased the concentrations of total cholesterol, TGs, VLDLs, and LDLs and increased the concentration of HDL-c in the DOX-induced rats. QU and QU-CHSNPs were both effective at ameliorating cholesterol levels. Other studies have shown the effectiveness of QU in minimizing dyslipidemia (Schimmel et al. 2004). Furthermore, QU has anti-hypercholesterolemia, antihypertensive, vasodilator, and anti-atherosclerotic properties (Aziz 2021). The presented results suggest that QU has a talent for acting as a cardioprotective agent against DOX-induced cardiotoxicity.
CK-MB, AST, and LDH activities
This investigation showed that DOX-induced cardiotoxicity was manifested by a significant elevation in serum CK-MB, AST, and LDH activities (Fig. 7), in agreement with previous research (p < 0.001) (Sharma et al. 2020; Ahmed et al. 2017). A possible explanation is the DOX induced damage to the cardiomyocytes membranes, releasing these entities from the cytoplasm into the plasma. Further, elevation of the cardiac enzyme function was found to be associated with histological degradation such as distinct necrosis of cardiomyocytes and inflammatory leukocytic infiltration into cardiac tissues (Ahmed et al. 2019; Chen et al. 2017). In this study, the QU and QU-CHSNPs treatments decreased the CK-MB, AST, and LDH in the DOX-injected rats. Our results significantly reinforce the conclusion of preceding research (Nimbal and Koti 2018), which showed that the QU pretreatments induced a substantial decline in the CK-MB, AST, and LDH activities when compared with the toxic control rats. This may be because of the protective effects of the QU in regulating the leakage of CK-MB, AST, and LDH. QU possesses free radical scavenging and antioxidant activities, which could justify its competence to defend the myocardium from DOX-induced damage by blocking the leakage of cardiac LDH, AST, and CK-MB isoenzymes (Raj et al. 2015). Based on these results, QU and QU-CHSNPs retain potent cardioprotective effects against DOX-induced heart injury.
The effect of QU and QU-CHSNPs on serum Troponin-I, AST, LDH, and CK-MB activities in normal and DOX-treated rats. The data are presented as mean SE. The number of samples in each group is six. F-probability: P < 0.001 values that share the same superscript symbol are not significantly different. Aspartate aminotransferase (AST), lactate dehydrogenase (LDH), creatine kinase-MB (CK-MB), DOX (doxorubicin), QU (quercetin), QU-CHS (QU-loaded CHSNPs), a, b, c, and d show similarity and non-similarity between groups. Groups that have the same superscript symbol(s) are not significantly different; groups that have different superscript symbols are significantly different
Cardiac troponin (c Tn-I)
The function of cTn-I is a sensitive and specific marker for myocardial cell injury. This contractile protein is not normally present in the serum and is released only after myocardial necrosis. Elevated troponin I levels predict the risks of both cardiac cell death and infarction following infarction (Nazmi et al. 2016). In this study, the development of cardiotoxicity in the DOX group was also evidenced by elevated c Tn-I level in the serum (p < 0.001) (Fig. 7), which was in accordance with previous research (Afroz 2016). However, the QU treatment reduced serum c TnI levels, which was consistent with previous research (Atas et al. 2015). These reductions in c TnI levels in the serum imply that QU and QU-CHSNPs protect rats from DOX-induced cardiotoxicity.
Cytokines (IL-1B, TNF-α and IL-10)
Inflammation participates in the pathogenesis of DOX-induced cardiotoxicity. It is fully cited that the overabundance of free radicals enhances the output of inflammatory mediators and triggers cardiac inflammatory processes (Yaseen et al. 2017). The progressive increase of pro-inflammatory and cytokine levels within heart tissues was identified as a potential pathological indication for DOX-induced cardiomyopathy. The results, in line with previous investigations, support the idea that inflammation plays a fundamental role in the pathogenesis of DOX-induced cardiotoxicity. The DOX group had a significant increase in serum TNF-α and IL-1B (p < 0.001), which is consistent with previous findings (Aziz et al. 2020; Shaker et al. 2018) (Fig. 8). The principal underlying mechanism promoting this elevation in inflammatory markers is not yet understood. However, it is possible that heightened levels of ROS, impaired tissue antioxidant capacity, and subsequent LPO are provoking factors for these variations.
Effect of QU and QU-CHSNPs on serum cytokines IL-1B, TNF-α and IL-10 level and serum apoptotic markers of caspase-3 of normal control and doxorubicin-administered rats. Data are expressed as Mean ± SE. Numbers of samples in each group is six. F-probability: P < 0.001 values, which share the same superscript symbol, are not significantly different. Interleukin (IL), TNF α tumor necrosis factor-alpha, DOX (doxorubicin), QU (quercetin), QU-CHS (QU-loaded CHSNPs), a, b, c, and d show similarity and non-similarity between groups. Groups that have the same superscript symbol(s) are not significantly different; groups that have different superscript symbols are significantly different
Indeed, it was declared that rises in inflammatory mediators interacted with heightened oxidative stress, which is considered to provoke inflammatory reactions through the activation of the NF-κB pathway, resulting in the transactivation of cytokines (Aziz et al. 2020). The administration of QU and QU-CHSNPs inhibited the elevated TNF-α in DOX-treated rats. In harmony with this finding, it has been shown that QU can inhibit NF-KB via its free radical scavenging and antioxidant activities (Raj et al. 2015). According to this result, QU and QU-CHSNPs inhibit the elevated IL-1β in DOX-treated rats.
The results of this study, on the other hand, show that DOX causes a significant decrease in serum levels of the anti-inflammatory cytokine IL-10 in DOX-rats (Fig. 8; (p < 0.001)), which is consistent with previous research (Yang 2021), whereas QU and QU-CHSNPs increase IL-10 levels. The existing research asserts that QU has anti-inflammatory properties and acts by suppressing pro-inflammatory cytokines, while the mechanisms by which QU increases IL-10 levels are not clear (Pecoraro et al. 2016).
Apoptotic markers
The oxidative stress generated by the DOX triggers many signaling pathways of cardiomyocyte apoptosis, including the activation of caspase-3 (Aziz et al. 2020). The caspase-3 protein is a cysteine-aspartic protease that is activated in apoptotic cells by the death ligand. The caspase-3 protein is thought to be a biomarker of cardiac tissue toxicity (Mkhize et al. 2017). The present study showed that DOX causes a significant elevation of caspase-3 levels (Fig. 8; p < 0.001), in agreement with previous research (Al-Kuraishy and Hussein 2017). Also, the results of this investigation agree with previous findings, which showed that DOX increases the levels of caspase-3, activates apoptosis by decreasing the expression of antiapoptotic proteins, and induces oxidative stress by increasing H2O2 production (Dash et al. 2015). The administration of QU and QU-CHSNPs reduced the elevated caspase-3. These results are in accordance with previous research, which supports the hypothesis that the protection presented against DOX-induced cardiotoxicity by treatment with QU may involve the suppression of cardiac apoptosis (Anghel et al. 2017).
Cardiac mRNA expression of Annexin-V, NrF2 and PPAR-γ
The Annexin isoforms II, V, and VI are a particular group of membrane-associated, Ca2 + -binding proteins that exist in heart tissue. Annexin-V is present in both cardiomyocytes and non-myocytes and may play a part in regulating cellular ion fluxes, organization, and secretion. To determine alterations in the Annexin-V isoform that might arise in heart failure, we measured the mRNA expression of this annexin. DOX administration caused a significant increase in cardiac Annexin-V (Fig. 9) compared with control (p < 0.001), indicating heart failure. A possible explanation for this might be that DOX can disturb the homeostasis of intracellular calcium cycling in the heart, in agreement with previous research (Chen et al. 2013). The DOX-rats treated with QU and QU-CHSNPs revealed a considerable decline in their cardiac Annexin-V ascent levels, consistent with the preceding study (Anghel et al. 2017). It is essential to notice that QU-CHSNPs decreased the Annexin-V elevation more than QU alone.
The effect of QU and QU-CHSNPs on cardiac mRNA expression of Annexin-V, NrF2, and PPAR in normal and doxorubicin-treated rats. The data are presented as mean SE. The number of samples in each group is six. F-probability: P < 0.001 values that share the same superscript symbol are not significantly different. NRF2 nuclear factor erythroid 2-related factor, peroxisome proliferator-activated receptor gamma 2, DOX (doxorubicin), QU (quercetin), QU-CHS (QU-loaded CHSNPs), a, b, c, and d show similarity and non-similarity between groups. Groups that have the same superscript symbol(s) are not significantly different; groups that have different superscript symbols are significantly different
Nuclear factor erythroid 2-related factor 2 (Nrf2), a transcriptional factor, retains crucial protective effects against oxidative stress. In the oxidative stress stage, Nrf2 is triggered, enters the cell nucleus, stimulates antioxidant gene expression, and reduces oxidative damage. Preceding research showed that DOX-induced oxidative stress is correlated with cardiac damage (Shaker et al. 2018). In our study, DOX administration decreased active nuclear Nrf2 in the cardiac tissues (Fig. 9; p < 0.001), consistent with previous research (Zhang et al. 2019). The upregulation of Nrf2 is a unique way to limit DOX-induced cardiac injury (Shaker et al. 2018). The current study proved that QU and QU-CHSNPS protect against DOX-induced cardiomyopathy by increasing NrF2 expression, which stimulates the synthesis of antioxidant defenses. Furthermore, QU has free radical scavenging and antioxidant properties (Aziz 2021).
The nuclear receptor peroxisome proliferation-activated receptor gamma (PPAR-γ) regulates the transcription of several genes that are primarily involved in fatty acid and energy metabolism (El-Agamy et al. 2019). PPAR-γ is found in a wide range of cell types, including cardiomyocytes and vascular cells. PPAR-γ activation reduces inflammatory cytokine production, and there is mounting evidence that PPAR- ligands have anti-inflammatory, antioxidative, and antiproliferative effects on cardiomyocytes (Ivanova et al. 2015). DOX treatment reduces PPAR-γ levels significantly (p < 0.001). These findings suggest that PPAR-γ may help protect rats from DOX-induced heart damage (Hasegawa et al. 2010). Furthermore, our findings showed that QU and QU-CHS treatment increased PPAR-γ levels (Fig. 9). Because of its anti-inflammatory properties, we concluded that quercetin increased PPAR-γ and decreased inflammation (Pecoraro et al. 2016).
Cardiac DNA fragmentation
In this investigation, the DNA fragmentation of cardiomyocytes was significantly high after the DOX administration (Fig. 10A, B; p < 0.001), which is a sign of oxidative stress. The reason is that DOX promotes mitochondrial ROS production, which in turn damages cardiomyocyte DNA and triggers apoptosis (Yan 2019; Francis and Nayak 2017). QU and QU-CHSNPs showed a reduction in DNA fragmentation. This is consistent with other studies (Mahmoud et al. 2020b), which found that QU reduces DOX cardiotoxicity by reducing oxidative stress, ROS levels, and DNA damage in order to maintain heart cell viability. Our results were harmonious with those of another investigation, which confirmed that QU limited the pernicious effects of DOX and cyclophosphamide on the kidney and liver by obstructing peroxidative destruction (Zanwar et al. 2013).
Effect of QU and QU-CHSNPs on cardiac DNA fragmentation of normal control and DOX-administered Rats. The data are presented as mean SE. The number of samples in each group is six. F-probability: P < 0.001 values that share the same superscript symbol are not significantly different. DOX (doxorubicin), QU (quercetin), QU-CHS (QU-loaded CHSNPs), show agarose gel electrophoresis of cardiac DNA fragmentation in normal (1), doxorubicin-administered groups (2), doxorubicin-administered groups treated with QU (3) and QU-CHS (4), and normal treated with QU-CHS (5). Lane M: DNA marker with 100 bp; Lanes 1, 4, and 5: DNA without streaking or laddering; Lane 2: show DNA streaking and laddering
Electron microscope and histopathology
Mitochondria are membrane-bound cells; their potential is saved in adenosine triphosphate (ATP). One of the key managers in DOX-induced cardiotoxicity is mitochondria. The reason is that DOX suppresses mitochondrial synthesis, stimulates fission, and impairs mitochondrial function, ending in heart collapse. In this investigation, the mitochondrial determinants of DOX cardiotoxicity were assessed, and the results showed that the size, shape, and integrity of the mitochondria were lost (Fig. 11B), while the QU-CHS preserved their membrane integrity but seemed to be compact (Fig. 11C) and not elongated like the control (Fig. 11A). Understanding the critical role of mitochondria in DOX-induced cardiomyopathy is critical to reducing the barriers that severely limit the clinical success of this life-saving anticancer therapy.
Considering the histological changes of the heart in normal rats, no histopathological alterations were observed, and a normal histological structure of the cardiac muscles could be found (Fig. 12A). Severe degenerative changes and necrosis of the cardiac muscles were accompanied by focal lymphocytic infiltration in the DOX-treated group (Fig. 12B). Treatment of DOX-administered rats with QU showed moderate hyalinosis of the cardiac muscles and focal lymphocytic infiltration (Fig. 12C). Treatment of the DOX-administered rats with QU-CHS was associated with mild-to-moderate degenerative changes of the cardiac muscles and minimal lymphocytic myocarditis (Fig. 12D). Additionally, the normal group treated with QU-CHS showed minimal changes to the cardiac muscles and lymphocytic myocarditis (Fig. 12E). In general, DOX administration caused severe histopathological lesions, which were characterized by severe inflammatory cell infiltration, hemorrhage, degeneration, and necrosis, and this was in agreement with previous research (Kocahan et al. 2017). In this investigation, QU was found to have an anti-inflammatory effect as it improved the histopathological features of DOX-induced cardiotoxicity and decreased inflammation, degeneration, and necrosis of the myocardium, in agreement with other studies (Iqbal et al. 2008).
Overall, through the current study, one can see that the protective effect of QU-CHSNPs was better than that of QU-free. An explanation for this may be that the QU encapsulation with CHSNPs could avoid enzymatic degradation in the gastrointestinal tract, limit the burst release, and sustain an adequate QU release rate pattern. Chitosan is a suitable absorption enhancer for poorly absorbable drugs because of its mucoadhesive properties, and it could boost QU absorption across the intestinal mucosa, increasing its bioavailability. As was noted, the QU-CHSNPs have a positive surface charge, so it is possible to enter cardiac cells more rapidly through the endocytosis process when compared to QU-free. Another explanation is that the CHSNPs with high surface area could increase the loaded QU dose and enhance its therapeutic action.
Conclusion
The results indicated that QU and QU-CHSNPs have adequate preventive activity against doxorubicin-induced cardiotoxicity, and the QU-CHSNPs were extremely dominant in inhibiting the DOX-induced cardiac toxicities. These protective effects may be mediated by lowering the elevated levels of cardiac function parameters, improving hyperlipidemia, reducing oxidative stress, enhancing antioxidant defence mechanisms in the heart, increasing the expression of PPAR-mRNA, activating Nrf2, lowering the elevated levels of Annexin-V and DNA fragmentation, controlling inflammation by lowering the levels of the pro-inflammatory cytokines TNF-, IL-1B, and Caspase-3, increasing the levels of the anti-inflammatory, and improving the histopathological changes in cardiac tissue. However, additional clinical studies are needed to assess the safety, benefits, and hazards of QU-CHSNPS in humans.
Availability of data and materials
The authors emphasize the availability of data and materials.
References
Abd El-Rahmanand SN, Suhailah S (2014) Quercetin nanoparticles: preparation and characterization. Indian J Drugs 2(3):96–103
Abdel-Aziz MM, Yosri M, Amin BH (2017) Control of imipenem resistant-Klebsiella pneumoniae pulmonary infection by oral treatment using a combination of mycosynthesized Ag-nanoparticles and imipenem. J Radiat Res Appl Sci 10(4):353–360
Abdel-Moneim A et al (2020) Novel polydatin-loaded chitosan nanoparticles for safe and efficient type 2 diabetes therapy: in silico, in vitro and in vivo approaches. Int J Biol Macromol 154:1496–1504
Afroz R et al (2016) Sundarban honey confers protection against isoproterenol-induced myocardial infarction in Wistar rats. BioMed Res Int. https://doi.org/10.1155/2016/6437641
Ahmed M (2013) The protective effect of ginger (Zingiber officinale) against adriamycin-induced hepatotoxicity in rats: histological study. Life Sci J 10(1):1412–1422
Ahmed OM, Ashour MB, Abd El-Fattah AS (2017) The preventive effects of navel orange peel ethanolic extract and naringin on doxorubicin-induced nephrocardiotoxicity in male albino rats. Indo Am J Pharm Res 7(07):109–117
Ahmed OM et al (2019) Effects of green tea infusion and epicatechin on doxorubicin-induced renocardiotoxicity in male albino rats. IJPSR 10(5):1000–1014
Al-Harthi SE et al (2014) Amelioration of doxorubicin-induced cardiotoxicity by resveratrol. Mol Med Rep 10(3):1455–1460
AlKuraishy H, Hussein R. 2017 Caspase-3 levels (CASP-3) in doxorubicin induced-cardiotoxicity role of metformin pretreatment. Research Journal of Oncology. https://doi.org/10.32474/OAJOM.2018.01.000120
Anghel N et al (2017) Acute cardiotoxicity induced by doxorubicin in right ventricle is associated with increase of oxidative stress and apoptosis in rats. Histol Histopathol 33(4):365–378
Arafa HM, Abd-Ellah MF, Hafez HF (2005) Abatement by naringenin of doxorubicin-induced cardiac toxicity in rats. J Egypt Natl Canc Inst 17(4):291–300
Atas E et al (2015) Cardiac troponin-I, brain natriuretic peptide and endothelin-1 levels in a rat model of doxorubicin-induced cardiac injury. J Cancer Res Ther 11(4):882
Ayla S et al (2011) Doxorubicin induced nephrotoxicity: protective effect of nicotinamide. Int J Cell Biol. https://doi.org/10.1155/2011/390238
Aziz TA (2021) Cardioprotective effect of quercetin and sitagliptin in doxorubicin-induced cardiac toxicity in rats. Cancer Manag Res 13:2349
Aziz MM et al (2020) Protective effects of olmesartan and l-carnitine on doxorubicin-induced cardiotoxicity in rats. Can J Physiol Pharmacol 98(4):183–193
Baghel SS et al (2012) A review of quercetin: antioxidant and anticancer properties. World J Pharm Pharm Sci 1(1):146–160
Balachandar A, Malarkodi K, Varalakshmi P (2003) Protective role of DLa-lipoic acid against adriamycin-induced cardiac lipid peroxidation. Hum Exp Toxicol 22(5):249–254
Beutler E, Duron O, Kelly M (1963) Evaluation of glutathione level in extra-cellular fluid. J Lab Clin Med 61:882–887
Burtis CA, Ashwood ER, Bruns DE. Tietz textbook of clinical chemistry and molecular diagnostics, 4th ed. WBSaunders Co. 2005
Cappetta, D., et al., Oxidative stress and cellular response to doxorubicin: a common factor in the complex milieu of anthracycline cardiotoxicity. Oxidative medicine and cellular longevity, 2017.
Castillo RL et al (2018) Quercetin prevents diastolic dysfunction induced by a high-cholesterol diet: role of oxidative stress and bioenergetics in hyperglycemic rats. Oxid Med Cell Longev. https://doi.org/10.1155/2018/7239123
Chen J-Y, Hu R-Y, Chou H-C (2013) Quercetin-induced cardioprotection against doxorubicin cytotoxicity. J Biomed Sci 20(1):1–11
Chen PY et al (2017) Protective effect of Co-enzyme Q10 On doxorubicin-induced cardiomyopathy of rat hearts. Environ Toxicol 32(2):679–689
Chen X et al (2019) Quercetin protects cardiomyocytes against doxorubicin-induced toxicity by suppressing oxidative stress and improving mitochondrial function via 14-3-3γ. Toxicol Mech Methods 29(5):344–354
Chennuru A, Saleem MT (2013) Antioxidant, lipid lowering, and membrane stabilization effect of sesamol against doxorubicin-induced cardiomyopathy in experimental rats. BioMed Res Int. https://doi.org/10.1155/2013/934239
Dash SK et al (2015) Self-assembled betulinic acid protects doxorubicin induced apoptosis followed by reduction of ROS–TNF-α–caspase-3 activity. Biomed Pharmacother 72:144–157
do Nascimento TC et al (2020) Microalgae carotenoids intake: influence on cholesterol levels, lipid peroxidation and antioxidant enzymes. Food Res Int 128:108770
Dong Q et al (2014) Quercetin attenuates doxorubicin cardiotoxicity by modulating B mi-1 expression. Br J Pharmacol 171(19):4440–4454
Doroshow JH, Locker GY, Myers C (1980) Enzymatic defenses of the mouse heart against reactive oxygen metabolites: alterations produced by doxorubicin. J Clin Invest 65(1):128–135
Dubois-Deruy E, Cuvelliez M, Fiedler J, Charrier H, Mulder P, Hebbar E, Pfanne A, Beseme O, Chwastyniak M, Amouyel P et al (2017) MicroRNAs regulating superoxide dismutase 2 are new circulating biomarkers of heart failure. Sci Rep 7:1–10
El-Agamy DS et al (2019) Pristimerin protects against doxorubicin-induced cardiotoxicity and fibrosis through modulation of Nrf2 and MAPK/NF-kB signaling pathways. Cancer Manag and Res 11:47
Elberry AA et al (2010) Cranberry (Vaccinium macrocarpon) protects against doxorubicin-induced cardiotoxicity in rats. Food Chem Toxicol 48(5):1178–1184
Ertuğ PU et al (2013) Comparative study of the quercetin, ascorbic acid, glutathione and superoxide dismutase for nitric oxide protecting effects in mouse gastric fundus. Eur J Pharmacol 698(1–3):379–387
Fan W et al (2012) Formation mechanism of monodisperse, low molecular weight chitosan nanoparticles by ionic gelation technique. Coll Surf B 90:21–27
Ferenczyova K, Kalocayova B, Bartekova M (2020) Potential implications of quercetin and its derivatives in cardioprotection. Int J Mol Sci 21(5):1585
Francis A, Nayak Y (2017) Modulation of doxorubicin-induced cardiotoxicity by averrhoa bilimbi extract. J Young Pharm. https://doi.org/10.5530/jyp.2017.9.14
Friedewald WT, Levy RI, Fredrickson DS (1972) Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem 18(6):499–502
Friedman RB, Young DS (1997) Effects of disease on clinical laboratory tests, 3rd edn. AACC press, Washington
Goetz RM et al (1999) Estradiol induces the calcium-dependent translocation of endothelial nitric oxide synthase. Proc Natl Acad Sci 96(6):2788–2793
Ha J-H et al (1997) Albumin release from bioerodible hydrogels based on semi-interpenetrating polymer networks composed of poly (ϵ-caprolactone) and poly (ethylene glycol) macromer. J Control Release 49(2–3):253–262
Habig W, Pabst M, Jakoby W (1974) Biological assay kit measures total GST activity. WJ Biol Chem 249:7130–7139
Hasegawa H, Takano H, Komuro I (2010) Therapeutic implications of PPAR in cardiovascular diseases. PPAR Res. https://doi.org/10.1155/2010/876049
Hashish FE et al (2021) Possible protective effects of quercetin on doxorubicin-induced cardiotoxicity in rats. Menoufia Med J 34(1):333
Hozayen WG, Abou Seif HS, Amin S (2014) Protective effects of ruitn and/or hesperidin against doxorubicin-induced hepatotoxicity. Int J Clin Nutr 2(1):11–17
Hussain T et al (2020) Flavonoids and type 2 diabetes: evidence of efficacy in clinical and animal studies and delivery strategies to enhance their therapeutic efficacy. Pharmacol Res 152:104629
Ibrahim MA et al (2009) Angiotensin-converting enzyme inhibition and angiotensin AT1-receptor antagonism equally improve doxorubicin-induced cardiotoxicity and nephrotoxicity. Pharmacol Res 60(5):373–381
Ibrahim SS, Barakat MA, Helmy H (2010) Modulating effect of carvedilol on doxorubicin-induced cardiomyopathy and hepatic damage. J Am Sci 6(12):20–32
Ibrahim SS et al (2011) Role of selenium in attenuating cardiac and hepatic damages induced by the antitumor agent, doxorubicin. Life Sci J 8:1–12
Iqbal M et al (2008) Protective effects of telmisartan against acute doxorubicin-induced cardiotoxicity in rats. Pharmacol Rep 60(3):382
Iqubal A et al (2018) Clinical updates on drug-induced cardiotoxicity. Int J Pharm Sci Res 9:16–26
Ivanova EA et al (2015) Peroxisome proliferator-activated receptor (PPAR) gamma in cardiovascular disorders and cardiovascular surgery. J Cardiol 66(4):271–278
Jaganjac M et al (2013) Reactive aldehydes–second messengers of free radicals in diabetes mellitus. Free Radical Res 47(sup1):39–48
Kakran M, Sahoo NG, Li L, Judeh Z (2015) Fabrication of quercetin nanoparticles by anti-solvent precipitation method for enhanced dissolution. In AGRIS since 223:59–64
Kelleni MT, Amin EF, Abdelrahman AM (2015) Effect of metformin and sitagliptin on doxorubicin-induced cardiotoxicity in rats: impact of oxidative stress, inflammation, and apoptosis. J Toxicol. https://doi.org/10.1155/2015/424813
Khan H et al (2021) Flavonoids nanoparticles in cancer: Treatment, prevention and clinical prospects. Seminars Cancer Biol. https://doi.org/10.1016/j.semcancer.2019.07.023
Kim SH et al (2017) Comparison of doxorubicin-induced cardiotoxicity in the ICR mice of different sources. Lab Anim Res 33(2):165–170
Kocahan S et al (2017) Protective effect of quercetin against oxidative stress-induced toxicity associated with doxorubicin and cyclophosphamide in rat kidney and liver tissue. Iran J Kidney Dis 11(2):124
Korkina LG, Afanas’ Ev IB (1996) Antioxidant and chelating properties of flavonoids. Adv Pharmacol 38:151–163
Li Y et al (2016) Quercetin, inflammation and immunity. Nutrients 8(3):167
Lovegrove JA, Stainer A, Hobbs DA (2017) Role of flavonoids and nitrates in cardiovascular health. Proc Nutr Soc 76(2):83–95
Luque-Alcaraz AG et al (2012) Characterization and antiproliferative activity of nobiletin-loaded chitosan nanoparticles. J Nanomater. https://doi.org/10.1155/2012/265161
Lustriane C et al (2018) Effect of chitosan and chitosan-nanoparticles on post harvest quality of banana fruits. J Plant Biotechnol 45(1):36–44
Maaliki D et al (2019) Flavonoids in hypertension: a brief review of the underlying mechanisms. Curr Opin Pharmacol 45:57–65
Mahmoud A et al (2020a) Characterization of Ulva fasciata ethanolic extract-mediated biosynthesized silver nanoparticles and evaluation of their nephrocardiopreventive effects in doxorubicin-injected wistar rats. Adv Anim Vet Sci 8(s2):98–111
Mahmoud H et al (2020b) Effects of rutin and quercetin on doxorubicin-induced renocardiotoxicity in male Wistar rats. Adv Anim Veterin Sci 8(4):2020370–2020384
Matook AI et al (2012) Potential protective role of quercetin against chronic doxorubicin induced cardiotoxicity in rats EL-mina med. Bull 23:2
Mkhize NVP, Qulu L, Mabandla MV (2017) The effect of quercetin on pro-and anti-inflammatory cytokines in a prenatally stressed rat model of febrile seizures. J Experiment Neurosci 11:1179069517704668
Montogomery H, Dymock J (1961) The determination of nitrite in water: colorimetric method of nitric oxide assay. Analyst 86:414
Mubarak S et al (2018) Cardioprotective effect of date palm against doxorubicin-induced cardiotoxicity. Asian J Pharm Clin Res 11(7):141–146
Muller R (2004) Drug nanocrystals of poorly soluble drugs. Encyclopedia Nanosci Nanotechnol 627:38
Nandakumar N et al (2011) Hesperidin, a natural citrus flavonoglycoside, normalizes lipid peroxidation and membrane bound marker enzymes in 7, 12-Dimethylbenz (a) anthracene induced experimental breast cancer rats. Biomed Prev Nutr 1(4):255–262
Nathiya S, Durga M, Devasena T (2014) Preparation, physico-chemical characterization and biocompatibility evaluation of quercetin loaded chitosan nanoparticles and its novel potential to ameliorate monocrotophos induced toxicity. Dig J Nanomater Bios 9:1603–1613
Nazmi AS et al (2016) Protective effects of Bombyx mori, quercetin and benazepril against doxorubicin induced cardiotoxicity and nephrotoxicity. J Saudi Chem Soc 20:S573–S578
Nimbal, S. and B. Koti, Article Details Effect of Ethanolic Extract Fractions of Rosa centifolia in Doxorubicin-induced Myocardial Toxicity in Albino Rats. 2018.
Ohkawa H, Ohishi N, Yagi K (1979) Assay for lipid peroxides in animal tissues by thiobarbituric acid reaction. Anal Biochem 95(2):351–358
Omóbòwálé TO et al (2018) Ameliorative effect of gallic acid on doxorubicin-induced cardiac dysfunction in rats. J Basic Clin Physiol Pharmacol 29(1):19–27
Paglia DE, Valentine WN (1967) Studies on the quantitative and qualitative characterization of erythrocyte glutathione peroxidase. J Lab Clin Med 70(1):158–169
Pecoraro M et al (2016) Inflammatory mediators in a short-time mouse model of doxorubicin-induced cardiotoxicity. Toxicol Appl Pharmacol 293:44–52
Pereira CG et al (2011) Drug-induced cardiac mitochondrial toxicity and protection: from doxorubicin to carvedilol. Curr Pharm Des 17(20):2113–2129
Pugazhendhi A et al (2018) Toxicity of doxorubicin (Dox) to different experimental organ systems. Life Sci 200:26–30
QuanJun Y et al (2017) Protective effects of dexrazoxane against doxorubicin-induced cardiotoxicity: a metabolomic study. PLoS ONE 12(1):e0169567
Raj LA et al (2015) Preparation and characterization of BSA and chitosan nanopartices for sustainable delivery system for quercetin. J Appl Pharm Sci 5(7):001–005
Rashedi J et al (2019) Anti-tumor effect of quercetin loaded chitosan nanoparticles on induced colon cancer in wistar rats. Adv Pharm Bull 9(3):409
Ritchie AW et al (2013) Reversed crystal growth of rhombohedral calcite in the presence of chitosan and gum arabic. CrystEngComm 15(47):10266–10271
Saad SY, Najjar TA, Alashari M (2004) Cardiotoxicity of doxorubicin/paclitaxel combination in rats: effect of sequence and timing of administration. J Biochem Mol Toxicol 18(2):78–86
Sakthivel K et al (2013) Cardioprotective and antioxidant potential of Scilla hyacinthina. J Biol Sci 13(5):313
Sarkar SD et al (2013) Physico-chemical/biological properties of tripolyphosphate cross-linked chitosan based nanofibers. Mater Sci Eng C 33(3):1446–1454
Schimmel KJ et al (2004) Cardiotoxicity of cytotoxic drugs. Cancer Treat Rev 30(2):181–191
Shaker RA et al (2018) Enoxaparin attenuates doxorubicin induced cardiotoxicity in rats via interfering with oxidative stress, inflammation and apoptosis. BMC Pharmacol Toxicol 19(1):1–10
Sharma A et al (2020) Modulation of Nrf2 by quercetin in doxorubicin-treated rats. Heliyon 6(4):e03803
Songbo M et al (2019) Oxidative stress injury in doxorubicin-induced cardiotoxicity. Toxicol Lett 307:41–48
Stachurski ZH (2011) On structure and properties of amorphous materials. Materials 4(9):1564–1598
Tacar O, Sriamornsak P, Dass CR (2013) Doxorubicin: an update on anticancer molecular action, toxicity and novel drug delivery systems. J Pharm Pharmacol 65(2):157–170
Tamilselvan N, Raghavan CV (2015) Formulation and characterization of anti Alzheimer’s drug loaded chitosan nanoparticles and its in vitro biological evaluation. J Young Pharm 7(1):28
Tietz NW, Finley PR, Pruden E (1995) Clinical guide to laboratory tests, vol 624. WB Saunders company, Philadelphia
Witt I, Trendelenburg C (1982) Gemeinsame Studie zur Erstellung von Richtwerten für klinisch-chemische Kenngrößen im Kindesalter. Journal of clinical chemistry and clinical biochemistry. Zeitschrift Fur Klinische Chemie Und Klinische Biochemie 20:35–242
Xu Y et al (2007) Preparation of dual crosslinked alginate–chitosan blend gel beads and in vitro controlled release in oral site-specific drug delivery system. Int J Pharm 336(2):329–337
Yan J et al (2019) Piperine Alleviates Doxorubicin-Induced Cardiotoxicity via Activating PPAR-γ in Mice. PPAR Res. https://doi.org/10.1155/2019/2601408
Yang H-B et al (2021) Selenium attenuates doxorubicin-induced cardiotoxicity through Nrf2-NLRP3 pathway. Biol Trace Element Res. https://doi.org/10.1007/s12011-021-02891-z
Yaseen AE-RA et al (2017) Potential protective effects of trimetazidine and quercetin on isoprenaline-induced myocardial infarction in rats. Menoufia Med J 30(4):1110
Young D (2001) Effects of disease on Clinical Lab. Tests, 4th ed AACC
Zanwar AA, Hegde MV, Bodhankar SL (2013) Protective role of concomitant administration of flax lignan concentrate and omega-3-fatty acid on myocardial damage in doxorubicin-induced cardiotoxicity. Food Sci Human Wellness 2(1):29–38
Zhang J-Y et al (2006) Preparation of amorphous cefuroxime axetil nanoparticles by controlled nanoprecipitation method without surfactants. Int J Pharm 323(1–2):153–160
Zhang X et al (2019) Rosmarinic acid alleviates cardiomyocyte apoptosis via cardiac fibroblast in doxorubicin-induced cardiotoxicity. Int J Biol Sci 15(3):556
Zhao L-M et al (2011) Preparation and application of chitosan nanoparticles and nanofibers. Braz J Chem Eng 28:353–362
Zhao X et al (2012) Recrystallization and micronization of taxol using the supercritical antisolvent (SAS) process. Ind Eng Chem Res 51(28):9591–9597
Zhao L et al (2018) MicroRNA-140-5p aggravates doxorubicin-induced cardiotoxicity by promoting myocardial oxidative stress via targeting Nrf2 and Sirt2. Redox Biol 15:284–296
Acknowledgements
The authors are thankful to all members at Materials Science and nanotechnology Department, Faculty of Postgraduate Studies for Advanced Sciences (PSAS), Beni-Suef University, Egypt and Dr. El-Shaymaa El-Nahass, Associate Prof. of Histopathology, Pathology Department, Faculty of Veterinary Medicine, Beni-Suef University and Egypt for help in examining and determining the lesions in the heart histological stained sections.
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB). The present study was supported by individual funding.
Author information
Authors and Affiliations
Contributions
Dr. E conceived the idea for biomedical application, Dr. E, AGS, and MA-G participated in the study design. Dr. E, AGS and ZEE prepared the materials. Dr. E, AGS, and MA-G conducted the bio section. Dr. E, AGS wrote the manuscript. Dr. BM revised the manuscript. All the authors revised the manuscript and approved the final version to be published, and agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The authors followed the ethics of research, approved and consented to participate in this study. The Institutional Animal Care Committee of Beni-Suef University, Egypt, approved the entire conducted procedures (BSU/FS/2019/2). All the authors consent and understand that their participation is voluntary and that they are free to withdraw at any time, without giving a reason and without cost. All authors understand that they will be given a copy of this consent form. All authors voluntarily agree to take part in this study.
Consent for publication
The authors consent this manuscript for publication.
Competing interests
The authors confirm no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Soliman, A.G., Mahmoud, B., Eldin, Z.E. et al. Optimized synthesis characterization and protective activity of quercetin and quercetin–chitosan nanoformula against cardiotoxicity that was induced in male Wister rats via anticancer agent: doxorubicin. Cancer Nano 14, 10 (2023). https://doi.org/10.1186/s12645-023-00158-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12645-023-00158-x