Abstract
Background
Chloroplast development is coordinately regulated by plastid- and nuclear-encoding genes. Although many regulators have been reported to be involved in chloroplast development, new factors remain to be identified, given the complexity of this process.
Results
In this study, we characterized a rice mutant lethal albinic seedling 1(las1)form of a 4-hydroxy-3-methylbut-2-enyl diphosphate reductase (OsHMBPP) that was targeted to the chloroplasts. The LAS1 mutation caused the albino lethal phenotype in seedlings. Transmission electron microscopy indicated that las1 were defective in early chloroplast development. LAS1 is preferentially expressed in leaves, implying its role in controlling chloroplast development. The expression levels of many chloroplast-encoded genes were altered significantly in las1. The expression levels of nuclear-encoded gene involved in Chl biosynthesis were also decreased in las1. We further investigated plastidic RNA editing in las1 and found that the edit efficiency of four chloroplast genes were markly altered. Compared with WT, las1 exhibited defective in biogenesis of chloroplast ribosomes.
Conclusions
Our results show that LAS1/OsHMBPP plays an essential role in the early chloroplast development in rice.
Similar content being viewed by others
Background
Chloroplasts are essential for plant development and growth, through manipulating the fixation of CO2 and biosynthesis of carbon skeletons and other physiological processes (Jarvis and López-Juez 2013). Accumulating evidence has shown the importance of chloroplast biogenesis and development during germination for plant vitality, seed set and growth (Lopez-Juez and Pyke 2005; Pogson and Albrecht 2011). Chloroplast biogenesis is initiated from proplastids through endosymbiosis from an ancestor of extant cyanobacteria and is dependent on the coordinated expression of genes encoded in both nuclear and plastid genomes (Lopez-Juez and Pyke 2005; Kessler and Schnell 2009). The development of chloroplasts differs between organs and species depending on the specialization of tissues and stage of development. For example, distinct phenotypes between cotyledons and true leaves were observed in variegated (var) and snowy cotyledon (sco) mutants in Arabidopsis, respectively (Albrecht et al. 2008; Liu et al. 2010; Zagari et al. 2017), i.e. chlorotic true leaves but green cotyledons in the var. and chlorotic or bleached cotyledons but green true leaves in the sco mutants. It is evident that nucleus-encoded polymerases (NEPs) and plastid-encoded polymerases (PEPs) involved in gene transcription, RNA maturation, protein translation and modification have great effect on the biogenesis of chloroplasts (Pogson and Albrecht, 2011; Yu et al. 2014).
Isoprenoids play essential roles in plant growth and development. In higher plants, biosynthesis of the basic isoprenoid units occurs by two different pathways: the mevalonate (MVA) pathway and the methyl-D-erythritol-4-phosphate (MEP) pathway. For decades, it was thought that the MVA pathway was solely responsible for isoprenoid biosynthesis. However, the alternative MEP pathway was discovered recently in bacteria, green algae, and higher plants (Rohmer et al. 1993; Cunningham Jr. et al. 2000; Rodriguez-Concepcion and Boronat, 2002; Rohmer, 2003). The MVA pathway occurs in the cytosol and is responsible for the synthesis of sterols, certain sesquiterpenes, and the side chain of ubiquinone (Disch et al. 1998). By contrast, the MEP pathway operates in the plastids and is involved in providing the precursors for monoterpenes, isoprene, chlorophylls, carotenoids, tocopherols, taxadiene, gibberellins, and abscisic acid (Zeidler et al. 1997; Eisenreich et al. 1998; Lichtenthaler, 1999). In recent years, the entire MEP pathway and almost all the enzymes involved have been identified in Escherichia coli. The activity of the corresponding enzyme and the roles they played in the pathway have been widely demonstrated (Lois et al. 1998; Herz et al. 2000; Hecht et al. 2001; Adam et al. 2002). The genes encoding each enzyme in the MEP pathway are highly conserved. Genes encoding enzymes involved in the MEP pathway are important for chlorophyll and carotenoid biosynthesis in plants. Knockout mutations of several genes in this pathway in Arabidopsis, Zea may and Nicotiana benthamiana display the albino phenotype (Budziszewski et al. 2001; Gutierrez-Nava et al. 2004; Page et al. 2004; Hsieh and Goodman, 2005; Lu et al. 2012). However, the genes involved in the MEP pathway in rice has not been reported yet and needs further study.
In this study, we obtained a rice mutant lethal albinic seedling 1 (las1) by the CRISPR/Cas9 system. Identification of CRISPR/Cas9 sites revealed sequence differences in the gene encoding a 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, which is involved in the isoprenoids biosynthesis. The mutant seedlings were albinic and eventually died from lacking of chlorophyll. Additionally, the transcript levels of genes associated with chlorophyll biosynthesis, and those associated with chloroplast biogenesis were severely affected in the las1 mutant. These findings implicate that rice LAS1 plays an important role in chloroplast biogenesis during early leaf development.
Results
Characterization of the las1 Mutant in Rice
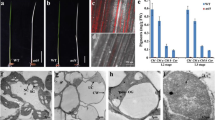
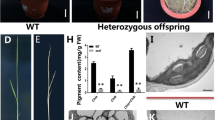
We isolated a rice mutant by the CRISPR/Cas9 system, which exhibited defects in leaf color, termed lethal albinic seedling 1(las1). The las1 mutant exhibited the albino phenotype and eventually died (Fig. 1a; Additional file 1: Figure S1). Consistent with their phenotype, the chlorophyll contents were significantly reduced in las1 compared with that of the wild type (Fig. 1b). Sequence confirmation indicated that the phenotype of las1 resulted from the LOC_Os03g52170 mutation encoding a 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, OsHMBPP (Fig. 1c). We identified five homozygous mutants and the phenotype of the heterozygous mutants were similar to that of WT (Additional file 1: Figure S1). Protein sequence alignments of the homozygous mutants and the wild type protein revealed that three mutants showed premature translational stops, and the line las1–1 encodes the least amino acids. So we chose the line las1–1 to represent the LAS1 mutant for further study. These data suggested that LAS1/OsHMBPP is essential for early chloroplast development in rice.
The phenotype and pigment contents of wild type and las1. a Morphology of wild type (WT) and las1 seedling (Bar = 5 cm). b Pigment contents of WT and las1 seedlings. Values represent the mean ± SD of three biological replicates. **p < 0.01. c Sequence confirmation of the LAS1 mutation. The 23-nt target sequence of the Cas9/sgRNA complex is underlined in red
Chloroplast Ultrastructure of the las1 Mutant
Chloroplast development depends on the successive synthesis and integration of Chl into photosynthetic complexes (Wang & Grimm, 2015). To further investigate the albino leaf phenotype of las1, we observed the chloroplast structure of las1 and wild type in leaves by TEM. The chloroplasts of the WT leaves were normal in shape, containing large starch granules and normal thylakoid membranes with stacks of grana (Fig. 2a-b). In contrast, chloroplasts were severely disrupted in the las1 leaves (Fig. 2c-d). Our results indicated that the albino leaf phenotype of las1 resulted from the abnormal development of chloroplasts.
Expression Pattern of OsHMBPP
According to the Rice eFP Browser (http://bar.utoronto.ca/efprice/cgi-bin/efpWeb.cgi), we found that OsHMBPP was expressed in all tissues, especially in leaves (Additional file 1: Figure S2). To verify these data, we examined the expression level of LAS1 in different organs of the wild type by qRT-PCR. OsHMBPP was highly expressed in the mature leaves and also detected in the sheaths, seeds, roots and panicles (Fig. 3a). Our results demonstrated that OsHMBPP is constitutively expressed in various tissues and mainly functions in the green tissues.
To further investigate whether the expression of OsHMBPP is modulated by photoperiod, we compared the day/night expression patterns during the 14-h light/10-h dark photoperiod conditions. Test samples were taken every 4 h. qRT-PCR showed the expression of OsHMBPP oscillated during a light/dark cycle and the highest transcript level was detected during light period (Fig. 3b).
LAS1/OsHMBPP Encodes a Conserved Chloroplast-Targeted Protein
LAS1/OsHMBPP encodes a HMBPP reductase, which is involved in catalyzing 2C-methyl-D-erythritol4-phosphate (MEP) converted to IPP and DMAPP in the nonMVA pathway in Arabidopsis thaliana and maize, and affect the development of chloroplasts (Hsieh and Goodman, 2005; Lu et al. 2012). Therefore, we measured the contents of IPP and DMAPP between WT and las1. IPP and DMAPP contents in the las1 seedlings were significantly reduced, indicating that the nonMVA pathway in las1 was impaired (Additional file 1: Figure S3). Based on amino acid sequence alignment, OsHMBPP shares high identity with its orthologs in other species, including Sorghum bicolor (89.7%), Arabidopsis (73.8%), and Zea mays (90.1%) (Fig. 4b). Given that OsHMBPP and Arabidopsis, HDR, share 73.8% of amino acid identity and that structural predictions of both proteins revealed three-dimensional similarities by I-TASSER algorithm (Fig. 4a). ChloroP analysis revealed that the OsHMBPP protein contains a predicted chloroplast transit peptide of 31 amino acid residues at its N-terminal (Additional file 1: Table S1). To further determine its subcellular localization, a transient expression system was performed in rice protoplasts. As expected, the fusion protein OsHMBPP-GFP was clearly co-localized with Chl auto-fluorescence (Fig. 5), indicating OsHMBPP was localized in chloroplasts.
Protein structure of OsHMBPP and sequence alignment of multiple amino acids from different organisms. a Structure prediction of the HMBPP reductase IspH (Escherichia coli), HDR (Arabidopsis), HDR(Zea mays) and OsHMBPP (Oryza sativa) by I-TASSER algorithm. b Amino acid sequence alignment of the 3 types of OsHMBPP homologs. Amino acids that were fully or partially conserved are shaded blue and green, respectively
Subcellular location of the OsHMBPP protein. GFP signals of OsHMBPP-GFP fusion protein was located in the chloroplasts by transient expression analyses in rice protoplasts. Green fluorescence shows GFP, red fluorescence shows chloroplast autofluorescence, and yellow fluorescence shows the merged fluorescence
Altered Expression of Chlorophyll Biosynthesis and Plastid-Encoded Genes
We examined the expression levels of nuclear genes associated with Chl biosynthesis. qRT-PCR analyses showed that some Chl biosynthesis related genes were significantly down-regulated in the las1 mutant (Fig. 6b). To investigate whether the LAS1 mutation affects transcription by PEP and NEP, we examined the transcript abundance of various plastidic genes in the las1 mutant by qRT-PCR. Plastid genes consist of three types of genes, which are transcribed by NEP, PEP, and both NEP and PEP, respectively (Yu et al. 2014). In las1, the expression levels of class III genes (e.g rpoA, rpoB, rpoC) were strongly increased, while class I genes (e.g psaA, psbA, rbcL) decreased (Fig. 6a). These results suggest that las1 was defective in PEP activity and influenced the optimal expression of plastidic genes in rice seedlings.
Expression analysis of genes involved in chloroplast development and chlorophyll biosynthesis. a Expression analysis of chloroplast development related genes in wild-type and las1. b Expression analysis of chlorophyll biosynthesis related genes in wild-type and las1. The relative expression level of each gene in WT plants was set to 1.0. Ubiquitin gene was used as a reference. Error bars indicate SD (n = 3)
Mutation in LAS1 Affects Four Plastdic Editing Sites and Chloroplast Ribosome Biogenesis
The translation deficiency in las1 suggests that LAS1 might be involved in posttranscriptional RNA metabolism processes. We next detected whether LAS1 was involved in the chloroplast RNA editing and RNA splicing. There are 21 chloroplast RNA editing sites in the 12 chloroplast genes (Corneille et al. 2000). Therefore, we analysed all 21 sites in the chloroplast cDNA in wild type and las1. We found that four C sites of chloroplast genes in las1 did not edit normally. The editing of rpl2-C2, ndhG-C(− 11), and atpA-C1159 were reduced in las1, but the editing of rpoB-C467 was increased (Fig. 7). There was no difference between the wild type and las1 in the rest 17 RNA editing sites (Additional file 1: Figure S4). There are 18 introns in the rice chloroplast genome (Kaminaka et al. 1999). Furthermore, we carried out RT-PCR with primers flanking the introns and compared the PCR products length between wild type and las1 by agarose gel. Splicing of the 17 chloroplast transcripts were not significantly impeded in las1 (Additional file 1: Figure S5). These data suggest that LAS1 affects the RNA editing efficiency of chloroplast genes, but not function in chloroplast RNA splicing.
Abnormal chloroplast development is often associated with inhibition of chloroplast ribosome synthesis (Ge et al. 2017; Cao et al. 2019), which is composed of 30 S small subunit and 50 S large subunit. We further examined whether the activity of chloroplast ribosome changed in las1. qRT-PCR analysis indicated that the expression levels of 16 S and 23 S were decreased whereas 18 S and 25 S were increased in las1, compared with the wild type (Fig. 8). The result indicated that las1 was defective in chloroplast ribosome biogenesis.
LAS1/OsHMBPP Interacts with the MORF Family Protein in Yeasts
The multiple organellar RNA editing factor (MORF) family proteins are essential for the chloroplast/mitochondrion RNA editing in plants (Bentolila et al. 2012; Takenaka et al. 2012; Zhang et al. 2017). There are nine MORF proteins and seven MORF proteins in Arabidopsis and rice, respectively. To investigate whether LAS1/OsHMBPP interacted with the MORF family proteins, we used the yeast two hybrid to detect the interaction. Notably, the results showed that only Os09g33480 had an interaction with OsHMBPP (Fig. 9a-b). Furthermore, we performed BiFC assays using N. benthamiana cells and found that OsHMBPP-nYFP and Os09g33480-cYFP could interact with each other (Fig. 9c). Os09g33480 belongs to the Arabidopsis MORF8 branch. Os09g33480 may be targeted to both mitochondria and chloroplasts similar to Arabidopsis MORF8. Meanwhile, WSP1, Os06g02600, Os09g04670, Os08g04450, Os11g11020, and Os03g38490 did not interact with OsHMBPP (Fig. 9a-b). Based on the Rice eFP Browser, Os09g33480 is ubiquitously expressed in many rice organs, especially in seeds, panicles and leaves (Additional file 1: Figure S6).
The interaction assay between OsHMBPP and MORF families in rice. a-bOsHMBPP was fused to the pGBKT7 vector (OsHMBPP-BD). Seven MORFs were fused to the pGADT7 vector. DDO, control medium (SD–Leu/−Trp); QDO, selective medium (SD–Leu/−Trp/−His/−Ade). c Interaction between OsHMBPP and Os09g33480 in BiFC assays. Bars = 100 μm
Expression Patterns of the MEP Pathway Genes in a Normal Day/Night Cycle
To further investigate whether expression of the MEP pathway genes is regulated by photoperiod, we compared the expression patterns of six genes during the 14-h light/10-h dark photoperiod conditions (Additional file 1: Figure S7). The expression patterns of CMK and DXS were similar during the 14-h-light/10-h-dark cycle. Peak levels of CMK and DXS were detected in the early period of the light cycle (8–12 h in the light). Oscillations in the expression of DXR, HDS, MCS, and CMS also occurred during the light/dark cycle. In contrast to CMK and DXS, peak levels of DXR and HDS expression appeared in the late period of the dark cycle (20-24 h). The expression of all MEP pathway genes is significantly repressed during the transition from light to dark (Additional file 1: Figure S7).
Discussion
Characterization of LAS1/OsHMBPP
Chloroplast defective development generally results in an albino or chlorotic leaf phenotype, which severely affects the biomass and even the survival of plants, particularly in crops. Isoprenoids play an essential role in chloroplast development. In this study, we isolated the las1 mutant, which displayed an albino leaf phenotype and was seedling lethal (Fig. 1). TEM observation demonstrated that the phenotype of las1 resulted from the defective development of thylakoids, eventually leading to the breakdown of entire chloroplasts (Fig. 2). LAS1 encodes 4-hydroxy-3-methylbut-2-enyl diphosphate reductase OsHMBPP in rice (Fig. 5).HMBPP functions in the isoprenoid biosynthesis via the MEP pathway (McAteer et al. 2001; Guevara-Garcia et al. 2005; Hsieh and Goodman, 2005). Isoprenoids are derived from a basic five-carbon unit, isopentenyl diphosphate (IPP) and allyl isomerdimethylallyl diphosphate (DMAPP). HMBPP is the rate limiting enzyme of this process, and regulates the biosynthesis of chlorophylls, ABA and GA. Mutation of HMBPP in maize and A. thaliana produced an albino phenotype with impaired chloroplasts containing large vesicles (Hsieh and Goodman, 2005; Lu et al. 2012). In rice, the OsHMBPP mutation resulted in a reduction in the content of IPP and DMAPP (Additional file 1: Figure S3), which led to decrease of the chlorophyll content and plant death. On the other hand, OsHMBPP interacted with the MORF protein Os09g33480(Fig. 9), and affected the RNA editing of chloroplast genes. These results indicate that the function of HMBPP is very conserved in different species, and is essential for the early chloroplast development in rice.
LAS1 Encodes a Chloroplast-Targeted Protein and Regulates the Early Chloroplast Development in Rice
Our results showed that LAS1 was constitutively expressed, with high levels of transcript in the leaves, implying that LAS1 might play an essential role in leaf development (Fig. 3a). Subcellular localization in rice protoplasts demonstrated that the LAS1 protein is routed to the chloroplasts (Fig. 4). The chloroplast is a semi-autonomous organelle, which contains about 100 genes, although more than 3000 proteins function within it (Leister, 2003). Thus, nucleus-encoded factors play essential roles in regulating chloroplast development, which requires the coordinated expression of both nucleus-encoded and chloroplast-encoded genes. The LAS1 mutation disrupts the transcripts of plastid and nuclear genes associated with chloroplast development (Fig. 6). The transcript accumulation of both PEP-and NEP-dependent genes(atpB) and PEP-transcribed plastid genes (psaA, psbA, rbcL) were severely suppressed (Fig. 6), suggesting that the accumulation of transcripts for PEP components did not result in the formation of functional PEP due to the disruption of transcription/ translation apparatus.
OsHMBPP Is Required for Chloroplast Genes Transcript Editing
RNA editing is an important event in post-transcriptional RNA process, which alters RNA sequences by converting specific target cytidines to uridine in transcripts of plastids and mitochondria (Takenaka et al. 2013). Abolishing editing in plastidic genes can disturb plastidic functionality and inhibit rice normal development and growth (Tang et al. 2017; Zhang et al. 2017; Liu et al. 2018; Cui et al. 2019). For example, in the wsp1 mutant, editing at ndhD-878, rps14–80, ndhG(− 11), and rpoB-467, 545, 560 are abolished, resulting in the defective chloroplast development. Meanwhile, the mutation of pentatricopeptide repeat protein gene DUA1 reduced the editing efficiency of rps-182 and rpoB-545, 560, leading to the seedling lethality under cold stress. Our findings showed that the editing efficiency of four chloroplast RNA editing sites in las1 is remarkably altered, compared with the wild type (Fig. 7), which may be an explanation for the phenotype of las1. Edited rpoB codon 467 is located in the Dispensable Region I and the editing of the rpoB-467 site is non-essential (Corneille et al. 2000). In rice, the rpoB-467 site is partial editing. Therefore, the altered editing of the rpoB-467 site was not the cause of the phenotype of las1. The chloroplast and mitochondria RNA editing events may be an indication of evolutionary adaptation (Chu and Wei, 2019). The rpl2, rpoB and atpA gene sequences of the chloroplast genomes of different species were analyzed from the perspective of evolution. The rpoB-467 and atpA-1159 encode a Ser in monocots, but encode either a Ser or a Leu in eudicots (Additional file 1: Figure S8). We found an interesting phenomenon that rpl2–2 encodes Thr in monocots, but it encodes Met in eudicots (Additional file 1: Figure S8). These results showed that the rpl2–2 sites of monocots and eudicots were highly differentiated. The ‘C’ at rpl2–2 of monocots may have been acquired during the evolution.
Plant MORF proteins are essential for RNA editing and splicing, but they are rarely reported in rice. Recently, A MORF2-like protein, WSP1, was identified as a rice RNA editing/splicing factor (Zhang et al. 2017). The WSP1 mutation affects the RNA editing of ndhG and rpoB, suggesting that WSP1 and OsHMBPP might cooperate together for RNA editing. Therefore, we tested the interaction between OsHMBPP, WSP1 and other six MORF proteins. Y2H assays showed that OsHMBPP and WSP1 did not interact together in vivo (Fig. 9). However, we confirmed that OsHMBPP can interact with Os09g33480 by Y2H and BIFC assays (Fig. 9a-c), but not interact with the other six MORFs in yeasts (Fig. 9a). Our results suggested that OsHMBPP may act as an organellar RNA editing factor via an editosome coupled with Os09g33480.
Conclusions
We isolated a novel seedling-lethal albino rice mutant las1 by the CRISPR/Cas9 system. Mutation in the HMBPP reductase is responsible for the phenotype of las1. Our results demonstrate that LAS1/OsHMBPP is important for plastid gene expression and normal chloroplast development. Also, we found the plastid RNA editing in las1 was altered, and LAS1/OsHMBPP interacted with the MORF protein Os09g33480 in vivo. Further studies on identifying the mutant phenotype of other MEP pathway genes, and how the rice MEP pathway enzymes are regulated.
Methods
Plant Materials and Growth Conditions
The las1 mutant was obtained from the pool of the japonica cultivar Nipponbare by the CRISPR/Cas9 system. The CRISPR/Cas9 binary vectors were constructed as previously described (Lu et al. 2017). The seedlings were grown in a growth chamber or in the field. The growth chamber condition was set as follow:14-h light at 30 °C and 10-h dark at 25 °C.
Measurement of Pigment Contents
Pigments were extracted from fresh leaf samples (0.2 g) from wild-type and las1 plants that were cut into small segments and incubated with 5 ml of 95% ethanol in the dark for 48 h. The supernatant was measured with a dual-beam ultraviolet spectrophotometer (TU1901, Beijing) at 470, 645, and 663 nm. Chl a, Chl b, and carotenoid (Car) contents were measured according to the methods by Porra et al. (1989). Three biological replicates were analyzed.
Transmission Electron Microscopy (TEM)
For TEM analysis, transverse sections of leaf samples were taken from las1 and WT seedlings. Leaf samples were fixed in 2.5% glutaraldehyde and then in 1% OsO4. Samples were prepared as described previously (Gothandam et al. 2005), and were observed using a Jeol 1230 electron microscope (Tokyo, Japan).
Protein Structure and Sequence Alignment Analysis
The predicted full-length LAS1 protein sequence, with 459 amino acids, was obtained from RGAP (http://rice.plantbiology.msu.edu/). The sequences used in the alignment analysis were obtained by a BLASTP search using the LAS1 protein sequence as the query at the National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/). The full-length amino acid sequences were aligned using the DNAMAN.
Measurement of IPP and DMAPP Contents
IPP and DMAPP contents were determined by the specific ELISA Kit from the Sci-tech innovation company (Qingdao, China). Baed on the manufacturer’s instructions, each sample was assayed five times.
Subcellular Localization
To investigate the subcellular localization of LAS1, we amplified the full-length cDNA of LAS1 without the termination codon (Additional file 1: Table S2). The fragment was cloned into the GFP vector pAN580. The LAS1-GFP vector was transformed into rice protoplasts (Wang et al. 2016), and the transformed protoplasts were observed with a Zeiss LSM700 laser scanning confocal microscope.
RNA Extraction and qRT-PCR Analysis
Total RNA was extracted and purified from different tissues using the RNApure Plant Kit (CWBIO) according to the manufacturer’s instructions. First strand cDNAs were synthesized from 2 μg total RNA using a PrimeScript 1st Strand cDNA Synthesis Kit (Takara). qRT-PCR was performed with a CFX96 Touch Real-time PCR Detection System using the SYBR Premix Ex TaqTM kit (Takara). The rice UBQ gene was used as an internal control. All primers for qRT-PCR are obtained as reported previously (Wang et al. 2016; Ge et al. 2017; Liu et al. 2018). The data were expressed as the mean ± SD of three biological replicates.
RNA Editing Analysis
All 21 known rice chloroplast RNA editing sites were assayed as reported (Corneille et al. 2000). Specific cDNA fragments with the editing sites were amplified between wild type and las1, and then sequenced. The cDNA sequences were compared to identify C to T changes resulting from RNA editing by DNAMAN software. All primers for RNA editing analysis are obtained as reported previously (Wang et al. 2017).
Yeast Two-Hybrid and Transient Expression Assays
Full-length coding sequences of LAS1 was cloned into pGBKT7 vector, and seven MORF genes (Os06g02600, WSP1, Os03g38490, Os09g33480, Os09g04670, Os11g11020, Os08g04450) were cloned into pGADT7 vector, resulting in LAS1-BD and MORFs-AD respectively (Additional file 1: Table S2). The assay was performed following the manufacturer’s instructions (Clontech).
For Bimolecular Fluorescent Complimentary assay, OsHMBPP-nYFP, Os09g33480-cYFP and corresponding empty vectors were transformed into Agrobacterium strain EHA105 and transfected leaves of N. benthamiana (Additional file 1: Table S2). The fluorescence was observed with a confocal scanning microscopy (ZEISS).
Availability of Data and Materials
All data supporting the conclusions of this article are provided within the article (and its additional files).
Abbreviations
- MVA:
-
Mevalonate
- MEP:
-
Methyl-D-erythritol-4-phosphate
- MORF:
-
Multiple organellar RNA editing factor
- Chl:
-
Chlorophyll
- rpl2:
-
Ribosomal protein L2
- TEM:
-
Transmission electron microscopy
- qRT-PCR:
-
Quantitative real-time PCR
- atpA:
-
ATP synthase CF1 alpha subunit
- PEP:
-
Plastid-encoded RNA polymerase
- NEP:
-
Nuclear-encoded polymerase
References
Adam P, Hecht S, Eisenreich W, Kaiser J, Grawert T, Arigoni D, Bacher A, Rohdich F (2002) Biosynthesis of terpenes: studies on 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate reductase. Proc Natl Acad Sci U S A 99:12108–12113
Albrecht V, Ingenfel A, Apel K (2008) Snowy cotyledon 2: the identification of a zinc finger domain protein essential for chloroplast development in cotyledons but not in true leaves. Plant Mol Biol 66:599–608
Bentolila S, Heller WP, Sun T, Babina AM, Friso G, Wijk KJ, Hanson MR (2012) RIP1, a Member of an Arabidopsis Protein Family, Interacts With the Protein RARE1 and Broadly Affects RNA Editing. Proc Natl Acad Sci USA 109:1453–1461.
Budziszewski GJ, Lewis SP, Glover LW, Reineke J, Jones G, Ziemnik LS, Lonowski J, Nyfeler B, Aux G, Zhou Q, McElver J, Patton DA, Martienssen R, Grossniklaus U, Ma H, Law M, Levin JZ (2001) Arabidopsis genes essential for seedling viability: isolation of insertional mutants and molecular cloning. Genetics 159:1765–1778
Cao P, Ren Y, Liu X, Zhang T, Zhang P, Xiao L, Zhang F, Liu S, Jiang L, Wan JM (2019) Purine nucleotide biosynthetic gene GARS controls early chloroplast development in rice (Oryza sativa L.). Plant Cell Rep 38:183–194
Chu D, Wei L (2019) The chloroplast and mitochondrial C-to-U RNA editing in Arabidopsis thaliana shows signals of adaptation. Plant Direct 3:e00169
Corneille S, Lutz K, Maliga P (2000) Conservation of RNA editing between rice and maize plastids: are most editing events dispensable? Mol Gen Genet 264:419–424
Cui X, Wang Y, Wu J, Han X, Gu X, Lu T, Zhang Z (2019) The RNA editing factor DUA1 is crucial to chloroplast development at low temperature in rice. New Phytol 221:834–849
Cunningham FX Jr, Lafond TP, Gantt E (2000) Evidence of a role for LytB in the nonmevalonate pathway of isoprenoid biosynthesis. J Bacteriol 182:5841–5848
Disch A, Hemmerlin A, Bach T, Rohmer M (1998) Mevalonate-derived isopentenyl diphosphate is the biosynthetic precursor of ubiquinone prenyl side chain in tobacco BY-2 cells. J Biochem 331:615–621
Eisenreich W, Schwarz M, Cartayrade A, Arigoni D, Zenk MH, Bacher A (1998) The deoxyxylulose phosphate pathway of terpenoid biosynthesis in plants and microorganisms. Chem Biol 5:R221–R233
Ge C, Wang L, Ye W, Wu L, Cui Y, Chen P, Pan J, Zhang D, Hu J, Zeng D, Dong G, Qian Q, Guo L, Xue D (2017) Single-point mutation of an histidine-aspartic domain-containing gene involving in chloroplast ribosome biogenesis leads to white fine stripe leaf in rice. Sci Rep 27:3298
Gothandam KM, Kim ES, Cho H, Chung YY (2005) OsPPR1, a pentatricopeptide repeat protein of rice is essential for the chloroplast biogenesis. Plant Mol Biol 58:421–433
Guevara-Garcia A, San Roman C, Arroyo A, Cortes ME, de la Luz G-NM, Leon P (2005) Characterization of the Arabidopsis clb6 mutant illustrates the importance of posttranscriptional regulation of the methyl-D-erythritol 4-phosphate pathway. Plant Cell 17:628–643
Gutierrez-Nava ML, Gillmor CS, Jimenez LF, Guevara-Garcia A, Leon P (2004) CHLOROPLAST BIOGENESIS genes act cell and noncell autonomously in early chloroplast development. Plant Physiol 135:471–482
Hecht S, Eisenreich W, Adam P, Amslinger S, Kis K, Bacher A, Arigoni D, Rohdich F (2001) Studies on the nonmevalonate pathway to terpenes: the role of the GcpE (IspG) protein. Proc Natl Acad Sci U S A 98:14837–14842
Herz S, Wungsintaweekul J, Schuhr CA, Hecht S, Luttgen H, Sagner S, Fellermeier M, Eisenreich W, Zenk MH, Bacher A, Rohdich F (2000) Biosynthesis of terpenoids: YgbB protein converts 4-diphosphocytidyl-2C-methyl-D-erythritol 2-phosphate to 2C-methyl-D-erythritol 2,4- cyclodiphosphate. Proc Natl Acad Sci U S A 97:2486–2490
Hsieh MH, Goodman HM (2005) The Arabidopsis IspH homolog is involved in the plastid non mevalonate pathway of isoprenoid biosynthesis. Plant Physiol 138:641–653
Jarvis P, López-Juez E (2013) Biogenesis and homeostasis of chloroplasts and other plastids. Nat Rev Mol Cell Biol 14:787–802
Kaminaka H, Morita S, Tokumoto M, Yokoyama H, Masumura T, Tanaka K (1999) Molecular cloning and characterization of a cDNA for an iron-superoxide dismutase in rice (Oryza sativa L.). Biosci Biotechnol Biochem 63:302–308
Kessler F, Schnell D (2009) Chloroplast biogenesis: diversity and regulation of the protein import apparatus. Curr Opin Cell Biol 21:494–500
Leister D (2003) Chloroplast research in the genomic age. Trends Genet 19:47–56
Lichtenthaler HK (1999) The 1-deoxy-D-xylulose-5-phosphate pathway of isoprenoid biosynthesis in plants. Annu Rev Plant Biol 50:47–65
Liu X, Lan J, Huang YS, Cao PH, Zhou CL, Ren YK, He NQ, Liu SJ, Tian YL, Nguyen T, Jiang L, Wan JM (2018) WSL5, a pentatricopeptide repeat protein, is essential for chloroplast biogenesis in rice under cold stress. J Exp Bot 69:3949–3961
Liu X, Rodermel SR, Yu F (2010) A var2 leaf variegation SUPPRESSOR locus, SUPPRESSOR OF VARIEGATION3, encodes a putative chloroplast translation elongation factor that is important for chloroplast development in the cold. BMC Plant Biol 10:287
Lois LM, Campos N, Putra SR, Danielsen K, Rohmer M, Boronat A (1998) Cloning and characterization of a gene from Escherichia coli encoding a transketolase-like enzyme that catalyzes the synthesis of D-1-deoxyxylulose 5-phosphate, a common precursor for isoprenoid, thiamin, and pyridoxol biosynthesis. Proc Natl Acad Sci U S A 95:2105–2110
Lopez-Juez E, Pyke KA (2005) Plastids unleashed: their development and their integration in plant development. Int J Dev Bio 49:557–577
Lu XM, Hu XJ, Zhao YZ, Song WB, Zhang M, Chen ZL, Chen W, Dong YB, Wang ZH, Lai JS (2012) Map-based cloning of zb7 encoding an IPP and DMAPP synthase in the MEP pathway of maize. Mol Plant 5:1100–1112
Lu Y, Ye X, Guo R, Huang J, Wang W, Tang JY, Tan LT, Zhu JK, Chu CC, Qian YW (2017) Genome-wide targeted mutagenesis in rice using the CRISPR/Cas9 system. Mol Plant 10:1242–1245.
McAteer S, Coulson A, McLennan N, Masters M (2001) The lytB gene of Escherichia coli is essential and specifies a product needed for isoprenoid biosynthesis. J Bacteriol 183:7403–7407
Page JE, Hause G, Raschke M, Gao W, Schmidt J, Zenk MH, Kutchan TM (2004) Functional analysis of the final steps of the 1-deoxy-D-xylulose 5-phosphate (DXP) pathway to isoprenoids in plants using virusinduced gene silencing. Plant Physiol 134:1401–1413
Pogson BJ, Albrecht V (2011) Genetic dissection of chloroplast biogenesis and development: an overview. Plant Physiol 155:1545–1551
Porra RJ, Thompson WA, Kriedemann PE (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. BBA-Bioenergetics 975:0–394
Rodriguez-Concepcion M, Boronat A (2002) Elucidation of the methylerythritol phosphate pathway for isoprenoid biosynthesis in bacteria and plastids. A metabolic milestone achieved through genomics. Plant Physiol 130:1079–1089
Rohmer M (2003) Mevalonate-independent methylerythritol phosphate pathway for isoprenoid biosynthesis: elucidation and distribution. Pure Appl Chem 75:375–388
Rohmer M, Knani M, Simonin P, Sutter B, Sahm H (1993) Isoprenoid biosynthesis in bacteria: a novel pathway for the early steps leading to isopentenyl diphosphate. Biochem J 295:517–524
Takenaka M, Zehrmann A, Verbitskiy D, Kugelmann M, Hartel B, Brennicke A (2012) Multiple organellar RNA editing factor (MORF) family proteins are required for RNA editing in mitochondria and plastids of plants. Proc Natl Acad Sci USA 109:5104–5109.
Takenaka M, Zehrmann A, Verbitskiy D, Hartel B, Brennicke A (2013) RNA editing in plants and its evolution. Annu Rev Genet 47:335–352.
Tang JP, Zhang WW, Wen K, Chen GM, Sun J, Tian YL, Tang WJ, Yu J, An HZ, Wu TT, Kong F, Terzaghi W, Wang CM, Wan JM (2017) OsPPR6, a pentatricopeptide repeat protein involved in editing and splicing chloroplast RNA, is required for chloroplast biogenesis in rice. Plant Mol Biol 95:345–357.
Wang P, Grimmm B (2015) Organization of chlorophyll biosynthesis and insertion of chlorophyll into the chlorophyll-binding proteins in chloroplasts. Photosynth Res 12:189–202
Wang Y, Ren Y, Zhou K, Liu L, Wang J, Xu Y, Zhang H, Zhang L, Feng Z, Wang L, Ma W, Wang Y, Guo X, Zhang X, Lei C, Cheng Z, Wan JM (2017) WHITE STRIPE LEAF4 encodes a novel P-type PPR protein required for chloroplast biogenesis during early leaf development. Front Plant Sci 8:1116
Wang Y, Wang C, Zheng M, Lyu J, Xu Y, Li X, Niu M, Long W, Wang D, Wang H, Terzaghi W, Wang Y, Wan JM (2016) WHITE PANICLE1, a val-tRNA synthestase regulating chloroplast ribosome biogenesis in rice, is essential for early chloroplast development. Plant Physiol 170:2110–2123
Yu QB, Huang C, Yang ZN (2014) Nuclear-encoded factors associated with the chloroplast transcription machinery of higher plants. Front Plant Sci 5:316
Zagari N, Sandoval-Ibañez O, Sandal N, Su J, Rodriguez-Concepcion M, Stougaard J, Pribil M, Leister D, Pulido P (2017) SNOWY COTYLEDON 2 promotes chloroplast development and has a role in leaf variegation in both lotus japonicus and Arabidopsis thaliana. Mol Plant 10:721–734
Zeidler J, Lichtenthaler H, May H, Lichtenthaler F (1997) Is isoprene emitted by plants synthesized via the novel isopentenyl pyrophosphate pathway? Z Naturforsch C 52:15–23
Zhang Z, Cui X, Wang Y, Wu J, Gu X, Lu T (2017) The RNA editing factor WSP1 is essential for chloroplast development in rice. Mol Plant 10:86–98
Acknowledgements
The authors would like to thank AiMi (www.aimieditor.com) for providing linguistic assistance.
Funding
This work was supported by National Natural Science Foundation of China (31900388), Natural Science Foundation of Jiangsu Province (BK20191055) and Natural Science Foundation of Colleges of Jiangsu Province (19KJB180009).
Author information
Authors and Affiliations
Contributions
LX designed this research project and performed gene expression analysis. C-PH measured the chlorophyll content and observed the chloroplast structure. T-DD managed all plant materials, did the RNA editing and splicing analysis, and performed yeast two hybrid. H-QQ and Y-YR constructed all the vectors and performed the rice and N. benthamiana transformation, and measured the contents of IPP and DMAPP. LX and C-PH wrote and revised the manuscript. All authors approved the manuscript.
Corresponding author
Ethics declarations
Ethics Approval and Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Figure S1.
The phenotype and sequence confirmation of las1. Figure S2. Expression pattern of OsHMBPP at different growth stages. Data was collected from the Rice eFP Browser. Figure S3. Determination of IPP and DMAPP between WT and las1. Figure S4. The rest 17 chloroplast RNA editing sites between WT and las1. Figure S5. Splicing Analysis of rice chloroplast genes in wild-type and las1. Figure S6. Expression pattern of Os09g33480 at different growth stages. Data was collected from the Rice eFP Browser. Figure S7. Expression patterns of rice nonMVA pathway genes in a 14 h light/ 10 h dark. Wild-type seedlings were grown in a growth chamber for 14 days after germination, and then the seedlings were sampled every 4 h. Error bars indicate SD (n = 3). Figure S8. RNA-edited amino acids alignment of the plastid rpl2, atpA and rpoB in monocot(1–6) and eudicot(7–12) . 1: Zea mays, 2: Oryza sativa, 3: Hordeum vulgare, 4: Sorghum bicolor, 5:Triticum aestivum, 6: Phoenix dactylifera, 7: Arabidopsis thaliana, 8:Brassica napus, 9: Nicotiana tabacum, 10: Glycine max, 11: Vitis vinifera, 12: Gossypium arboreum. Table S1. Chloroplast signal peptide prediction. Table S2. Primers used in real-time PCR and vector construction.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liu, X., Cao, P.H., Huang, Q.Q. et al. Disruption of a Rice Chloroplast-Targeted Gene OsHMBPP Causes a Seedling-Lethal Albino Phenotype. Rice 13, 51 (2020). https://doi.org/10.1186/s12284-020-00408-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12284-020-00408-1