Abstract
People with type 2 diabetes have a tenfold higher prevalence of hypomagnesemia, which is suggested to be caused by low dietary magnesium intake, medication use, and genetics. This study aims to identify the genetic loci that influence serum magnesium concentration in 3466 people with type 2 diabetes. The GWAS models were adjusted for age, sex, eGFR, and HbA1c. Associated traits were identified using publicly available data from GTEx consortium, a human kidney eQTL atlas, and the Open GWAS database. The GWAS identified a genome-wide significant locus in TAF3 (p = 2.9 × 10−9) in people with type 2 diabetes. In skeletal muscle, loci located in TAF3 demonstrate an eQTL link to ATP5F1C, a gene that is involved in the formation of Mg2+-ATP. Serum Mg2+ levels were associated with MUC1/TRIM46 (p = 2.9 × 10−7), SHROOM3 (p = 4.0 × 10−7), and SLC22A7 (p = 1.0 × 10−6) at nominal significance, which is in combination with the eQTL data suggesting that they are possible candidates for renal failure. Several genetic loci were in agreement with previous genomic studies which identified MUC1/TRIM46 (Pmeta = 6.9 × 10−29, PQ = 0.81) and SHROOM3 (Pmeta = 2.9 × 10−27, PQ = 0.04) to be associated with serum Mg2+ in the general population. In conclusion, serum magnesium concentrations are associated with genetic variability around the regions of TAF3, MUC1/TRIM46, SHROOM3, and SLC22A7 in type 2 diabetes.
Similar content being viewed by others
Introduction
Magnesium (Mg2+) is an essential ion that is involved in more than 600 enzymatic reactions, including DNA synthesis and energy metabolism [1]. Consequently, Mg2+ levels are tightly regulated by the interplay of kidney, intestine, bone, and soft tissues [1]. The serum concentration of Mg2+ is partly explained by genetic factors with a heritability estimate ranging from 15 to 39% [2]. GWAS performed in the healthy population identified genetic variants in MUC1, TRPM6, and SHROOM3 loci to be associated with serum Mg2+ concentration [3,4,5].
The prevalence of hypomagnesemia is between 10 and 45%, which is 10 times higher in people with type 2 diabetes mellitus compared to the healthy population [6,7,8]. Serum Mg2+ is inversely associated with HbA1c in people with type 2 diabetes. The inverse association of serum Mg2+ with insulin levels is found in people with and without diabetes [7, 9, 10]. Since hypomagnesemia is associated with poor glycemic control and insulin resistance, it leads to a higher incidence of type 2 diabetes and worsens existing type 2 diabetes [11, 12]. This mechanism may explain why genetic variants in Mg2+-related ion channels have been linked to diabetes risk [13]. For example, genetic variants in TRPM6 are associated with the development of gestational diabetes and type 2 diabetes [14, 15] and SNPs in CNNM2, SLC41A2, and TRPM6 were associated with the risk of diabetes through serum Mg2+ levels [11].
Several causes for hypomagnesemia have been described in people with type 2 diabetes including low dietary Mg2+ intake, dyslipidemia, medication use, and genetics. [3, 7, 11, 14] Given the multifactorial pathophysiological causes of type 2 diabetes, including insulin resistance, low-grade chronic inflammation, and dyslipidemia, additional genetic factors may contribute to hypomagnesemia in type 2 diabetes [16]. People with type 2 diabetes often have hypermagnesiuria, which is a fractional excretion of Mg2+ (FEMg) above 4%, which is suggested to be caused by reduced Mg2+-channel TRPM6 activity [14, 15, 17, 18]. Hypermagnesiuria in the presence of hypomagnesemia suggests that urinary Mg2+ wasting resulting from reduced reabsorption in the kidneys is a major cause of hypomagnesemia in people with type 2 diabetes [6, 19]. However, it is unknown whether genetic variants in people with type 2 diabetes contribute to urinary Mg2+ wasting. Gene variants that determine serum Mg2+ levels in people with type 2 diabetes have never been systematically studied. The genetic variation of serum Mg2+ in type 2 diabetes compared to the healthy population can differ and help prevent the development of associated adverse clinical outcomes, such as heart failure and microvascular disease [20]. Therefore, we aimed to determine the genetic variations in serum Mg2+ specifically for people with type 2 diabetes.
Materials and methods
Study population
The Hoorn DCS is a prospective observational study of individuals with diabetes defined as (1) at least one symptom of excessive thirst, polyuria, weight loss, hunger, or pruritus combined with fasting plasma glucose ≥ 7.0 mmol/L or random plasma glucose ≥ 11.1 mmol/L, or (2) two elevated plasma glucose concentrations on two different occasions in the absence of symptoms. The baseline biobanking was done in the years 2008–2009 and 2012–2014. Further details of this cohort are published by van der Heijden et al. [21].
Ethical approval was obtained from the VU University Medical Center Ethical Review Committee (09/07/2009, ref: NL27783.029.09), Amsterdam. All participants gave written informed consent before participation in the Hoorn DCS biobank.
Serum magnesium measurement
Serum samples of 4445 people were measured for Mg2+, of which 45 were stored/collected in EDTA tubes or had insufficient volumes and were therefore excluded. Furthermore, 52 people were excluded due to a different type of diabetes. From the 4348 participants that were left, 882 participants were excluded because GWAS data was not available. Mg2+ was measured in 2019 at the Laboratory Medicine Department (Radboud University Medical Center) using a calibrated standardized colorimetric assay with a coefficient of variation of 1.98% (Cobas C8000; Roche Diagnostics, Risch-Rotkreuz, Switzerland). Additional information is published elsewhere by Oost et al. [20]. Missing data was for all variables < 5%.
Statistical methods for genome-wide association study
Genotypes were determined with the Illumina Human Core Exome array. Principal component analysis was performed with plink. Genotypes were imputed using the Michigan Imputation Server based on the Haplotype Reference Consortium (HRC) panel.
GWAS on serum Mg2+ concentration was performed using rvtest (version 20170613) on an additive model on 39127679 SNPs. Principal components (PCs) were included to adjust for the potential effects of population stratification. SNPs with an allele frequency below 0.05 or with low imputation confidence (info score < 0.3) were filtered out. After filtering, 5407047 remained in the final set. A p value below 5 × 10−8 was considered genome-wide significant. Three models were explored to evaluate the association between genetics and serum Mg2+: base model adjusted for age, sex, and the first three PCs (1–3), model 1 additionally adjusted for eGFR, and model 2 additionally adjusted for eGFR and HbA1c. Models 1 and 2 were adjusted for eGFR combined with HbA1c because eGFR is a measure of kidney function and HbA1c is a measure of the severity of type 2 diabetes. Additionally, serum Mg2+. Hence, the variability of glycemic genetic factors may cause fluctuations in serum Mg2+.
After identification of the lead SNPs, expression quantitative trait loci (eQTL) and associated traits were assessed using publicly available data from the genotype-tissue expression (GTEx) consortium (v8), the human kidney eQTL atlas from the Susztak lab (meQTL_S443-eQTM_S414) and the Open GWAS database (v7.5.12) [22,23,24]. For comparison to previous GWAS, we downloaded the identified top hits identified by Meyer et al. [3]. Loci were meta-analyzed using GWAMA (v2.1) based on the current data and Meyer et al. Meta-analyzed results and Cochran’s Q test were obtained for overlapping loci. For ATP2B1 we used a proxy SNP because the original SNP was not available in our own dataset.
A power calculation was performed using the R package genpwr with the following settings: N = 3466, sd_y = 0.0789, MAF = 0.05, 0.10, effect size = 0.005, 0.01, 0.015, 0.02, alpha = 0.05, additive model.
Results
In 3466 participants, the mean serum Mg2+ was 0.80 ± 0.08 mmol/L and 330 individuals (9.5%) showed hypomagnesemia (serum Mg2+ < 0.7 mmol/L). All of these people were of European descent. Other participant characteristics are reported in Table 1.
Magnesium GWAS
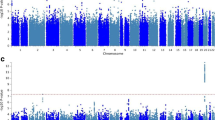
Figure 1 shows the Manhattan plot for associations between SNPs and serum Mg2+ concentration after adjustment for age, sex, and PC 1–3. The Q-Q plots of the observed versus expected p value distributions for associations between the SNPs and Mg2+ are provided in Supplementary Figure S1. The lowest p value within the region (lead SNP) that reached genome-wide significance (p < 5 × 10−8) was located in the TAF3 gene (rs7894336, chromosome (chr)10, p = 2.86 × 10−9). Other lead SNPs with nominal significance (equal to or below p < 10−6) were located near MUC1/TRIM46 (rs11264341, chr1, p = 2.9 × 10−7), SHROOM3 (rs10019833, chr4, p = 4.0 × 10−7), and SLC22A7 (rs2270860, chr6, p = 1.0 × 10−6). Regional association plots (LocusZoom) for chr1, 4, 6, and 10 are shown in Fig. 2A–D.
Genome-wide –log10(p-value) plots from association analyses with serum Mg2+ concentration in 3466 people with type 2 diabetes in the Hoorn, DCS study. Adjusted for age, sex, and PC1-3. One locus (TAF3, rs7894336) reached genome-wide significance (P < 5 × 10−8), indicated by the red horizontal line. Three loci (TRIM46, rs11264341; SHROOM3, rs10019833, and SLC22A7, rs2270860) reached nominal significance (P < 10−6), indicated by the blue horizontal line. DCS=Diabetes Care System, Mg2+ = magnesium, PC = principal component, T2D = type 2 diabetes
Regional locus association plot showing −log10 (P values) for the serum Mg2+-associated regions near (A) TRIM46 and MUC1 gene on chr1, (B) SHROOM3 and CCDC158 gene on chr4, (C) SLC22A7 and ZNF318 gene on chr6, and (D) TAF3 gene on chr10. Top panel shows the P value (−log10) for each SNP. The colors of the points indicate the linkage disequilibrium with the top signal indicated by the diamond. The blue line represents the recombination rate. The middle panel shows the location of the SNP indicated by a blue line and the lower panel shows the genes in the region. Chr = chromosome, Mg2+ = magnesium
Adjusting the serum Mg2+-SNP associations for eGFR did not change the association with serum Mg2+ levels (Supplementary Figure S2A and Supplementary Table S1). Adjusting the Mg2+-SNP associations for eGFR and HbA1c, demonstrated a stronger significance only at loci SLC22A7 (rs2270860, chr6) from p = 1.3 × 10−6 to p = 5.6 × 10−8 (Supplementary Figure S2B and Supplementary Table S1). Loci identified in a previous GWAS did show similar effect sizes. Although they were not significant in DCS, six out of the nine loci were significant when meta-analyzed (Supplementary Table S2). Heterogeneity between cohorts was observed for two loci (SHROOM3 and TRPM6, Q_P-value = 0.035~0.043), but this was based on differences in the magnitude of effect and not the direction of effect.
eQTL studies using GTEx data
The lead genetic variant rs7894336 (chr10, TAF3) had no expression quantitative trait loci (eQTL) associations. SNPs in or near the TAF3 locus were associated with ATP5F1C expression in skeletal muscle (rs10795574, chr10, p = 9.4 × 10−7) (Supplementary Table S3). On chromosome 10, ATP5F1C is located near TAF3 (Fig. 2D). ATP5F1C encodes the gamma subunit of the catalytic core F1 of the mitochondrial ATP synthase and creates a proton gradient for ATP synthesis [25]. Mg2+ is known to bind to the catalytic F1 unit to produce the high-energy terminal bond of ATP (Mg2+-ATP), which is essential in glycolysis [1, 26].
rs11264341 (chr1 near MUC1/TRIM46) had significant associations with GBAP1 expression in multiple tissues, including artery tibial (p = 2.1 × 10−30), artery aorta (p = 1.76 × 10−25), heart left ventricle (p = 7.0 × 10−25) and lung (p = 2.7 × 10−20). Other associations of rs11264341 with MUC1 and THBS3 were most significant in esophagus mucosa tissue (p = 1.8 × 10−21) and whole blood (p = 7.6 × 10−20), respectively. rs10019833 (chr4, SHROOM3) had significant associations with FAM47E in thyroid tissue (p = 2.1 × 10−27), STBD1 in nerve tibial tissue (p = 5.8 × 10−21) and CCDC158 in artery tibial tissue (p = 3.1 × 10−18). rs2270860 (chr6, SLC22A7) had significant associations with SLC22A7 in testis (p = 2.5 × 10−17), ZNF318 in adrenal gland tissue (p = 1.6 × 10−9) and CRIP3 in left ventricle heart tissue (p = 3.0 × 10−7). An overview of the information on the lead SNPs within each chromosomal region is presented in Table 2 and eQTL links from the genotype-tissue expression (GTEx) consortium in Table 3.
eQTL and DNA methylation studies using the human kidney eQTL atlas from the Susztaklab and the Open GWAS database
Since serum Mg2+ concentration is primarily controlled by the kidney, significant genetic variability was mapped to human traits in kidney tissue using the human kidney eQTL atlas from the Susztak lab [24]. In kidney tissue, no eQTLs associations of rs7894336 (chr10, TAF3) were shown. The three lead SNPs with nominal significance (all p ≤ 10−6) did show significant cis associations in human kidney tissue. rs11264341 (chr1 near MUC1/TRIM46) had a significant association with MUC1 (rs11264341, chr1, p = 1.7 × 10−13), rs10019833 (chr4, SHROOM3) had a significant association with STBD1 (chr4, p = 5.5 × 10−7) and rs2270860 (chr6, SLC22A7) had a significant association with CRIP3 (rs2270860, chr6, p = 1.1 × 10−5) in human kidney tissue. The human kidney methylation quantitative trait loci atlas of the Susztak-lab showed associations of nominal significant lead SNPs with EFNA3, MUC1, STBD1, SHROOM3, POLH, and SLC22A7 (Supplementary Table S4). The Open GWAS database showed the lead genetic variant rs7894336 (chr10, TAF3) association with phosphate. The nominal significant lead SNP rs11264341 (chr near MUC1/TRIM46) was associated with urea, while rs10019833 (chr4, SHROOM3) and rs2270860 (chr6, SLC22A7) were associated with cystatin C (Supplementary Table S5) [23].
Discussion
We report the first GWAS of serum Mg2+ in 3466 individuals with type 2 diabetes. We did have 80% power to identify variants with a MAF of 5% and an effect of 0.012 and 80% power to identify variants with a MAF of 10% and an effect of 0.009. The association of the TAF3 locus with the serum Mg2+ concentration reached genome-wide significance in the base model and the eGFR- and eGFR/HbA1c-adjusted models. MUC1/TRIM46, SHROOM3, and SLC22A7 loci were associated with serum Mg2+ concentration at nominal significance. We linked serum Mg2+-associated SNPs with MUC1, STBD1, and CRIP3 in kidney tissue. The lead SNP in the TAF3 region was associated with ATP5F1C in skeletal muscle.
TAF3 is a general transcription factor involved in histone modification and gene expression of the tumor suppressor p53 [29]. Genetic variability in TAF3 was linked to ATP5F1C expression in muscle. ATP5F1C encodes the gamma-subunit of the mitochondrial ATP Synthase, which is essential in the formation of Mg2+-ATP. In skeletal muscle, we also identified an eQTL for RPL7L1, which is predicted to enable RNA binding activity and structural constituent of the ribosome. This suggests that genetic variability in serum Mg2+ may be related to genes involved in transcription, translation, and ATP synthesis. Interestingly, genetic variability in GATA3, a region located near TAF3, was associated with serum calcium (Ca2+) levels in the general population (5). However, we did not find a genetic link to GATA3 based on eQTL data.
In this study, we also found nominal significant genetic variability in MUC1/TRIM46, of which the lead SNP (rs11264341, chr1) was negatively associated with serum uric acid levels in a previously published GWAS [30]. Interestingly, increased Mg2+ intake is associated with a decreased risk of hyperuricemia [31]. Furthermore, the accumulation of uric acid can lead to kidney stone formation, oxidative stress, insulin resistance, and increased type 2 diabetes incidences [32]. Although the same lead SNP (rs11264341) in MUC1/TRIM46 did not provide evidence of a causal relationship between serum uric acid and incident diabetes, in people without a history of diabetes [33]. In our study, there was an eQTL link of the lead SNP with GBAP1 in multiple tissues and with MUC1 in kidney tissue. The association of serum Mg2+ with the MUC1/TRIM46 locus is also found in all previously published GWAS on serum Mg2+ concentration performed in the general population [3,4,5]. MUC1 is a membrane-bound glycosylated phosphoprotein that is attached to the apical surface of epithelial cells in the intestinal tract and plays a critical in mucosal defense by preventing the binding of pathogens [34].
Genetic variability in SHROOM3 (chr4) is associated with serum Mg2+ levels in GWAS based on the general population [3, 5]. Furthermore, intronic SHROOM3 genetic variants in multiple GWAS have been associated with chronic kidney disease (CKD) and kidney function markers: eGFR and creatinine levels [35, 36]. In kidney tissue, the lead SNP is linked to STBD1 expression. STBD1 encodes starch binding domain 1, which plays an important role in the transport of glycogen to lysosomes [37]. STBD1 is identified in a GWAS meta-analysis that prioritized target genes for kidney diseases [24]. Animal studies have shown that disruption of SHROOM3 causes podocyte effacement and impairment of the glomerular filtration barrier [38], demonstrating that this genetic loci is important for kidney health. On chromosome 6, we identified genetic variability in SLC22A7 in the association with serum Mg2+. Still, a lot is unknown about the function of SLC22A7, but previous studies report that it is involved in the transport of cyclic nucleotide cGMP, renal excretion, and possibly creatine reabsorption in renal proximal tubular cells [39, 40]. The cGMP signaling pathway does induce Mg2+ release [41], and creatinine clearance is an important measure of renal function.
According to the Open GWAS database, all genetic variants identified in this study were associated with markers of kidney function; like phosphate, urea, and cystatin C [23]. The kidney plays a role in the volume and mineral balance and therefore is an essential regulator of the serum Mg2+ concentration [42]. Nevertheless, we exclude kidney function as an explanation for the association between the SNPs and serum Mg2+, since adjusting the serum Mg2+-SNP associations for eGFR did not change the association.
Previous studies have identified serum Mg2+-associated genetic variants (rs1114413 or rs113607577 or rs3824347, chr9) near the TRPM6 gene, but in the current study, we did not replicate this finding [3, 4, 43]. A possible explanation is that our cohort consists mainly of insulin-resistant individuals. In healthy people, insulin would regulate TRPM6 channel activity [15], but this may be impaired in diabetes. Based on the eQTL associations and Open GWAS database, all genetic variants are linked to disturbance in Mg2+ absorption and markers of renal failure. Interestingly, our eGFR-adjusted analyses did not alter the associations of the identified genetic variations with serum Mg2+. However, a large majority of the cohort (approximately 25%) already had a low eGFR (< 60 mL/min/1.73 m2) [20], suggesting that adjustment for eGFR does not have pronounced effects on the association of these genetic variants with serum Mg2+. This would suggest that the genetic variants identified are associated with markers of renal failure, which may reduce Mg2+ absorption in the kidneys.
This study has several strengths and limitations. One limitation of this study is that all eQTL and mQTL associations are obtained from databases that used tissue samples from the general population. There is a possibility that these associations are not present in tissue samples from people with type 2 diabetes. A second limitation is that 3,466 participants are a relatively small population for a GWAS. Despite the relatively small population size, many associations were below nominal significance p < 10−6, suggesting that the current study has sufficient power to find stronger signals. The population size was unfortunately too small to perform a causal analysis (i.e., Mendelian randomization analysis) to assess the relationship between serum Mg2+ and health complications of type 2 diabetes. Larger type 2 diabetes GWAS studies that measured serum Mg2+ are warranted to confirm our results and to perform causal analysis, however, to the best of our knowledge these are not available yet.
In this study, we have discovered novel loci that are associated with serum Mg2+ in people with type 2 diabetes. Our results suggest that genetic variation in or near TAF3, MUC1/TRIM46, SHROOM3, and SLC22A7 are associated with the regulation of serum Mg2+ concentrations which may be partially explained by renal function, in type 2 diabetes. Genetic variation may explain why certain individuals with type 2 diabetes are at risk of developing hypomagnesemia.
Availability of data and materials
The data supporting the conclusions of this research are reported within the article and its supplementary files, including the summary statistics files. Individual data cannot be made public due to privacy issues.
References
de Baaij JHF, Hoenderop JGJ, Bindels RJM. Magnesium in man: implications for health and disease. Physiol Rev. 2015;95(1):1–46.
Hunter DJ, De Lange M, Snieder H, MacGregor AJ, Swaminathan R, Thakker RV, Spector TD. Genetic contribution to renal function and electrolyte balance: a twin study. Clin Sci. 2002;103(3):259–65.
Meyer TE, Verwoert GC, Hwang SJ, Glazer NL, Smith AV. Genome-wide association studies of serum magnesium, potassium, and sodium concentrations identify six loci influencing serum magnesium levels. PLoS Genetics. 2010;6(8):1–11.
Tin A, Köttgen A, Folsom AR, Maruthur NM, Tajuddin SM, Nalls MA, Evans MK, Zonderman AB, Friedrich CA, Boerwinkle E, et al. Genetic loci for serum magnesium among African-Americans and gene-environment interaction at MUC1 and TRPM6 in European-Americans: The Atherosclerosis Risk in Communities (ARIC) study. BMC Genetics. 2015;16(1):1–9.
Chang X, Li J, Guo Y, Wei Z, Mentch FD, Hou C, Zhao Y, Qiu H, Kim C, Sleiman PMA, et al. Genome-wide association study of serum minerals levels in children of different ethnic background. PLoS One. 2015;10(4):8–15.
Adiwinoto RP, Adiwinoto RD, Prajitno JH. Magnesium deficiency associated with diabetic retinopathy in type 2 diabetes mellitus: a meta-analysis. Int J Public Health Sci. 2021;10(3):565–73.
Kurstjens S, de Baaij JHF, Bouras H, Bindels RJM, Tack CJJ, Hoenderop JGJ. Determinants of hypomagnesemia in patients with type 2 diabetes mellitus. Eur J Endocrinol. 2017;176(1):11–9.
Ahmed F, Mohammed A. Magnesium: the forgotten electrolyte—a review on hypomagnesemia. Med Sci. 2019;7(4):56–56.
Humphries S, Kushner H, Falkner B. Low dietary magnesium is associated with insulin resistance in a sample of young, nondiabetic black Americans. Am J Hypertens. 1999;12(8 I):747–56.
Reddy SB, Pavuluri P, Chandan ST, Shetty R. Correlation between magnesium levels and HbA1C in controlled and uncontrolled type 2 diabetes. Int J Contemp Med Res. 2018;5(11):11–4.
Kieboom BCT, Ligthart S, Dehghan A, Kurstjens S, de Baaij JHF, Franco OH, Hofman A, Zietse R, Stricker BH, Hoorn EJ. Serum magnesium and the risk of prediabetes: a population-based cohort study. Diabetologia. 2017;60(5):843–53.
Li W, Jiao Y, Wang L, Wang S, Hao L, Wang Z, Wang H, Zhang B, Ding G, Jiang H. Association of Serum Magnesium with Insulin Resistance and Type 2 Diabetes among Adults in China. Nutrients. 2022;14(9):1–11.
Chan KHK, Chacko SA, Song Y, Cho M, Eaton CB, Wu WCH, Liu S. Genetic variations in magnesium-related ion channels may affect diabetes risk among African American and Hispanic American women. J Nutr. 2015;145(3):418–24.
Song Y, Hsu YH, Niu T, Manson JAE, Buring JE, Liu S. Common genetic variants of the ion channel transient receptor potential membrane melastatin 6 and 7 (TRPM6 and TRPM7), magnesium intake, and risk of type 2 diabetes in women. BMC Med Genet. 2009;10(4):1–12.
Nair AV, Hocherb B, Verkaart S, Van Zeeland F, Pfab T, Slowinski T, Chen YP, Schlingmann KP, Schaller A, Gallati S, et al. Loss of insulin-induced activation of TRPM6 magnesium channels results in impaired glucose tolerance during pregnancy. Proc Natl Acad Sci U S A. 2012;109(28):11324–9.
Smushkin G, Vella A. What is type 2 diabetes? Medicine. 2010;38(11):597–601.
Lee CT, Lien YHH, Lai LW, Chen JB, Lin CR, Chen HC. Increased renal calcium and magnesium transporter abundance in streptozotocin-induced diabetes mellitus. Kidney Int. 2006;69(10):1786–91.
Kurstjens S, Bouras H, Overmars-Bos C, Kebieche M, Bindels RJM, Hoenderop JGJ, de Baaij JHF. Diabetes-induced hypomagnesemia is not modulated by metformin treatment in mice. Sci Rep. 2019;9:1770–1770.
Takayanagi K, Shimizu T, Tayama Y, Ikari A, Anzai N, Iwashita T, Asakura J, Hayashi K, Mitarai T, Hasegawa H. Downregulation of transient receptor potential M6 channels as a cause of hypermagnesiuric hypomagnesemia in obese type 2 diabetic rats. Am J Physiol Renal Physiol. 2015;308(12):F1386–97.
Oost LJ, van der Heijden AAWA, Vermeulen EA, Bos C, Elders PJM, Slieker RC, Kurstjens S, van Berkel M, Hoenderop JGJ, Tack CJ, et al. Serum magnesium is inversely associated with heart failure, atrial fibrillation, and microvascular complications in type 2 diabetes. Diabetes care. 2021;44(8):1–9.
Van Der Heijden AAWA, Rauh SP, Dekker JM, Beulens JW, Elders P, T’Hart LM, Rutters F, Van Leeuwen N, Nijpels G. The Hoorn Diabetes Care System (DCS) cohort. A prospective cohort of persons with type 2 diabetes treated in primary care in the Netherlands. BMJ Open. 2017;7(5):1–8.
Consortium GT. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369(6509):1318–30.
Elsworth B, Lyon M, Alexander T, Liu Y, Matthews P, Hallett J, Bates P, Palmer T, Haberland V, Smith GD et al: The MRC IEU OpenGWAS data infrastructure. BioRxiv. 2020. https://doi.org/10.1101/2020.08.10.244293.
Liu H, Doke T, Guo D, Sheng X, Ma Z, Park J, My TH, Nadkarni GN, Abedini A, Miao Z, et al. Epigenomic and transcriptomic analyses define core cell types, genes and targetabale mechanisms for kidney disease. Nat Genet. 2022;54(7):950–62.
Jonckheere AI, Smeitink JAM, Rodenburg RJT. Mitochondrial ATP synthase: Architecture, function and pathology. J Inherit Metab Dis. 2012;35(2):211–25.
Chen C, Saxena AK, Simcoke WN, Garboczi DN, Pedersen PL, Ko YH. Mitochondrial ATP synthase: Crystal structure of the catalytic F 1 unit in a vanadate-induced transition-like state and implications for mechanism. J Biol Chem. 2006;281(19):13777–83.
Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581(7809):434–43.
Ibrahim SM, Mix E, Bo T, Koczan D, Gold R, Rolfs A. Gene expression profiling of the nervous system in murine experimental autoimmune encephalomyelitis. Brain. 2001;124:1927–38.
Laubert SM, Takahiro N, Wu X, Ferris A, Tang Z, Hughes SH, Roeder RG. H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation. Cell. 2013;152(5):1021–36.
Köttgen A, Albrecht E, Teumer A, Vitart V, Krumsiek J, Hundertmark C, Pistis G, Ruggiero D, O’Seaghdha CM, Haller T, et al. Genome-wide association analyses identify 18 new loci associated with serum urate concentrations. Nat Genet. 2013;45(2):145–54.
Zhang Y, Qiu H. Dietary magnesium intake and hyperuricemia among us adults. Nutrients. 2018;10(3):1–12.
Xiong Q, Liu J, Xu Y. Effects of uric acid on diabetes mellitus and its chronic complications. Int J Endocrinol. 2019;2019:9691345.
Keerman M, Yang F, Hu H, Wang J, Wang F, Li Z, Yuan J, Yao P, Zhang X, Guo H, et al. Mendelian randomization study of serum uric acid levels and diabetes risk: Evidence from the Dongfeng-Tongji cohort. BMJ Open Diabetes Res Care. 2020;8(1):1–10.
McAuley JL, Linden SK, Chin WP, King RM, Pennington HL, Gendler SJ, Florin TH, Hill GR, Korolik V, McGuckin MA. MUC1 cell surface mucin is a critical element of the mucosal barrier to infection. J Clin Investig. 2007;117(8):2313–24.
Köttgen A, Glazer NL, Dehghan A, Hwang S-J, Katz R, Li M, Yang Q, Gudnason V, Launer LJ, Harris TB, et al. Multiple novel loci are associated with indices of renal function and chronic kidney disease. Nat Genet. 2009;41(6):712–7.
Gorski M, Li M, Hoffmann MM, Huang C. Association of eGFR-related loci identified by GWAS with incident CKD and ESRD. Jacques S Beckmann. 2011;7(9):1–8.
Sixin J, Wells CD, Roach PJ. Starch-binding domain-containing protein 1 (Stbd1) and glycogen metabolism: identification of the Atg8 family interacting motif (AIM) in Stbd1 required for interaction with GABARAPL1. Biochem Biophys Res Commun. 2011;413(3):420–5.
Yeo NC, O’Meara CC, Bonomo JA, Veth KN, Tomar R, Flister MJ, Drummond IA, Bowden DW, Freedman BI, Lazar J, et al. Shroom3 contributes to the maintenance of the glomerular filtration barrier integrity. Genome Res. 2015;25(1):57–65.
Cheryl DC, Takafumi K, Shima JE, Urban TJ, Yee SW, More SS, Giacomin KM. Organic anion transporter 2 (SLC22A7) is a facilitative transporter of cGMP. Mol Pharmacol. 2008;73(4):1151–8.
Shen H, Liu T, Morse BL, Zhao Y, Zhang Y, Qiu X, Chen C, Lewin AC, Wang XT, Liu G, et al. Characterization of organic anion transporter 2 (SLC22A7): a highly efficient transporter for creatinine and species-dependent renal tubular expression. Drug Metab Dispos. 2015;43(7):984–93.
Yamanaka R, Shindo Y, Hotta K, Suzuki K, Oka K. NO/cGMP/PKG signaling pathway induces magnesium release mediated by mitoKATP channel opening in rat hippocampal neurons. FEBS Lett. 2013;587(16):2643–8.
Blaine J, Chonchol M, Levi M. Renal control of calcium, phosphate, and magnesium homeostasis. Clin J Am Soc Nephrol. 2015;10(7):1257–72.
Corre T, Arjona FJ, Hayward C, Youhanna S, de Baaij JHF, Belge H, Nägele N, Debaix H, Blanchard MG, Traglia M, et al. Genome-wide meta-analysis unravels interactions between magnesium homeostasis and metabolic phenotypes. J Am Soc Nephrol. 2018;29(1):335–48.
Acknowledgements
We would like to thank the participants and the research staff of the Diabetes Care System West-Friesland.
Funding
This work was supported by the Dutch Diabetes Research Foundation [2017-81-014] received by J.H.F.d.B. Furthermore, the GWAS in the DCS cohort was supported by grants received by L.M.H. from the Foundation for the National Institutes of Health Accelerating Medicines Partnership [HART17AMP] and the Innovative Medicines Initiative Joint Undertaking under grant agreement [115317], resources of which are composed of financial contribution from the European Union’s Seventh Framework Programme [FP7/2007-2013] and European Federation of Pharmaceutical Industries (EFPIA) companies’ in-kind contribution. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
L.J.O., R.C.S. and L.M.H. performed the data interpretation. L.J.O., R.C.S., L.M.H. and J.H.F.d.B contributed to study design. L.J.O., R.C.S, L.M.H and J.H.F.d.B wrote the manuscript. L.J.O. analyzed the serum Mg2+ concentrations. R.C.S., L.M.H, M.T.B. and J.W.J.B. contributed to cohort data collection. J.G.J.H. and J.H.F.d.B. supervised the study. All authors reviewed and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was performed in line with the principles of the Declaration of Helsinki. Ethical approval was obtained from the VU University Medical Center Ethical Review Committee (09/07/2009, ref: NL27783.029.09), Amsterdam. All participants gave written informed consent before participation in the Hoorn DCS biobank.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Supplementary Figure S1.
Q-Q plots showing the distribution of observed versus expected −log10(pvalues) for Mg2+ in (A) base model - corrected for age, sex and PC1-3 - genomic inflation factor 1.003277, (B) model 1- corrected for age, sex, eGFR, PC1-3 - genomic inflation factor 1.003411, and (C) model 2 - corrected for age, sex, eGFR, HbA1c, PC1-3 - genomic inflation factor 1.005874, in the Hoorn DCS cohort. Supplementary Figure S2. Genome-wide –log10(p-value) plots from association analyses with serum Mg2+ concentration in 3,466 people with type 2 diabetes in the Hoorn, DCS study. (A) Adjusted for age, sex, PC 1-3 and eGFR and (B) Adjusted for age, sex, PC 1-3, eGFR and HbA1c. Genome-wide significance (P<5×10−8) is indicated by the red horizontal line. The blue line presents significance p<10−6. DCS=Diabtetes Care Ssytem, eGFR=estimated glomerular filtration rate, HbAtc=hemoglobin Atc, PC=principal component. Supplementary Table S1. Adjusted models of associations between serum Mg2+ concentrations and the lead regional genome-wide significant SNPs. Supplementary Table S2. Loci that display similar effect sizes and an identical direction of the effect on serum Mg2+ levels in people with type 2 diabetes (DCS cohort) and in a previous study focused on the general population. Supplementary Table S3. Loci associated with genetic variability according to GTEx consortium in skeletal muscle tissue. Supplementary Table S4. Loci associated with genetic variability according to human kidney meQTL and eQTM association analyses. Supplementary Table S5. Traits associated with genetic variability according to the Open GWAS database
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Oost, L.J., Slieker, R.C., Blom, M.T. et al. Genome-wide association study of serum magnesium in type 2 diabetes. Genes Nutr 19, 2 (2024). https://doi.org/10.1186/s12263-024-00738-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12263-024-00738-5