Abstract
The suboptimal efficacies of existing anti-malarial drugs attributed to the emergence of drug resistance dampen the clinical outcomes. Hence, there is a need for developing novel drug and drug targets. Recently silver nanoparticles (AgNPs) constructed with the leaf extracts of Euphorbia cotinifolia were shown to possess antimalarial activity. Therefore, the synthesized AgNPs from Euphorbia cotinifolia (EcAgNPs) were tested for their parasite clearance activity. We determined the antimalarial activity in the asexual blood stage infection of 3D7 (laboratory strain) P. falciparum. EcAgNPs demonstrated the significant inhibition of parasite growth (EC50 of 0.75 µg/ml) in the routine in vitro culture of P. falciparum. The synthesized silver nanoparticles were seen to induce apoptosis in P. falciparum through increased reactive oxygen species (ROS) ROS production and activated programmed cell death pathways characterized by the caspase-3 and calpain activity. Also, altered transcriptional regulation of Bax/Bcl-2 ratio indicated the enhanced apoptosis. Moreover, inhibited expression of PfLPL-1 by EcAgNPs is suggestive of the dysregulated host fatty acid flux via parasite lipid storage. Overall, our findings suggest that EcAgNPs are a non-toxic and targeted antimalarial treatment, and could be a promising therapeutic approach for clearing malaria infection.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
P. falciparum, a highly virulent member of the Plasmodia family, is the causative agent of severe malaria infection, resulting in higher mortality and morbidity worldwide [1]. The severity of malaria infection is influenced by the interaction between the pathogen and the immune status of host [2]. The prevalence rate of malaria infection is particularly higher in socioeconomically challenged regions, wherein the optimal conditions for mosquito vectors facilitate the transmission of the disease. Also, the concurrent threat of malaria and COVID-19 co-infection has further exacerbated the public health challenges in malaria infection sensitive regions [3]. The complex life cycle of malaria parasite, spawning both mosquito vector and human host, complicates the understanding of its pathophysiology and biology [4]. Clinical manifestations of malaria are the results of the complex biological processes orchestrated by P. falciparum within the human host [5].
The emergence of resistance to almost all the anti-malarials poses challenge to treat P. falciparum infection, and absence of effective malaria vaccine worsens the situation [6,7,8,9,10]. Chemically synthesized metal based nanoparticles could be a viable therapeutic option for treating malaria infection and circumventing the drug resistance [11,12,13,14,15]. These nanoscale drug carriers deliver antimalarial drugs [16,17,18,19] and candidate antigens [20, 21] in a target specific and sustained manner, and exposed parasite with drugs for longer durations. This increased the possibility of parasite clearance and minimizes the evolution of drug resistance. Our group and others have been exploring the delivery potential of novel drug delivery systems (NDDS) in drug [22,23,24,25] and vaccine therapeutics [20, 21, 26, 27].
Euphorbia cotinifolia, known for its evergreen leaves and therapeutic properties, has been explored for its potential in treating respiratory ailments, asthma, bronchitis, and viral infections as well as rodent malaria infection [28, 29]. The leaf extract of E. cotinifolia has demonstrated regulatory effects on hay fever, tumor, throat spasms, and mucosal accumulation in the nasal and pharyngeal passages [30]. The presence of active ingredients such as flavonoids, quercetin and empfero in E. cotinifolia confer the antimicrobial, antiviral and antioxidant properties [31]. Recent research has focused on the biosynthesis of silver nanoparticles (AgNPs) using plant extract, including E. odoratum that showed antimicrobial and larvicidal activity [32, 33]. In addition, the nanostructures composed of Ag/Ag20 were explored for their antimicrobial and anti-biofilm properties [34].
Metal nanoparticles are shown to play a crucial role in malaria therapeutics [35, 36]. Therefore, we biosynthesized and characterized silver nanoparticles derived from the leaf extract of E. cotinifolia (EcAgNPs) and assessed the formulation for their parasite clearance activity in the routine culture of the asexual (BS) infection of P. falciparum. Besides, we investigated the mechanism underlying the anti-malarial activity of the EcAgNPs, that include the activation of metacaspase-2 and calpain. The latter leads to the clearance of P. falciparum from within huRBCs. EcAgNPs were found to disrupt the activity of P. falciparum isophospholipase (PfLPL-1), and hence inhibiting fatty acid flux and lipid storage, an essential process for the growth and development of parasite. Also, DNA fragmentation observed in the EcAgNPs treated parasitized RBCs suggests that synthesis of EcAgNPs could be viable therapeutic strategy for treating malaria infection. Collectively, our finding suggested that EcAgNPs alter the antioxidant profile and induce oxidative stress in P. falciparum. And EcAgNPs treatment increased the caspase activity that leads to the apoptosis of P. falciparum within the huRBCs confirmed by the alterations seen in the transcript levels of Bax/Bcl-2 levels. In the end, EcAgNPs formulation presents a promising option for the treatment of P. falciparum infection as well as regulating the emergence of drug tolerant P. falciparum.
2 Experimental: materials and methods
2.1 Materials and reagents
A young, mature E. cotinifolia leaf was taken from the garden. Himedia Laboratories in Mumbai, India provided the silver nitrate (AgNO3), chloroauric acid (HAuCl4), and sodium hydroxide (NaOH). Sigma Aldrich (Bangalore, India), Himedia provided all other reagents, medium, and chemicals unless otherwise specified.
2.1.1 Plant ethics
Euphorbia cotinifolia commonly called "carribbean copper plant" was collected in Molecular Biology Unit, Institute of Medical Sciences, Banaras Hindu University, Varanasi, Uttar Pradesh, India. The plant material was identified by Dr. Dilip Verma, BAMS—Medicine, BHU Varanasi. This plant was commonly available and hence we did not deposit it to any herbarium. Also, we do not have any plant ethics committee in India and carried out experiments.
2.2 Preparation of plant extract
Mature E. cotinifolia leaves were thoroughly cleaned with water and rinsed with Milli-Q water before being grinded in a mixer. Each 10-g portion of powdered leaves was boiled in 100 ml of deionized water for one hour at 55 °C. The extract was filtered through a Whatman No. 1 filter paper and stored at 4 °C until use.
2.3 Biosynthesis of silver nanoparticles from E. cotinifolia (EcAgNPs)
The silver NPs (AgNPs) were synthesized from E. cotinifolia (Ec) leaf extract using a modified protocol [37]. Fresh leaves (10% w/v) were homogenized in 100 ml of 20 mM Tris–HCl buffer, pH 8.0, for 5 min at 4 °C.. The extract was centrifuged at 2000 g for 15 min at 4 °C and stored at 4 °C until needed for the synthesis of green NPs. AgNPs were synthesized by mixing 1 ml of E. cotinfolia leaf extract (0.01 mg/ml) with 90 ml of a prepared aqueous solution of 1 mM silver nitrate. The AgNPs solution was diluted to 100 ml with 20 mM Tris–HCl buffer (pH 8) and incubated at room temperature. Aliquots were withdrawn at different time intervals for characterization by ultraviolet-visible (UV–Vis) spectroscopy (300–600 nm) (Hitachi 3220 UV spectrophotometer). Silver nitrate (AgNO3) and E. cotinfolia leaf extract served as controls in all spectral analysis. Synthesis of AgNPs was investigated with respect to the concentration and pH of E. cotinfolia leaf extract, using 20 mM sodium acetate at pH 4.0 and 20 mM Tris–HCl at pH 4.0 and in the range of 8.0–9.0. The resulting AgNPs solution was centrifuged at 2000 g for 15 min at 4 °C to separate the NPs. The pellet was washed five times with distilled water, air dried, and used for characterization.
2.4 Characterization of synthesized green silver nanoparticles (EcAgNPs)
The UV–VIS spectrum was used to optimize the temperature, pH, and time of synthesis for characterizing biosynthesized EcAgNPs. The prepared nanoparticles were characterized by determining the zeta potential of their surface using Malvern zeta sizer. The formulated NPs were deposited onto the microscope glass slide and dried in a hot air oven at 50 °C and analysed for X-ray diffraction (X-RD) patterns. This process was repeated to create a layer, and the X-ray diffractometer was set to a range of 20 to 80 degrees with scanning rate (0.050/s). Diffusion reflectance infrared Fourier transform (DFTIR) spectra were measured using a Shimadzu IR Affinity-1 FTIR spectrophotometer. Samples were scanned five times with a resolution of 4 cm−1 between 400 and 4000 cm−1 and 200 kV. The accelerating voltage was used to capture the images by the transmission electron microscope (TEM). Additionally, the size and selected area electron diffraction patterns of the prepared NP formulation were analysed.
2.5 In vitro parasite culture
The laboratory strain (3D7) of P. falciparum was obtained from National Institute of Malaria Research (NIMR), New Delhi. The 3D7 strain of P. falciparum (Pf3D7) was cultured in complete RPMI 1640 medium supplemented with 2 g/L glucose, 2 g/L sodium bicarbonate, 300 mg/L glutamine, 25 µg/ml gentamicin and 100 µM hypoxanthine, 0.5% albumax II, and human RBCs at 2–5% hematocrit under a gas mixture of 5% CO2, 5% O2, and 90% N2 at 37 °C following the established protocols with minor modifications [38, 39]. The ring stage of the parasite was synchronized by treatment with 5% sorbitol [40]. Giemsa-stained thin blood smears were examined under a light microscope to assess the parasite growth.
2.6 P. falciparum sensitivity to EcAgNPs
The antimalarial activity of EcAgNPs was determined by the SYBR Green I based emission assay on the routine culture of asexual BS infection of P. falciparum [41]. Briefly, 100 µl of asynchronous culture of infected erythrocytes (95% ring) with 0.8–1% parasitaemia at 1% hematocrit was added to each well of 96-well plates containing two-fold serial dilutions of EcAgNPs. Eight wells served as positive controls (without drug), and four wells served as negative controls (without parasite and drug). DMSO (0.5%) and Chloroquine (0.5 µM) were used as a vehicle and positive control. Ring stage parasites (50 µl) at 0.5% parasitemia were added to each well and incubated for 72 h at 37 °C. The culture medium was then replaced with the fresh complete medium containing different concentrations of EcAgNPs, DMSO (vehicle control) and chloroquine (positive control). After incubation, 100 µl of lysis buffer (20 mM Tris–Cl, 5 mM EDTA, 0.008% saponin, 0.08% Triton X981 100, pH 7.5, and SYBR green 1) was added to each well according to the manufacturer’s recommended dilution. Plates were incubated for 30 min at 37 °C, and fluorescence was measured at excitation (485 nm) and emission (530 nm) wavelength using a fluorescence reader. The background fluorescence from the CQ control was subtracted from that of DMSO and wells containing test compounds. IC50 was calculated based on the DNA content of the parasite using MS-Excel template.
2.7 Estimation of parasitemia (%)
The thin blood smears were drawn from EcAgNPs and drug treated parasites, fixed, and stained with Giemsa. The stained thin blood smears were then examined under the 100× oil immersion light microscope (Nikon, Japan). In each smear, 10,000 RBCs were scanned, and the % parasitemia was determined using the formula:
Parasite growth inhibition assay was conducted in a 96-well flat bottom plate with 10 µl of the EcAgNPs and 90 µl of parasite culture at 0.5% parasitemia and 2% hematocrit. EcAgNPs treated parasites were incubated for 48 h, and % parasitemia was determined from the Giemsa-stained thin blood smears. Morphological evaluation of treated and untreated parasites showed the inhibition of parasite development with EcAgNPs treatment compared to that seen with the untreated control [6, 42].
2.8 Schizont maturation inhibition assay to determine parasite development
EcAgNPs (1 mg/ml) were reconstituted in 1% DMSO to prepare a stock solution, which was further diluted to prepare different working concentrations in complete medium. The prepared concentrations of EcAgNPs were incubated for 48 h. In a 96-well plate, 90 μl of complete medium was added in each well, along with working concentrations of the EcAgNPs (0.6, 1.2, 2.5, 5 µg/ml) and drugs to be tested. Chloroquine (10 μM) and artesunate (2 μM) served as the positive controls in this experiment. Synchronized ring-stage parasites were added at 1.5% parasitemia and 1.8% final hematocrit, and plates were incubated for 48 h in the CO2 incubator. Thin blood smears were drawn from each well, stained with Giemsa, and inhibition of schizont maturation in comparison to the control was determined as follows:
Also, the thin smears drawn from EcAgNPs and chloroquine (10 µM) treated parasites were read to evaluate the differential count (%) of rings, trophozoites and schizonts per 1000 RBCs.
2.9 Anti-oxidant activity of EcAgNPs
2.9.1 Reactive oxygen species (ROS) estimation
The intracellular ROS levels were quantified in the blood stage infection of P. falciparum. Parasite cultures were synchronized to the trophozoite and schizont stages. The method involved alternative synchronization with 5% D-sorbitol followed by a 50–90% Percoll gradient centrifugation to enrich the population of trophozoite and schizonts. EcAgNPs (1 mg/ml stock solution) were diluted to the working concentrations and added to parasitized erythrocytes in the complete medium. 100 μl of parasite from 1% parasitemia at 0.2% hematocrit was seeded in a 96-well black plate to quantify the ROS production. The parasitized RBCs were stained with dichlorofluorescein diacetate (H2DCFDA) (20 μM) followed by treatment with various concentrations of EcAgNPs for 1 h. Further, fluorescence was measured to quantify the oxidation of H2DCFDA to the highly fluorescent DCF. Fluorescence of treated parasites was measured at 493 nm/522 nm using a Synergy™ H1-Biotek multi-plate reader. The control cells were added with PBS only [43].
2.9.2 Glutathione 5-transferase (GST) assay
GST activity was determined by measuring the conjugation of l-chloro-2,4-dinitrobenzene (CDNB) with glutathione (GSH) using spectrophotometer. P. falciparum at 0.2–0.5% parasitemia and 2% hematocrit in complete RPMI-1640 was treated with 0.6–5 μg/ml EcAgNPs for 6 h. After further incubation for 24 h, untreated P. falciparum served as a negative control. The assay mixture (1 ml) containing 0.5 mM CDNB, 1 mM GSH and 100 mM potassium phosphate buffer at pH 6.5. The phosphate buffer (PBS)-CDNB mixture was pre-incubated for 10 min at 37 °C, and the reaction was initiated by adding GSH, followed by the addition of purified enzyme. The increase in absorbance at 340 nm was measured for 5 min at 37 °C against a blank containing the reaction mixture without enzyme [44].
2.9.3 Glutathione assay
GSH concentration was determined following the published protocol with slight modifications [45]. Parasite culture at 0.2–0.5% parasitemia and 2% hematocrit in complete medium were treated with 0.6–5 μg/ml EcAgNPs for 6 h. After further incubation for 24 h, untreated P. falciparum served as a negative control. Parasite lysate (200 μl) was mixed with an equal volume of 5% meta-phosphoric acid and centrifuged at 3000 rpm for 10 min at 4 °C. The reaction mixture (1 ml) consisted of 100 μl supernatant, 800 μl of 0.1 M potassium phosphate buffer (pH 8.0), and 0.02 ml of 4% DTNB [5,5′-dithiobis-(2-nitrobenzoic acid)]. Following incubation at room temperature for 15 min, the absorbance of the developed coloured product was measured at 412 nm. The GSH concentration was determined by plotting a standard graph using the known concentrations of GSH [45].
2.9.4 Thioredoxin reductase (TrxR)
Parasite at 0.2–0.5% parasitemia and 2% hematocrit in complete medium treated with 0.6–5 μg/ml EcAgNPs for 6 h. After washing, parasite was further incubated for 24 h, with untreated parasite served as a negative control. TrxR activity was measured spectrophotometrically based on nicotinamide adenine dinucleotide phosphate (NADPH) mediated reduction of DTNB [46]. The assay mixture (1 ml) comprising 100 μl parasite lysate in 0.1 M potassium phosphate buffer with 2 mM EDTA, supplemented with a 10 μM NADPH and 50 μM DTNB. The reaction was initiated by introducing NADPH, and the increase in absorbance was read at 412 nm for 3-min at 25 °C. Enzyme activity was defined as one unit, representing the NADPH-dependent generation of 2 μmol of 2-nitro-5-thiobenzoate (ε412nm 13.6 mM−1 cm−1) per minute.
2.9.5 NADPH oxidase activity
We determined the NADPH oxidase activity by the protocol published with slight modifications [47, 48]. 0.2–0.5% parasitemia and 2% haematocrit in complete medium treated with 0.6–5 μg/ml EcAgNPs for 6 h and then incubated for an additional 24 h after washing. Untreated parasites served as a negative control. Parasite lysate was subjected to a reaction mixture (of 1 ml) containing 50 mM phosphate buffer (pH 7.0), 0.25% SDS, and 1 mM MgCl2. After centrifugation at 2000 rpm for 10 min at 4 °C, the supernatant (100 μl) was mixed with 80 μM cytochrome c and 2 mM sodium azide. The reaction was initiated by 10 μM NADPH, and the absorbance of coloured product was measured at 550 nm [47].
2.9.6 Detection of caspase like activity
Caspase-like activity was determined using Caspase-3 assay kit (Molecular probe, USA) in parasitized huRBCs at 2% parasitemia treated with various concentrations of EcAgNPs for 24 h. Parasitized RBCs were lysed with saponin to obtain cell-free parasite, with untreated parasites serving as an experimental control. The lysate was gently sonicated, and a reaction buffer (parasite lysate and caspase substrate Z-DEVD-R110 in a 1:1 ratio) was prepared. The reaction mixture was incubated for 30 min at room temperature in the dark. Fluorescence was measured using a microplate reader (SynergyHT Biotic) at excitation and emission wavelength of 460 nm and 530 nm, respectively, to detect the cleavage of the Z-DEVD-R110 substrate.
2.9.7 Determination of calpain activity
Calpain activity was measured following a published protocol [49] with slight modifications. Calpain activity was assessed using 35 µl of parasite cell lysate,7 µl fluorogenic substrate (Carbobenzoxy-Phe-Arg-4-methylcoumaryl-7-Amide), and 700 µl reaction buffer (50 mM Tris–HCl, pH 7.5, 5 mM MgCl2, 1 mM DTT, and 1 mM ATP) at 37 °C for 90 min. The hydrolysing activity of the enzyme was determined by fluorimetric quantification of substrates Suc-LLVY-AMC at excitation and emission wavelength of 380 nm and 440 nm, respectively, using a fluorescence spectrophotometer (Synergy HT Biotic).
2.10 Expression of lysophospholipase 1 (LPL1) of P. falciparum
2.10.1 RNA isolation and cDNA synthesis
For the expression of PfLPL1, parasite culture at 0.2–0.5% parasitemia and 2% hematocrit in complete medium were treated with 0.6–5 μg/ml EcAgNPs prepared in 0.4% DMSO for 6 h and DMSO served as an experimental control. Treated parasites were washed with PBS and lysed with saponin (0.075%) to remove the red blood cells. The pellet was washed three times with ice-cold PBS to remove the debris of RBCs. The cell suspension was centrifuged at 9000 rpm at 4 °C for 5 min each. RNA was isolated by the RNeasy mini kit (Qiagen). Total RNA was quantified with a Nanodrop 2000 (Thermo Fisher) before being adjusted to 30 ng/μl for the cDNA synthesis.
2.10.2 Real-time quantitative PCR (RT-qPCR)
cDNA was synthesized from the total RNA with the random hexamers using the cDNA synthesis kit (Cat No: GE27-9261-01, Sigma). The cDNA was used to carry out real-time quantitative PCR to calculate the fold regulation of relative mRNA expression of PfLPL1, Bax and Bcl-2 (primer sequence; Table 1). The reaction conditions used were as follows; Melting temperatures (Tm), 55-650C. RT-qPCR was performed using Roche LightCycler® 480 System in a 25 μl reaction volume with the PCR Master Mix Power SYBR™ Green Kit. Three independent biological samples were assayed in duplicates. 18S rRNA sequence was used as the house-keeping gene for data normalization, and melting curves were analysed at each reaction cycle. The relative fold change was determined using the 2 − ΔΔCt method [50].
2.10.3 DNA fragmentation of parasitized RBCs
The terminal deoxynucleotidyl transferase (TdT) mediated nick end labelling kit (QIA33, Sigma Aldrich, USA) was used to carry out the TUNEL assay following the manufacturer’s recommendations. P. falciparum infected and EcAgNPs treated RBCs were washed with PBS and fixed with 1% p-formaldehyde in 1X PBS for 30 min on ice. Cells were permeabilized with 0.1% triton X-100 in 0.1% sodium citrate and then treated with the AlexaFluor488. Flow cytometry and microscopic analysis were performed by TdT enzyme labelling as reported elsewhere [51]. DAPI stained slides were observed under the fluorescence microscope, and the EcAgNPs treated parasite, untreated unlabelled parasites were treated with DNase to remove any unwanted DNA of RBCs.
2.10.4 Assessment of programmed cell death (PCD) by Annexin V-FITC assay
1 × 107 parasitized RBCs (pRBC) were treated with 0.05% saponin and washed with PBS to assess the death of parasitized RBCs following the EcAgNPs treatment. Cells were harvested by trypsin/EDTA, and the pellet was washed with 1X PBS. The cells were then suspended in 100 µl of 1X Annexin binding buffer, stained with Annexin-FITC and propidium iodide (PI) for 15–20 min in the dark, analysed by flow cytometry within 1 h of treatment using Annexin V- FITC Assay kit (Cat No: APOAF, Sigma) [52].
2.10.5 RBCs hemolysis
The hemolysis assay was conducted following the published protocol [53]. A suspension of EcAgNPs nanoparticles (stock concentration = 1 mg/mL) was prepared. The cultures of pRBCs were maintained at 1.5–2% parasitemia at 1% hematocrit in a 37 °C incubator with a 95/5% (vol/vol) air in the CO2 incubator. All assays were performed under these conditions of parasitemia and hematocrit. A hemolysis buffer (5 mmol/L sodium phosphate, 1 mmol/L EDTA, pH 8.0) served as the positive control (Ctrl +), while 1X PBS and 5 mM glucose were taken as the negative controls (Ctrl-). A suspension of nanoparticles (stock concentration = 1 mg/mL) prepared in sterile isotonic 1X PBS with 5 mM glucose was added to a diluted pRBC culture (0.1 mL) at various concentrations (0.6, 1.25, 2.5, 5 μg/ml) for 24 & 48 h of incubation at 37 °C and 41 °C (using a Thermomixer) to determine the hemolytic activity of pRBCs. Samples were then centrifuged for 1 min, and the absorbance of hemoglobin released into the supernatant was measured using a UV–VIS spectrophotometer (Boitek Synergy HTX Multi-Mode Reader) at 600 nm with a standard wavelength of 405 nm.
% hemolysis was calculated using the formula:
2.11 Statistical analysis
The data are shown as mean ± SD, and statistical analysis was carried out by one-way and two-way analysis of variance (ANOVA) with Tukey–Kramer multiple comparison post-test using the GraphPad InStat™ software (GraphPad Software Inc., San Diego, California). The statistical variations are presented as *p < 0.05, **p < 0.01, ***p < 0.001, and ns = non-significant (p > 0.05). Data presented is Mean ± SEM of at least three independent experiments (n = 3), ***P < 0.0001, **P < 0.001 *P < 0.05.
3 Results
3.1 Formulation and characterization of EcAgNPs
3.1.1 UV–VIS spectroscopy
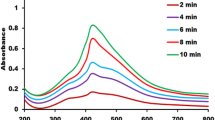
The plant E. cotinifolia serves as a bio-reducing agent for the green synthesis of AgNPs (EcAgNPs). The synthesized AgNPs were marked and characterized by the development of a dark brown colour. This colouration process is a morphological manifestation of AgNPs article synthesis (Fig. 1A). The reaction conditions were optimized to bio-synthesize AgNPs using the aqueous extract of E. cotinifolia leaves. The stability of synthesized green nanoparticle formulation was confirmed by the UV–visible spectroscopy (UV–VIS), which exhibited a spectral peak at 430 nm. On the contrary, chemically synthesized AgNPs using sodium borohydride (NaBH4) showed an absorption maxima at 398 nm. No discernible spectral peak was observed with the plant extract (Fig. 1B). The zeta potential was determined to ascertain the electrostatic charge potential within the electrical double layer surrounding the formulated NPs [54]. Higher negative zeta potential measured by the zeta sizer (-29.4 mV) of green NPs constructed by E. cotinifolia confirmed the stability of synthesized green nanoparticles formulation. And, this stability is attributed to the presence of phytochemicals in the plant extract (Fig. 1C).
Preparation of green silver nanoparticles synthesized from E. cotinifolia leaves (EcAgNPs). A a) Biosynthesized EcAgNPs colloid b) E. cotinifolia plant extract c) silver nanoparticle (AgNO3) solution. Characterization of biosynthesized EcAgNPs through B Ultraviolet–visible (UV–VIS) absorption spectrum of plant (E. cotinifolia; Ec) extracts (pink), AgNPs solution (Red) and EcAgNPs solution (black) C Determination of surface zeta potential to confirm the stability of EcAgNPs D The FTIR spectral analysis of synthesized green nanoparticles. The comparative analysis of peaks observed with the classical AgNPs (black) and EcAgNps (red). Three prominent peaks; a broad peak around 3438 cm−1 corresponds to O–H stretching, Peaks at 1828 cm−1 represent carbonyl groups and 1548 cm−1 originates from stretching vibration of ester phenolic compounds (n = 3)
3.1.2 Fourier transform infrared spectroscopy (FTIR) analysis
Potential interaction between silver (Ag) ions and biomolecular functional groups present in E. cotinifolia, including EFL-9, Helioscopinolide, quercetin-3-O-β-d-glucoside, 4αβ,9α,16,20-tetrahydroxy-14(13 → 12)-abeo-12αH-1,6-tigliadiene-3,13 dione,helioscopianoid P and H and euphol) was investigated through FTIR spectroscopy [28]. These functional groups have been reported to confer various health benefits [28]. The mature leaves of E. cotinifolia are known to be rich in diterpenoids and flavonoids and possess antioxidant, antibacterial, and antiviral properties [55]. The FTIR spectra of EcAgNPs green nanoparticles showed overlapping peaks with those of the plant extract, signifying their crucial role in the capping and effective stabilization of nanoparticles (Fig. 1D). The analysis of EcAgNPs revealed the presence of carbonyl groups and phenolic compounds in the spectral analysis. Our data indicated three prominent peaks; a broad peak around 3438 cm−1 corresponding to O–H stretching, peaks at 1828 cm−1 representing carbonyl groups, and 1548 cm−1 originating from stretching vibration of ester phenolic compounds. The presence of these compounds suggested their role in stabilizing the AgNPs formulation. A comparative FTIR analysis was essentially needed to understand the capping process, involving both the dry plant extracts and associated AgNPs (Fig. 1D).
3.1.3 X-ray diffraction (XRD) analysis
Size of formulated silver nanoparticles was estimated by utilizing Scherrer's formula, aligning the presence of silver phase with the whole width at half maximum of high intensity diffraction peak as is evident with the series of enlarged line profiles.
D represents the maximum crystalline domain size perpendicular to the reflecting plane, λ is the x-ray wavelength, β is the diffraction peak, and ϴ is the diffraction angle. The hydrodynamic size of "green nanoparticles" for EcAgNPs was found to average approximately 30 nm, as reported previously [56]. The diffraction peak of EcAgNPs showed characteristics of a metallic face-centered structure (Fig. 2A). The stability of silver nanoparticles in aqueous media was determined by a high negative Zeta potential (−29 mv) (Fig. 1C). Moreover, the robust dispersion of EcAgNPs in aqueous medium prevented the aggregation of synthesized nanoparticles confirmed by the absorbance (Fig. 1D). Energy dispersive X-ray spectroscopy (EDX) was performed to determine the compositions of different elements during synthesis of the EcAgNPs formulation [57]. A prominent peak was observed for silver metal, with smaller peaks corresponding to other metals (C, O, Ca) in the green nanoparticle formulation (Fig. 2B).
Characterization of synthesized green nanoparticles. The Biosynthesized green EcAgNPs were characterized by A X-ray diffraction pattern, B Energy dispersive X ray spectrum of biosynthesized green nanoparticles, C Structural characterization of the biosynthesized silver nanoparticles by the transmission electron microscopy (TEM). D Estimated size of EcAgNPs was 10–15 nm, and the histogram of size distribution of biosynthesized EcAgNPs (n = 3)
3.1.4 Morphological characterization of green nanoparticle
Morphology and structural characteristics of biosynthesized silver nanoparticles were analysed by transmission electron microscopy (TEM) to confirm the size of prepared green nanoparticles (EcAgNPs) (Fig. 2C). The EcAgNPs maintained a spherical shape throughout the formulation process. The average particle size, as determined by the TEM analysis, was estimated to be 50 nm, suggesting the presence of a staking layer with a size distribution spawning from 20 to 50 nm. This small size of the prepared nano-formulations (EcAgNPs) mitigates issues associated with uptake and retention by the reticulo-endothelial system (RES) [58] (Fig. 2C). The estimated % frequency of nanoparticle with smaller size was observed higher upon counting the cells (Fig. 2D).
3.1.5 Antimalarial activity of E. cotinifolia silver NPs (EcAgNPs)
P. falciparum was maintained in the complete medium at 0.5% parasitemia (3% hematocrit) for 24 h and treated with tenfold successive dilutions of EcAgNPs to determine parasite inhibition and half-maximal effective concentration (EC50) by the SYBR green based emission assay (Fig. 3A). Half-maximal effective concentration (EC50) by the SYBR Green fluorescent probe estimated to be 0.75 µg for EcAgNPs. Based on EC50, four concentrations (0.6, 1.2, 2.5, 5 µg/ml) were opted for further cell based assays with EcAgNP. The assays were conducted with lower (than EC50) concentration, medium, and eightfold higher (5 µg/ml) concentration to confirm the antimalarial activity of EcAgNPs (Fig. 3A–C). The antimalarial activity of prepared EcAgNPs was assessed in the routine in vitro culture of chloroquine (CQ)-sensitive laboratory strain (3D7) of P. falciparum using a SYBR green emission-based drug sensitivity assay. Parasites were treated with E. Cotinifolia leaf extract (50, 100, 200 µg), AgNPs (0.6, 1.25, 5 µg) and EcAgNPs (0.6, 1.25, 5 µg) and incubated for 6 h. The treated P. falciparum was then washed with the in incomplete medium and cultured in the complete medium with the same EcAgNPs concentrations for 72 h. The incubation period (24, 48 h) dependent parasite growth inhibition was seen with the EcAgNPs formulations (Fig. 3B). Our data indicated that the reduction in parasite growth was significantly higher (****P < 0.0001) with EcAgNPs formulation following the 24 and 48 h of incubation. % reduction in parasitemia confirming the inhibited parasite growth was seen more pronounced at 5 µg EcAgNPs as compared to that seen with the higher concentration (200 µg) of E. cotinifolia (Ec) and 5 µg AgNPs (Fig. 3B). This data suggested the importance of chemically synthesized green nanoparticles to confer the antimalarial activity of EcAgNPs. These findings prompted us to perform the ring stage survival assay (RSSA) by determining the % decrease in parasite re-invasion (Fig. 3C). The synchronized ring stages were treated with different concentrations (0.6, 1.2, 2.5, 5 µg/ml) of EcAgNPs (Fig. 3C). Results with 5 µg EcAgNPs showed a decrease in the ring stage of parasite development compared to untreated control confirming inhibited parasite replication and transformation to the schizonts (Fig. 3C). This data is important as parasite re-invasion is crucial for the replication and growth of parasite [59, 60].
In vitro antimalarial activity of green silver nanoparticles biosynthesized from E. cotinifolia (EcAgNO3) in the asexual blood stage infection of P. falciparum A Sensitivity of 3D7 strain of P. falciparum was determined by the SYBR green based drug sensitivity assay and half-maximal effective concentration (EC50) of EcAgNO3 calculated is 0.75 μg/ml at the different concentrations of EcAgNPs tested. B Assessment of % growth (parasitemia) inhibition when P. falciparum was treated with incrementally increasing the concentration from 0.6 to 5 μg/ml of EcAgNPs. 0.6, 1.2, 2.5, 5 µg/ml concentrations were designed based on EC50. Assays were performed with the lower (than EC50) concentration, medium and eightfold higher (5 µg/ml) concentration for confirming the antimalarial potential of EcAgNPs. Parasites were treated with E. Cotinifolia leaf extract (50, 100, 200 µg), AgNPs (0.6, 1.25, 5 µg) and EcAgNPs (0.6, 1.25, 5 µg) C Inhibited reinvasion (% decrease in the parasite re-invasion) as shown by the decrease in the ring stage parasitemia after the treatment with EcAgNPs at different NPs concentrations (0.6, 1.2, 2.5, 5 µg/ml) for 6 h followed by washing with the RPMI-1640. Parasite reinvasion was determined by reading the Giemsa stained thin blood smears drawn on 8, 16, 24, 32, 40 h of incubation D) Inhibition in schizont maturation was determined when parasite was incubated with EcAgNPs (0.6, 1.2, 2.5, 5 µg/ml), Chloroquine (CQ;10 μM) and artesunate (AS;2 μM) for 48 h. Untreated parasite served as negative control whereas AS and CQ as positive control. E) Differential counting of ring, trophozoites and schizonts in EcAgNPs (0.6, 1.2, 2.5, 5 µg/ml) and CQ (10 µM) treated parasite per 1000 RBCs counted (n = 3). ****P < 0.0001 (highly significant), ***P < 0.001 (moderately significant) **P < 0.001 (significant), *P < 0.05 (less significant),
3.1.6 Inhibition of schizont maturation
We next decided to confirm the inhibition of schizont maturation (Fig. 3D) from trophozoites when the parasite was treated with EcAgNPs, artemisinin (AS), and (CQ) for 48 h, wherein AS and CQ serve as the positive controls. A concentration dependent and significant inhibition (****P < 0.0001) in schizont maturation was observed with the decrease proportional to the EcAgNPs concentration. The highest inhibition was seen with 5 µg/ml EcAgNPs, compared to that with the lowest concentration of EcAgNPs (0.1 µg/ml) and this inhibition was comparable to that with AS and CQ (Fig. 3D).
3.1.7 Differential counting of developmental forms of P. falciparum
The clearance of parasite and inhibition of schizont maturation following treatment with CQ (10 µM) and EcAgNPs (0.6, 1.2, 2.5, 5 µg/ml) led us to carry-out the differential counting of developmental forms (rings, trophozoites, schizonts) of P. falciparum (Fig. 3E). We assessed the % differential ratios of rings, trophozoites, schizonts per 1000 RBCs. After 48 h of incubation with EcAgNPs and CQ, a significant (***P < 0.001) decrease in the developmental forms counting was observed, and was proportional to the EcAgNPs concentrations tested. This decrease was compared to that seen with the untreated control and CQ treated formulations (Fig. 3E). The highest decrease, seen with 5 µg/ml EcAgNPs, was comparable to CQ treatment (Fig. 3E). Therefore, this data suggests that EcAgNPs can clear all developmental forms and might be a viable option for malaria treatment.
The inhibition of parasite growth and development led us to examine the number of schizonts on smear drawn from parasite culture receiving treatment with 5 µg EcAgNPs as compared to the untreated control (Fig. 3). This indicates the DNA concentration in the parasite and confirms parasite viability. The ability of EcAgNPs to permeate across P. falciparum cell membranes in order to check the growth and development of the parasite was demonstrated (Fig. 3A–C). The thin blood smears drawn on treated parasites were stained with Giemsa, and morphological alterations, particularly distortion in the ring stage of parasites at 6 & 12 h of P. falciparum development, were observed in comparison to the untreated control (SI Fig. S1). This data is consistent with published [61, 62] findings confirming inhibited reinvasion of parasite by Giemsa stained parasites. Here, we confirm that smears drawn from parasite receiving treatment with 5 µg/ml EcAgNPs for 24 and 48 h, compared with untreated control (SI Fig. S1), show inhibited growth of the ring stage parasite and distorted morphology of EcAgNPs treated P. falciparum following 48 h of treatment. This again suggests for the antimalarial activity of EcAgNPs (SI Fig. S1).
3.1.8 Measurement of reactive oxygen species (ROS) and antioxidant enzymes activity
We next decided to dissect the mechanism underlying the parasite clearance and showing antimalarial activity of EcAgNPs. The generation of ROS was assessed using dichlorofluorescein diacetate (DCFDA) stained P. falciparum treated with AgNPs and EcAgNPs at 0.6, 1.2, 2.5 and 5 µg/ml (Fig. 4A).
Generation of reactive oxygen species (ROS) and antioxidant enzymes activity. P. falciparum stained with dichlorofluorescein diacetate (DCFDA) dye to quantify the ROS production. A Increased ROS production by AgNO3 and biosynthesized AgNO3 nanoparticles (EcAgNPs) on ROS production in DCFDA stained parasitized huRBCs (PfhuRBCs). ROS production was increased in proportion to the increased fluorescence with the increasing AgNPs and EcAgNPs concentration as shown by the DCF positive cells B The flow cytometry histograms based analysis showed an increase in DCF positive cells at different time points (1–10 h) when parasite was treated with the different concentrations (0.6, 1.2, 2.5, 5 µg/ml) of EcAgNPs. Antioxidant enzymes such as Glutathione (C), Thioredoxin reductase D and NADPH oxidase E activity was determined in parasitized RBCs at ~ 1.8% parasitemia following 6 h treatment of various AgNPs and EcAgNP concentrations in the prepared homolysate. The levels of GSH are expressed as μM/mg protein whereas other enzymes activities are indicated as U/ml where 1 unit is equal to µM/cm/min. Data expressed as the Mean ± SEM of three independent experiments (n = 3). ****P < 0.0001 (highly significant), ***P < 0.001 (moderately significant) **P < 0.001 (significant), *P < 0.05 (less significant),
The increase in the fluorescence was observed with increasing concentrations of AgNPs and EcAgNPs. The DCFDA assay is based on the diffusion of DCFDA into the cells, followed by its de-acetylation by the cellular esterases to form a non-fluorescent compound, which is then oxidized by ROS into 2’, 7’-dichlorofluorescein (DCF). DCF is highly fluorescent and can be detected by fluorescence spectroscopy with excitation and emission wavelength of 485 and 535 nm, respectively [63]. Also, we quantified the number of DCF positive cells to determine the ROS production. Our data suggested that increase in the ROS production was proportional to the increasing concentrations of AgNPs and EcAgNPs (0.6, 1.2, 2.5, 5 µg/ml) (Fig. 4A). A significantly higher (nearly 1.5-fold) (****P < 0.0001) ROS production (indicated by % DCF positive cells) was observed with EcAgNPs at 5 µg/ml compared to that with AgNPs (Fig. 4A). Additionally, our data suggested higher fluorescence was emitted by the 5 µg/ml EcAgNPs 10 h post-treatment. Based on these findings, we confirmed the concentration and duration of incubation (with P. falciparum) dependent increase in the ROS production was seen with EcAgNPs (Fig. 4B).
This increased ROS production by the EcAgNPs led us to assess the activity of redox enzymes, including glutathione S-transferase (GST) (Fig. 4C), glutathione reductase (GR), and TrxR (Fig. 4D), in the cytosolic fraction of P. falciparum receiving treatment with EcAgNPs (0.6, 1.2, 2.5, 5 µg/ml). We observed a decrease in the activity of redox enzymes (GSH and TrxR) (Fig. 4C, D) proportional to the increasing (0.6, 1.2, 2.5, 5 µg/ml) concentrations of EcAgNPs. Treatment with AgNPs and EcAgNPs was compared to that with the untreated infection control, and both formulations were shown to decrease GSH (Fig. 4C) and TrxR (Fig. 4D) at all the concentrations tested. This decrease was seen significantly pronounced (****P < 0.0001) with EcAgNPs at 5 µg/ml when compared to that with AgNPs treated parasite (Fig. 4C, D).
Furthermore, the activity of NADPH oxidase, a multi-subunit enzyme comprising membrane and cytosolic components was determined. NADPH oxidase plays a key role in clearing parasite infection. Since NADPH oxidase activity of the host compromised during malaria infection, the lack of a functional NADPH oxidase leads to the generation of phagocyte-derived ROS, and hence contributes to the malaria pathogenesis [64]. Treatment with EcAgNPs and AgNPs formulations showed increased levels of NADPH oxidase activity at all the concentrations tested (Fig. 4E). Significantly (****P < 0.0001) higher NADPH oxidase activity may clear P. falciparum infection (Fig. 4E).
3.1.9 EcAgNPs treatment mediates the endoplasmic reticulum (ER) stress in P. falciparum
We evaluated the ROS production during ER stress by assessing protein disulfide isomerase (PDI) activity in the cytosol and ER fraction of EcAgNPs treated parasites, compared to untreated experimental control (Fig. 5A). PDI activity facilitates disulphide bond formation during ER stress [65]. A concentration dependent increase in PDI activity was observed when cytosolic and ER fractions of P. falciparum were treated with the increasing EcAgNPs concentrations (0.6, 1.2, 2.5, 5 µg/ml). Our data indicated a sixfold increase in the PDI activity in the ER fraction compared to the cytosolic fraction at the 5 µg/ml treatment with EcAgNPs. The untreated infection control served as a negative control (Fig. 5A). These data suggest that EcAgNPs can modulate ER stress. Further, we assessed calpain (calcium-dependent cysteine protease) activity in the cytosolic fractions to determine calcium levels. Calpain mediates haemoglobin degradation and leads to the increased ER stress, heat shock, and altered the levels of oxidative enzymes in EcAgNPs treated parasite. Our data showed increased fluorescence intensity (Cps) at a 5 µg/ml treatment of EcAgNPs when compared to the untreated control (Fig. 5B). PDI is an important enzyme for designing anti-malarial drugs and vaccine targets, ensuring the correct folding of proteins required for parasite propagation [66]. Calpain activation facilitates the cleavage of Bcl2 into its active form/membrane targeted death ligand (tBID), which targets the mitochondrial outer membrane and the apoptotic protein Bcl2 [67]. Our data suggest that P. falciparum adapts through the unfolded protein response (UPR) mechanisms to survive immune response onslaught and changing environmental conditions (such as pH and temperature changes, oxidative stress, and nutrient deficiency) during ER stress [68]. We next determined caspase-3 activity by measuring fluorescence units to present calpain activity and confirmed the death of P. falciparum (Fig. 5C). We observed significantly (**P < 0.001) higher fluorescence at a 2.5 µg/ml EcAgNPs treatment compared to a relatively less significant (*P < 0.05) at a 5 µg/ml EcAgNPs treated in comparison to the untreated control (Fig. 5C).
The endoplasmic reticulum (ER) stress is delineated in terms of Protein disulfide isomerase (PDI) activity to assess the calcium release, A PDI activity was assessed in the subcellular fractions (cytosol and ER) at 650 nm by the spectrophotometer B Calpain activity was determined in cytosolic fraction of P. falciparum lysate following the treatment with 5 µg/ml of EcAgNPs. The fluorescence intensity (CPS) was determined by quantifying the fluorogenic substrate Suc-LLVY-AMC at 380 nm excitation/440 nm emission spectrum using the Synergy HT Biotic fluorescence spectrophotometer. The untreated infection control (Red line) was kept as a negative control. Black line shows the 5 µg/ml EcAgNPs treated P. falciparum, C Detection of the caspase-3 activity in the parasitized RBCs following the treatment with EcAgNPs at different concentrations (0.6-5 µg/ml), D The qualitative evaluation of the DNA fragmentation by TUNEL assay in EcAgNPs treated routine culture of asexual blood stage infection of P. falciparum, E quantification of the tunnel positive cells following the EcAgNPs treatment F) Bax/Bcl-2 ratio. Higher transcript regulation of Bax/Bcl-2 ratio at 5 µg/ml of EcAgNPs treated parasite indicates the apoptosis. G) EcAgNPs treatment for 12 h (4.4%) and 24 h (16.91%) induced programmed death of P. falciparum was determined by the Annexin-V staining. ****P < 0.0001 (highly significant), ***P < 0.001 (moderately significant) **P < 0.001 (significant), *P < 0.05 (less significant)
3.1.10 DNA fragmentation
We next conducted a TUNEL assay to assess the DNA fragmentation in P. falciparum infected erythrocytes receiving treatment with the varying concentrations (0.6, 1.2, 2.5, 5 µg/ml) of EcAgNPs (Fig. 5D). Qualitatively observation of DNA fragmentation in the nucleated schizonts of P. falciparum were made after treating parasites with different concentrations of EcAgNPs for 12 h (Fig. 5D). Additionally, TUNEL positive cells were quantified to confirm the DNA fragmentation (Fig. 5E) since the DNA of parasitized RBCs was seen fragmented by the EcAgNPs treatment. This data suggests that EcAgNPs are capable of inducing the death of parasitized RBCs.
3.1.11 Apoptosis like death of parasitized RBCs
The lower relative mRNA expression of apoptotic family protein (Bcl-2) [69] and higher expression of the core regulator of apoptosis, Bax [70], indicated the regulated apoptosis of parasitized RBCs (Fig. 5F, SI Fig. S2). The significantly highest (****P < 0.0001) (Fig. 5F) ratio of Bax and Bcl-2 at 5 µg/ml EcAgNPs confirmed the apoptosis of P. falciparum infected RBCs.
We then detected DNA fragmentation in parasitized RBCs to confirm the induction of apoptosis in P. falciparum culture treated with 5 µg/ml EcAgNPs for 12 and 24 h using Annexin V/PI staining (Fig. 5G). Our data indicated the induced death of parasitized RBCs by the EcAgNPs treatment. Higher parasite clearance was observed following 24 h of exposure to NPs compared to that seen with the 12 h. Hence, our data suggested higher apoptosis of parasitized RBCs based on the incubation of P. falciparum culture with the EcAgNPs. We observed higher % killing of parasitized RBCs at 24 h (16.91%) compared to 12 h (4.41%). In the end, we infer that the significant parasite clearance was achieved with EcAgNPs formulation compared to the untreated control (Fig. 5G). Also, the PI staining confirmed the apoptosis of parasitized RBCs (Fig. 5G).
3.1.12 Expression of lysophospholipase 1 (LPL1) of P. falciparum
The LPL1 of P. falciparum is known to maintain the lipid homeostasis in P. falciparum [71, 72]. Therefore, we decided to investigate the expression of LPL1 in the parasite treated with different EcAgNPs concentrations (0.6, 1.2, 2.5, 5 µg/ml) (Fig. 6). The concentration-dependent decreased (**P < 0.001) expression of PfLPL1 was quantified from the NPs treated P. falciparum [71] (Fig. 6A). This study highlights the complex interplay between EcAgNPs, ER stress, calcium homeostasis, and lipid metabolism in P. falciparum. The lower fold change (**P < 0.001) in the relative mRNA expression of LPL1 at 5 µg/ml EcAgNPs compared to that seen with the other concentrations confirmed the disrupted lipid homeostasis in P. falciparum (Fig. 6B). The house-keeping (β-actin) gene was used as an internal control (Fig. 6B).
Detection of the gene expression of lipoprotein lipase-1 (LPL-1) of P. falciparum through RT-PCR. RNA was extracted from all groups (control, 0.6, 1.2, 2.5, and 5 µg/ml) by loading with Triazol and cells were lysed using saponin enrichment to carry-out qRT-PCR to determine the expression of A PfLPL-1 through agarose gel electrophoresis of PCR product B fold change of relative mRNA expression of PfLPL-1 C % hemolysis of RBCs following the treatment with EcAgNPs (0.6, 1.2, 2.5, and 5 µg/ml) for 24 and 48 h. ****P < 0.0001 (highly significant), ***P < 0.001 (moderately significant) **P < 0.001 (significant), *P < 0.05 (less significant)
3.1.13 Toxicity of EcAgNPs (RBC hemolysis)
To assess the toxicity of EcAgNPs, we performed a hemolysis assay on RBCs and compared with AgNPs (data not shown). We observed 4 times lesser RBCs hemolysis with AgNPs as compared to that with EcAgNPs at all the concentrations tested. Parasitized RBCs were treated with all the EcAgNPs concentrations (0.6, 1.2, 2.5, 5 µg/ml) for 24 and 48 h. Hemolysis buffer served as a positive control, and PBS as a negative control (Fig. 6C). The results showed significantly lower (****P < 0.0001) RBC hemolysis when parasitized RBCs were incubated with EcAgNPs for 24 and 48 h. Hemolysis seen at 48 h was non-significantly (****P < 0.0001) higher at 5 µg/ml compared to that seen with lower concentrations (0.6, 1.2, 2.5 µg/ml) (***P < 0.001) following 24 h incubation. These data suggest that green AgNPs do not exhibit toxicity and could be safe for human use (Fig. 6C).
4 Discussion
The ever-emerging field of nanotechnology has been at the forefront of devising the therapeutic options to address the infectious diseases. Hence, plant extract-based synthesis of silver nanoparticles (AgNPs) has been emerging and could be instrumental in the fighting the malaria infection. Therefore, utilizing extracts from Artemisia species [35], Terminalia bellirica leaves [73], Azadirachta indica (neem) [53], Azadirachta indica seed [74], and Salvia officinalis (sage) [75] to produce AgNPs that target P. falciparum, P. berghei, and P. chabaudi, the causative agents of human and rodent malaria, respectively. AgNPs have shown remarkable antimalarial properties, with the mechanisms ranging from the inhibition of parasite growth and the induction of oxidative stress to the protection of liver tissue against malaria-induced injury. The eco-friendly synthesis methods, which leverage the natural reducing agents present in plant extracts, not only ensure the hemocompatibility of the AgNPs but also underscore the potential of phytochemicals in yielding effective and non-toxic therapeutic agents. The stability of these AgNPs over time and their compatibility with blood cells, coupled with their protective effects on hepatic tissue, place them as viable candidates for alternative or complementary antimalarial therapies.
Hence, present study reports the successful biosynthesis of silver nanoparticles (AgNPs) using a leaf extract from Euphorbia cotinifolia (EcAgNPs) via a green synthesis method (EcAgNPs). The synthesized EcAgNPs were subjected to comprehensive characterization using X-ray diffraction (XRD), F-TIR spectroscopy, and TEM [76]. This analysis revealed the stability and spherical morphology, with an average size range of 20–30 nm for the EcAgNPs. FTIR spectroscopy identified phenolic compounds within the E. cotinifolia extract, which were shown to play a critical role in stabilizing the biosynthesized AgNPs. The presence of capping agents and functional groups from the plant's phyto-constituents was also confirmed. Moreover, the XRD analysis confirmed the crystalline structure of the synthesized nanoparticles [77]. The antimalarial properties of EcAgNPs were evaluated through drug sensitivity assay in the routine in vitro culture of asexual BS of P. falciparum. The lower half maximal effective concentration of EcAgNPs (EC50; 0.75 µg/ml) indicated the high efficacy of EcAgNPs in inhibiting the growth and development of P. falciparum. The reduction in parasite burden, as evidenced by the decreased total parasitemia at various concentrations of AgNPs and EcAgNPs confirmed the antimalarial activity of EcAgNPs [78]. This data validated the previously published findings [79] and confirmed the antimicrobial activity of EcAgNPs. Also, our data confirmed that EcAgNPs selectively target and kill the parasite without adversely affecting the shape or size of human RBCs [80]. Thus, this observation could serve as a biomarker for anti-malarial drug efficacy [80, 81].
Building upon the established antimalarial potential of biosynthesized AgNPs, we delved into the underlying mechanism of their antimalarial activity [82]. The cellular metabolism naturally produces ROS, which are effectively scavenged by the antioxidants. However, EcAgNPs facilitate an increase in ROS production and diminish mitochondrial activity, leading to apoptosis-like death of parasitized huRBCs [83]. This further disrupts the redox homeostasis due to the dysregulated antioxidant enzyme activity, essentially required for the growth and development of P. falciparum.
The ER stress is a critical factor in maintaining redox homeostasis, allowing P. falciparum to withstand and endure the host's cellular defence onslaught [84, 85]. The higher production of ROS triggered by EcAgNPs inhibits the activity of the antioxidant enzyme system [86, 87]. As a result, the compromised functionality of these antioxidants fails to mop up the free radicals, and hence escalate the ROS levels. The latter therefore program the death of P. falciparum within the huRBCs.
Further, our findings reveal the unique ability of EcAgNPs to target the TGR/Prx2 cascade at lower concentrations, as evidenced by their capability to kill the parasite. This work contributes to the understanding of specific pathways and targets that may be influenced by EcAgNPs in P. falciparum. The oxidative stress, characterized by increased lipid and protein oxidation, higher ROS levels, and impaired glutathione metabolism, is indicated by a decreased GSH/GSS ratio [88]. This stress phenomenon ultimately leads to an increased production of free radicals that impair antioxidant activity. Our findings, in line with others [89], confirm that oxidative stress is a pivotal mechanism in clearing malaria infection. This knowledge may guide us develop targeted and effective antimalarial treatment strategies for developing interventional approaches for treating P. falciparum infection. Consistent with previous research [90] showing that lower concentration of andrographolide decreases antioxidant enzyme activity in filarial parasites due to the elevated ROS, our data confirm that increased ROS levels drive apoptosis in the parasitized RBCs [91]. The oxidative stress induced by EcAgNPs treatment leads to apoptosis and parasite clearance, as a result of dysfunctional antioxidants (GSH, GST, and TrxR). This mechanism of action is similar to that of artesunate, which is known to reduce the antioxidant levels, causing DNA fragmentation, and increased intercellular ROS levels in P. falciparum [92]. Our work suggests that EcAgNPs and artesunate may share a common mode of action in killing the parasite.
The comprehensive bio-assays and caspase activity evaluations suggest that EcAgNPs play a significant role in inducing apoptosis of P. falciparum infected huRBCs. We observed programmed cell death induced by EcAgNPs at various concentrations, which was accompanied by a concentration-dependent increase in caspase activity, leading to the clearance of P. falciparum (Figs. 1, 2, 3, 4, 5, 6). These findings are consistent with and support previous reports of caspase-3-like activity indicating apoptosis in other protozoan parasites, such as P. berghei ookinetes [93] and Trypanosoma cruzi [94]. Furthermore, the enhanced activity of caspase-3, induced by calcium, had a notable impact on the morphological changes associated with nuclear apoptosis. The biochemical characterization of apoptosis, including DNA fragmentation analysis by the TUNEL assay, was corroborated by Annexin V staining, confirming the induced apoptosis of parasitized RBCs. Our data showed a significantly increased TUNEL-positive cells, which was directly proportional to the increasing concentration of EcAgNPs treatment (Fig. 5D). The dose-dependent DNA fragmentation observed in P. falciparum supports the conclusion that there is a considerable increase in the number of apoptotic parasitized RBCs. This study provides further confirmation of the antimalarial activity of EcAgNPs. Additionally, the bax/bcl2 ratio, indicating their high expression levels, confirms the induction of apoptosis in parasitized RBCs [95, 96].
To understand the programmed apoptosis induced by EcAgNPs in P. falciparum, Annexin-V/propidium iodide staining was used to determine nuclear nucleus position in relation to apoptotic characteristics. Treatment with EcAgNPs resulted in a significant increase in the percentage of apoptotic cells, along with a modest rise in the number of necrotic cells (Fig. 5G). Specifically, the proportion of apoptotic cells increased with 5 µg EcAgNPs after 12 h of treatment and continued to rise during 24 h treatment period (Fig. 5G). Our EcAgNPs formulation demonstrated the ability to disrupt the activity of lysophospholipase (PfLPL-1), a crucial enzyme for parasite development, and hence confirmed thee antimalarial activity of the EcAgNPs.
In summary, the green synthesis of AgNPs using an E. cotinifolia extract present a sustainable, eco-friendly, and non-toxic alternative to conventional chemical synthesis methods (Fig. 7). We anticipate that further optimization of the synthesis process will enhance the yield, stability, and scalability of EcAgNPs production. This work enhances the potential of EcAgNPs for large-scale manufacturing and potential clinical applications. Moreover, the targeted and controlled delivery of EcAgNPs is crucial for developing therapeutic interventions against malaria. Encapsulating novel drugs with EcAgNPs for targeted and controlled drug delivery could minimize the risk of emergence of drug tolerant parasites. Also, EcAgNPs might serve as a suitable partner drug in artemisinin based combination therapy (ACTs). In the end, exploration of optimized synthesis methods, combination therapies, and other therapeutic applications of E. cotinifolia is promising for the development of an effective treatment for malaria. In the wake of unavailability of an effective malaria vaccine and dried pipelines of antimalarials, malaria poses a formidable challenge. Thus, development of these plant-derived AgNPs represents a significant stride towards sustainable and effective malaria treatment strategies. However, further investigations are indeed imperatively required to elucidate the mechanisms underlying their action to assess the safety and efficacy of these AgNPs in clinical settings.
P. falciparum clearance from the in vitro routine culture was induced by the biogenic green nanoparticles (EcAgNPs) synthesized from Euphorbia cotinifolia. Modulation of ROS generation, ER Stress, and apoptotic-like processes via the employed intricate mechanisms. Bio-synthesized green nanoparticles (EcAgNPs) were produced and characterized using Euphorbia cotinifolia plant latex extracts. These nanoparticles demonstrated the ability to modulate the generation of ROS by dysregulating the activity of antioxidants such as glutathione (GSH) and thio-redoxin reductase. Treatment with these nanoparticles led to an augmentation in the activity of NADPH oxidase, thereby regulating Protein Disulfide Isomerase (PDI) activity. This process activated the Endoplasmic Reticulum (ER) stress and calpain activity, subsequently inducing the release of calcium from the ER. The green nanoparticles treatment increased the Bax/Bcl-2 ratio, resulting in the release of Cytochrome and activating caspase-3 to induce apoptosis of P. falciparum infected huRBCs
5 Conclusion and future perspectives
Present work suggests the therapeutic value of chemically synthesized silver nanoparticles of Euphorbia cotinifolia. The synthesized EcAgNPs showed the antimalarial activity, as confirmed by the increased ROS production and disrupting the redox equilibrium of parasite leading to induced apoptosis of P. falciparum hosting RBCs. Furthermore, the modulation of NAPDH oxidase, antioxidant enzyme system, and ER stress and inhibited expression of PfLPL- are the key assays showing the clearance of parasite by the EcAgNPs. This work warrants further investigation to optimize the EcAgNPs synthesis as well as its use as a partner drug in the artemisinin based combination therapy. Furthermore, we are in the process of validating the antimalarial activity of EcAgNPs in the rodent model of P. berghei infection followed by the challenge (huRBCs reconstituted immunodeficient mice receiving infection with P. falciparum) model of P. falciparum infection. In the end, we believe the green synthesis of EcAgNPs might be a therapeutic options for malaria treatment.
Data availability
Data is provided within the manuscript and supplementary information files.
Abbreviations
- AgNPs:
-
Silver nanoparticles
- BS:
-
Blood stage
- CDNB:
-
L-chloro-2,4-dinitrobenzene
- DCF:
-
2’, 7’-Dichlorofluorescein
- DTNB:
-
5,5′-Dithiobis-(2-nitrobenzoic acid
- EcAgNPs:
-
Euphorbia cotinifolia Silver nanoparticles
- ER:
-
Endoplasmic reticulum
- FTIR:
-
Fourier transform infrared spectroscopy
- GR:
-
Glutathione reductase
- GSH:
-
Glutathione
- GST:
-
Glutathione S-transferase
- huRBCs:
-
Human red blood cells
- NADPH:
-
Nicotinamide adenine dinucleotide phosphate
- PDI:
-
Protein disulfide isomerase
- pRBC:
-
Parasitized RBCs
- RBCs:
-
Red blood cells
- ROS:
-
Reactive oxygen species
- TEM:
-
Transmission electron microscopy
- TrxR:
-
Thioredoxinreductase
- XRD:
-
X-ray diffraction
References
Weiland AS. Recent advances in imported malaria pathogenesis, diagnosis, and management. Curr Emerg Hosp Med Rep. 2023;11(2):49–57.
Chandley P, et al. Host-parasite interactions during Plasmodium infection: implications for immunotherapies. Front Immunol. 2022;13:1091961.
Hussein R, et al. Impact of COVID-19 and malaria coinfection on clinical outcomes: a retrospective cohort study. Clin Microbiol Infect. 2022;28(8):1152-e1.
Venugopal K, et al. Plasmodium asexual growth and sexual development in the haematopoietic niche of the host. Nat Rev Microbiol. 2020;18(3):177–89.
Knackstedt SL, et al. Neutrophil extracellular traps drive inflammatory pathogenesis in malaria. Sci Immunol, 2019. 4(40).
Tyagi RK, et al. High-level artemisinin-resistance with quinine co-resistance emerges in P. falciparum malaria under in vivo artesunate pressure. BMC Med. 2018;16(1):181.
Sanmoung, W, et al. Application of loop-mediated isothermal amplification combined with lateral flow assay visualization of Plasmodium falciparum kelch 13 C580Y mutation for artemisinin resistance detection in clinical samples. Acta Trop. 2023: 106998.
Shibeshi MA, Kifle ZD, Atnafie SA. Antimalarial drug resistance and novel targets for antimalarial drug discovery. Infect Drug Resist. 2020;13:4047–60.
Wang S, et al. A review of malaria molecular markers for drug resistance in Plasmodium falciparum and Plasmodium vivax in China. Front Cell Infect Microbiol. 2023;13:1167220.
Nguyen TD, et al. Preventing antimalarial drug resistance with triple artemisinin-based combination therapies. Nat Commun. 2023;14(1):4568.
Lava M, et al. Characterization, anticancer, antibacterial, anti-diabetic and anti-inflammatory activities of green synthesized silver nanoparticles using Justica wynaadensis leaves extract. 2021. 46: 5942–7.
Nagaraj B., et al. Synthesis of plant mediated gold nanoparticles using flower extracts of Carthamus tinctorius L. (safflower) and evaluation of their biological activities. 2012;7: 1289–1296.
Basavegowda N, et al. Antioxidant and anti-tyrosinase activities of palladium nanoparticles synthesized using Saururus chinensis. 2016. 27: 733–44
Shabbir MA, et al. Synthesis of iron oxide nanoparticles from Madhuca indica plant extract and assessment of their cytotoxic, antioxidant, anti-inflammatory, and anti-diabetic properties via different nanoinformatics approaches. 2023;8(37): 33358–66.
Nzilu DM, et al. Green synthesis of copper oxide nanoparticles and its efficiency in degradation of rifampicin antibiotic. 2023;13(1): 14030.
Chaves JB, et al. Potential of nanoformulations in malaria treatment. Front Pharmacol. 2022;13:999300.
Neves Borgheti-Cardoso L, et al. Promising nanomaterials in the fight against malaria. J Mater Chem B. 2020;8(41):9428–48.
Alven S, Aderibigbe BA. Nanoparticles formulations of artemisinin and derivatives as potential therapeutics for the treatment of cancer, Leishmaniasis and Malaria. Pharmaceutics, 2020;12(8).
Kirtane AR, et al. Nanotechnology approaches for global infectious diseases. Nat Nanotechnol. 2021;16(4):369–84.
Chaudhari, R., et al. Transdermal immunization of elastic liposome-laden recombinant chimeric fusion protein of P. falciparum (PfMSP-Fu(24)) mounts protective immune response. Nanomaterials (Basel), 2021;11(2).
Tyagi RK, et al. Elastic liposome-mediated transdermal immunization enhanced the immunogenicity of P. falciparum surface antigen, MSP-119. Vaccine. 2015;33(36):4630–8.
Rajwar TK, et al. Opportunity in nanomedicine to counter the challenges of current drug delivery approaches used for the treatment of malaria: a review. J Drug Target. 2023;31(4):354–68.
Urban P, Ranucci E, Fernandez-Busquets X. Polyamidoamine nanoparticles as nanocarriers for the drug delivery to malaria parasite stages in the mosquito vector. Nanomedicine (Lond). 2015;10(22):3401–14.
Kumar H, et al. Galactose-anchored gelatin nanoparticles for primaquine delivery and improved pharmacokinetics: a biodegradable and safe approach for effective antiplasmodial activity against P. falciparum 3D7 and in vivo hepatocyte targeting. Mol Pharm. 2017;14(10):3356–69.
Amolegbe SA, et al. Mesoporous silica nanocarriers encapsulated antimalarials with high therapeutic performance. Sci Rep. 2018;8(1):3078.
Marepally S, Sahu T, Tyagi RK. Editorial: Nanomedicine in infectious diseases: drug delivery and vaccines. Front Pharmacol. 2022;13:928572.
Tyagi RK, Garg NK, Sahu T. Vaccination strategies against malaria: novel carrier(s) more than a tour de force. J Control Release. 2012;162(1):242–54.
Salehi B, et al. Euphorbia-derived natural products with potential for use in health maintenance. Biomolecules; 2019;9(8).
Muluye AB, et al. Anti-malarial activity of the root extract of Euphorbia abyssinica (Euphorbiaceae) against Plasmodium berghei infection in mice. Malar J. 2019;18(1):261.
Kemboi D, et al. A review of the ethnomedicinal uses, biological activities, and triterpenoids of euphorbia species. Molecules, 2020;25(17).
Xia M, et al. Anti-inflammatory and anxiolytic activities of Euphorbia hirta extract in neonatal asthmatic rats. AMB Express. 2018;8(1):179.
Elemike EE, et al. Phytosynthesis of silver nanoparticles using aqueous leaf extracts of Lippia citriodora: antimicrobial, larvicidal and photocatalytic evaluations. Mater Sci Eng C Mater Biol Appl. 2017;75:980–9.
Elemike EE, et al. Green synthesis of Ag/Ag(2)O nanoparticles using aqueous leaf extract of eupatorium odoratum and its antimicrobial and mosquito larvicidal activities. Molecules 2017;22(5).
Elyamny S, et al. One-pot fabrication of Ag@Ag(2)O core-shell nanostructures for biosafe antimicrobial and antibiofilm applications. Sci Rep. 2021;11(1):22543.
Avitabile E, et al. The potential antimalarial efficacy of hemocompatible silver nanoparticles from Artemisia species against P. falciparum parasite. PLoS ONE. 2020;15(9):e0238532.
Kojom Foko LP, et al. A systematic review on anti-malarial drug discovery and antiplasmodial potential of green synthesis mediated metal nanoparticles: overview, challenges and future perspectives. Malar J. 2019;18(1):337.
Asif M, et al. Green synthesis of silver nanoparticles (AgNPs), structural characterization, and their antibacterial potential. Dose Response. 2022;20(1):15593258221088708.
Sudhakar R, et al. Bazedoxifene, a postmenopausal drug, acts as an antimalarial and inhibits hemozoin formation. Microbiol Spectr. 2022;10(3):e0278121.
Trager W, Jensen JB. Human malaria parasites in continuous culture. Science. 1976;193(4254):673–5.
Lambros C, Vanderberg JP. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J Parasitol. 1979;65(3):418–20.
Johnson JD, et al. Assessment and continued validation of the malaria SYBR green I-based fluorescence assay for use in malaria drug screening. Antimicrob Agents Chemother. 2007;51(6):1926–33.
Arnold L, et al. Further improvements of the P. falciparum humanized mouse model. PLoS ONE. 2011;6(3):e18045.
Fu Y, et al. Dual labeling with a far red probe permits analysis of growth and oxidative stress in P. falciparum-infected erythrocytes. Cytometry A. 2010;77(3):253–63.
Habig WH, Pabst MJ, Jakoby WB. Glutathione S-transferases: the first enzymatic step in mercapturic acid formation. J Biol Chem. 1974;249(22):7130–9.
Ellman GL. Tissue sulfhydryl groups. Arch Biochem Biophys. 1959;82(1):70–7.
Holmgren A, Bjornstedt M. Thioredoxin and thioredoxin reductase. Methods Enzymol. 1995;252:199–208.
Heyneman RA, Vercauteren RE. Activation of a NADPH oxidase from horse polymorphonuclear leukocytes in a cell-free system. J Leukoc Biol. 1984;36(6):751–9.
Lesanavicius M, et al. Reactions of plasmodium falciparum ferredoxin:NADP(+) oxidoreductase with redox cycling xenobiotics: a mechanistic study. Int J Mol Sci, 2020;21(9).
Russo I, et al. A calpain unique to alveolates is essential in Plasmodium falciparum and its knockdown reveals an involvement in pre-S-phase development. Proc Natl Acad Sci USA. 2009;106(5):1554–9.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–8.
Chou ES, et al. A high parasite density environment induces transcriptional changes and cell death in Plasmodium falciparum blood stages. FEBS J. 2018;285(5):848–70.
Engelbrecht D, Coetzer TL. Sunlight inhibits growth and induces markers of programmed cell death in Plasmodium falciparum in vitro. Malar J. 2015;14:378.
Hawadak J, et al. In vitro antiplasmodial activity, hemocompatibility and temporal stability of Azadirachta indica silver nanoparticles. Artif Cells Nanomed Biotechnol. 2022;50(1):286–300.
Tsamesidis I, et al. Effect of silica based nanoparticles against plasmodium falciparum and leishmania infantum parasites. J Xenobiot. 2021;11(4):155–62.
Jayalakshmi B, Raveesha KA, Amruthesh KN. Isolation and characterization of bioactive compounds from Euphorbia cotinifolia. Future J Pharmaceutical Sci. 2021;7(1):9.
Satyavani K, et al. Biomedical potential of silver nanoparticles synthesized from calli cells of Citrullus colocynthis (L.) Schrad. J Nanobiotechnol. 2011;9:43.
Scimeca M, et al. Energy dispersive X-ray (EDX) microanalysis: a powerful tool in biomedical research and diagnosis. Eur J Histochem. 2018;62(1):2841.
Zhang M, et al. Influencing factors and strategies of enhancing nanoparticles into tumors in vivo. Acta Pharm Sin B. 2021;11(8):2265–85.
Joy S, et al. Basal and starvation-induced autophagy mediates parasite survival during intraerythrocytic stages of Plasmodium falciparum. Cell Death Discov. 2018;4:43.
Sharma P, et al. Oleuropein activates autophagy to circumvent anti-plasmodial defense. iScience. 2024;27(4):109463.
Yu X, et al. Ring-stage growth arrest: metabolic basis of artemisinin tolerance in Plasmodium falciparum. iScience. 2023;26(1):105725.
Platon L, Menard D. Plasmodium falciparum ring-stage plasticity and drug resistance. Trends Parasitol;2023.
Coronado LM, et al. Blood stage plasmodium falciparum exhibits biological responses to direct current electric fields. PLoS ONE. 2016;11(8):e0161207.
Potter SM, et al. Phagocyte-derived reactive oxygen species do not influence the progression of murine blood-stage malaria infections. Infect Immun. 2005;73(8):4941–7.
Inacio P, et al. Parasite-induced ER stress response in hepatocytes facilitates Plasmodium liver stage infection. EMBO Rep. 2015;16(8):955–64.
Angrisano F, et al. Protein disulfide isomerases: a way to tackle malaria. Trends Parasitol. 2023;39(8):622–5.
Dominguez-Fernandez T, et al. A calpain-like protein is involved in the execution phase of programmed cell death of entamoeba histolytica. Front Cell Infect Microbiol. 2018;8:339.
Zuzarte-Luis V, Mota MM. Parasite sensing of host nutrients and environmental cues. Cell Host Microbe. 2018;23(6):749–58.
Czabotar PE, et al. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat Rev Mol Cell Biol. 2014;15(1):49–63.
Pawlowski J, Kraft AS. Bax-induced apoptotic cell death. Proc Natl Acad Sci U S A. 2000;97(2):529–31.
Asad M, et al. An essential vesicular-trafficking phospholipase mediates neutral lipid synthesis and contributes to hemozoin formation in Plasmodium falciparum. BMC Biol. 2021;19(1):159.
Kumar B, et al. Metacaspase-3 of plasmodium falciparum: an atypical trypsin-like serine protease. Int J Biol Macromol. 2019;138:309–20.
Singh S, et al. Green synthesized silver nanoparticles of Terminalia bellirica leaves extract: synthesis, characterization, in-silico studies, and antimalarial activity. Artif Cells Nanomed Biotechnol. 2024;52(1):238–49.
Murugan K, et al. In vivo and in vitro effectiveness of Azadirachta indica-synthesized silver nanocrystals against Plasmodium berghei and Plasmodium falciparum, and their potential against malaria mosquitoes. Res Vet Sci. 2016;106:14–22.
Metwally DM, et al. Silver nanoparticles biosynthesized with salvia officinalis leaf exert protective effect on hepatic tissue injury induced by Plasmodium chabaudi. Front Vet Sci. 2020;7:620665.
Zhang XF, et al. Silver nanoparticles: synthesis, characterization, properties, applications, and therapeutic approaches. Int J Mol Sci., 2016. 17(9).
Garg NK, et al. Nanostructured lipid carrier mediates effective delivery of methotrexate to induce apoptosis of rheumatoid arthritis via NF-kappaB and FOXO1. Int J Pharm. 2016;499(1–2):301–20.
Zhang P, et al. Application of silver nanoparticles in parasite treatment. Pharmaceutics 2023;15(7).
Palau M, et al. In vitro antibacterial activity of silver nanoparticles conjugated with amikacin and combined with hyperthermia against drug-resistant and biofilm-producing strains. Microbiol Spectr. 2023;11(3):e0028023.
Deng X, et al. Reduced deformability of parasitized red blood cells as a biomarker for anti-malarial drug efficacy. Malar J. 2015;14:428.
Chotivanich K, et al. The mechanisms of parasite clearance after antimalarial treatment of Plasmodium falciparum malaria. J Infect Dis. 2000;182(2):629–33.
Mishra A, et al. Evaluation of antiplasmodial activity of green synthesized silver nanoparticles. Colloids Surf B Biointerfaces. 2013;111:713–8.
Singh S, et al. Dual role of an essential HtrA2/Omi protease in the human malaria parasite: Maintenance of mitochondrial homeostasis and induction of apoptosis-like cell death under cellular stress. PLoS Pathog. 2022;18(10):e1010932.
Chaudhari R, Sharma S, Patankar S. Glutathione and thioredoxin systems of the malaria parasite Plasmodium falciparum: Partners in crime? Biochem Biophys Res Commun. 2017;488(1):95–100.
Siddiqui G, et al. Peroxide antimalarial drugs target redox homeostasis in plasmodium falciparum infected red blood cells. ACS Infect Dis. 2022;8(1):210–26.
Kavishe RA, Koenderink JB, Alifrangis M. Oxidative stress in malaria and artemisinin combination therapy: pros and cons. FEBS J. 2017;284(16):2579–91.
Atamna H, Ginsburg H. Origin of reactive oxygen species in erythrocytes infected with Plasmodium falciparum. Mol Biochem Parasitol. 1993;61(2):231–41.
Murphy MP, et al. Guidelines for measuring reactive oxygen species and oxidative damage in cells and in vivo. Nat Metab. 2022;4(6):651–62.
Tiwari S, et al. Redox interactome in malaria parasite Plasmodium falciparum. Parasitol Res. 2021;120(2):423–34.
Yadav S, Ahmad F, Rathaur S. Antifilarial efficacy of andrographolide: ex vivo studies on bovine filarial parasite Setaria cervi. Comp Biochem Physiol C Toxicol Pharmacol. 2022;261:109442.
Muruganandham M, et al. Antibacterial, antifungal, antioxidant, and cytotoxicity activities of the aqueous extract of Syzygium aromaticum-mediated synthesized novel silver nanoparticles. 2023;12(1).
Phyo AP, et al. Antimalarial activity of artefenomel (OZ439), a novel synthetic antimalarial endoperoxide, in patients with Plasmodium falciparum and Plasmodium vivax malaria: an open-label phase 2 trial. Lancet Infect Dis. 2016;16(1):61–9.
Al-Olayan EM, et al. Complete development of mosquito phases of the malaria parasite in vitro. Science. 2002;295(5555):677–9.
Kosec G, et al. Metacaspases of Trypanosoma cruzi: possible candidates for programmed cell death mediators. Mol Biochem Parasitol. 2006;145(1):18–28.
Vucicevic K, et al. Association of bax expression and Bcl2/Bax ratio with clinical and molecular prognostic markers in chronic lymphocytic leukemia. J Med Biochem. 2016;35(2):150–7.
Somu P, et al. The Interplay between heat shock proteins and cancer pathogenesis: a novel strategy for cancer therapeutics. Cancers (Basel), 2024;16(3).
Acknowledgements
Savitri Tiwari would like to acknowledge the Delhi University south campus, New Delhi for allowing to use their FACS facility. Also, Savitri acknowledges ICMR for providing funds to carry out this work. Rajeev Tyagi expresses his thanks to MIL facility of CSIR-Institute of Microbial Technology (IMTECH), Chandigarh. Rajeev Tyagi would like to offer sincere thanks to DBT, New Delhi and the Government of India for financially supporting this study in the form of the Ramalingaswami Re-entry Fellowship-2019 (D.O. NO. BT/HRD/35/02/2006) Sanction order (BT/RLF/Re-entry/27/2018). Also, Indian Council of Medical Research (ICMR), New Delhi extramural grant (35/1/2020-Nano/BMS) generously supporting this study is duly acknowledged. Nikunj Tandel would like to thank the Nirma University and the Indian Council of Medical Research (ICMR), New Delhi, Government of India, for providing a fellowship to carry out his research (ICMR SRF No.2020-7623/CMB-BMS).
Funding
Department of Biotechnology, Ministry of Science and Technology, India, BT/RLF/Re-entry/27/2018, Indian Council of Medical Research, 35/1/2020-Nano/BMS.
Author information
Authors and Affiliations
Contributions
Conceptualization, ST, RKT, design and execution of the experiments, ST, RK, SD, NT, SM materials supply, PS, NRC, SN, NT, SP, SM writing – original draft preparation, RKT, ST, writing – review and editing, NT, RKT.
Corresponding author
Ethics declarations
Ethical approval
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1
. SI Figure 1 Evaluation of morphological changes and growth arrest at ring stage on the Giemsa stained thin blood smears drawn from the parasite after 6 and 12 h following the treatment with 5 µg/ml EcAgNPs. SI Figure 2 Relative mRNA expression of Bax and Bcl-2 in the P. falciparum treated with EcAgNPsindicating the induction of apoptosis.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Tiwari, S., Kumar, R., Devi, S. et al. Biogenically synthesized green silver nanoparticles exhibit antimalarial activity. Discover Nano 19, 136 (2024). https://doi.org/10.1186/s11671-024-04098-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s11671-024-04098-2