Abstract
Background
The transcription factor TWIST1 plays an important role in the epithelial–mesenchymal transition (EMT) process and in the migration, invasion and metastasis of cancer cells. OCT4, which is a homeobox transcription factor, has an important role in the self-renewal potential of cancer cells. Our aim here is to elucidate impact of ectopic expression of TWIST1 on OCT4 gene expression in esophageal squamous cell carcinoma (ESCC).
Methods
The ESCC line was KYSE30. GP293T cells were transfected with purf-IRES-GFP and pGP plasmids to produce recombinant viral particles. A semi-confluent KYSE30 culture was transduced with the prepared retroviral particles. mRNA extraction and cDNA synthesis were performed from normal KYSE30 cells and those ectopically expressing TWIST1. Expressional analysis of TWIST1 and OCT4 were performed with relative comparative real-time PCR.
Results
Ectopic expression of TWIST1 in KYSE30 cells was related to its significant overexpression: nearly nine-fold higher in GFP-hTWIST1 KYSE-30 cells than in control GFP cells. This induced expression of TWIST1 caused significant upregulation of OCT4 in GFP-hTWIST1 KYSE-30 cells: nearly eight-fold higher. In silico analysis predicted the correlation of TWIST1 and OCT4 through ETS2.
Conclusions
Overexpressed TWIST1 can be correlated with upregulation of the cancer stem cell marker OCT4 and the protein may play critical regulatory role in OCT4 gene expression. Since OCT4 is involved in the self-renewal process, the results may suggest a new linkage between TWIST1 and OCT4 in the cell biology of ESCC, highlighting the probable role of TWIST1 in inducing self-renewal.
Similar content being viewed by others
Introduction
Esophageal cancer (EC) is the eighth most common cancer and the sixth leading cause of cancer-related death worldwide [1]. Geographical factors, local culture and ethnicity have an influence on the incidence rate of EC in different regions [2].
Esophageal squamous cell carcinoma (ESCC) is the sixth most common cancer among men and ninth among women worldwide. Although surgery is therapeutically useful in the early stages of ESCC, most patients are diagnosed in the late stages of the disease, when common therapeutic methods, including surgery, chemotherapy and radiotherapy, are not effective enough to inhibit recurrence. Therefore, a more effective targeted therapy is needed to increase ESCC patient survival [3].
TWIST1 belongs to a class of transcription regulators with a basic helix-loop-helix (bHLH) DNA-binding domain. It identifies a hexanucleotide consensus sequence called E-box (CANNTG) in the promoter region of target genes [4]. Having recognized the E-boxes, TWIST1 can regulate downstream gene expression.
TWIST1 is also known to be involved in the complex process of epithelial–mesenchymal transition (EMT), which plays a role in the migration of cells during their development. EMT is also believed to have an important role in tumor invasion and metastasis, with TWIST1 upregulation enhancing this ability in different types of cancer cells [5, 6] including melanoma, nasopharyngeal carcinoma, ESCC, and breast, uterine, prostate, pancreatic, gastric and cervical cancers [7,8,9]. TWIST1 overexpressing cells show a significantly elevated level of cancer stem cell-like traits, such as tumorsphere formation, ALDH1 and CD44 gene expression, and activated β-catenin and Akt pathways [10].
OCT4 belongs to the POU domain family, the members of which play an important role during embryonic development. It is a multifunctional factor involved in stem cell self-renewal and differentiation, and in carcinogenesis [11]. OCT4 expression has been proved in mouse and human inner cell mass (ICM) cells, embryonic stem cells, germ cells, embryonic carcinoma cells and embryonic germ cells (pluripotent cells). It is activated mostly in undifferentiated stem cells [12, 13].
As the main stemness state marker, OCT4 is expressed in over 93% of ESCCs [14]. The expressions of stem cell markers such as OCT4 and survivin are closely related to the surgical stages of the disease and correlate with poor survival of patients. Since OCT4- or survivin-positive tumors are associated with much poorer prognosis than OCT4- or survivin-negative tumors [15], there may be a correlation between these stem cell markers and other markers of poor prognosis in ESCC, such as TWIST1.
Our aim was to evaluate the effect of TWIST1 upregulation on OCT4 gene expression in an ESCC cell line, KYSE30, and to evaluate a probable new route relating the contributions of these two important genes to ESCC development.
Materials and methods
In silico sequence analysis
The mRNA and gene sequences of OCT4 were obtained from Genebank (accession numbers NM_203289 and NC_000006.12, respectively). The sequence analysis was conducted using CLC Main Workbench version 7 (CLC bio).
Cell lines and culture conditions
KYSE-30 and GP293T cell lines were purchased from the Pasteur Institute and respectively cultured in RPMI1640 or DMEM containing 10% fetal bovine serum (FBS) and 1% pen-strep at 37 °C in a 95% humidity atmosphere with 5% CO2.
Retroviral transduction and overexpression study
The GP293T cell line was transfected with 5 μg of plasmid purf-IRES-GFP (pruf-IRES-GFP-hTWIST1) and 4 μg of pGP plasmid in 500 ml of DMEM without supplements, using X-tremeGENE HP DNA reagent (Roche Diagnostics GmbH), as described previously [16, 17]. Infectious particles were harvested from the supernatant and filtered through a 0.45-μm Nalgene filter (Nalgene Labware).
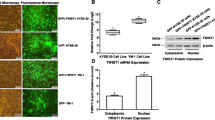
A semi-confluent KYSE-30 culture (1 × 105 cells/6-well in RPMI-60 + 10% FBS) was transduced with prepared recombinant retroviral particles. To determine the transfection accuracy, inverted fluorescence microscopy (Olympus IX-70) was used to observe stably transduced highly expressing GFP (control) and GFP-hTWIST1 KYSE-30 cells (>95% positive).
Comparative real-time PCR
Total RNA was extracted from 3 × 103 of both GFP-hTWIST and GFP-control KYSE-30 cells and transduced KYSE-30 cells using an RNeasy Mini Kit (QIAGEN). After cDNA synthesis using an Easy cDNA Reverse Transcription Kit (Fermentas), relative comparative real-time PCR of TWIST1 and OCT4 mRNA expression was performed using SYBR green PCR Master Mix (Fermentas) on a Stratagene Mx-3000P Real-Time Thermocycler (Stratagene) with the primer sets shown in Table 1. The thermal profile was 10 min at 95 °C followed by 40 cycles of 15 s at 95 °C, 30 s at 57 °C, and 45 s at 72 °C. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used to normalize gene expression, and PCR efficiencies for GAPDH and OCT4 were verified by generating standard curves [18].
Prediction of gene–gene interaction
The probable predictive connection between OCT4 and TWIST1, including annotation, cotreatment expression and protein–protein interaction was obtained using the biograph database (http://biograph.be/).
Statistical analysis
Data analyses were performed using the SPSS 19.9 statistical package. The correlation between gene expressions was assessed using either the χ2 or Fisher exact tests and Pearson’s correlation. p < 0.05 was considered statistically significant.
Results
Sequence analysis of OCT4 promoter
The sequences of the OCT4 transcription unit and its upstream region were examined for the existence of probable E-boxes. Seven different E-boxes were found in a 2-kb region upstream of the OCT4 transcription start site. Interestingly, two of the E-boxes were located close to the transcriptions start site, in positions −60 and −380. Other E-boxes were scattered from −609 to −1740 (Fig. 1). Furthermore, there were 85 E-boxes in the OCT4 transcription unit, of which 20 were located in exonic regions, while the remainder were distributed in the introns (Table 2).
Forced expression of TWIST1 upregulated OCT4 expression
After the transduction of KYSE30 cells, a functional study was performed to evaluate the expression of TWIST1 and OCT4. TWIST1 was overexpressed nearly nine-fold in GFP-hTWIST1 KYSE30 cells compared to the control. This level of TWIST1 overexpression caused a significant increase in OCT4 gene expression: nearly eight-fold in GFP-hTWIST1 KYSE30 cells compared to the control (Fig. 2a). The phase contrast and fluorescent microscopy images of GFP-hTWIST1 and GFP control KYSE30 cells are shown in Fig. 2b.
Enforced expression of TWIST1 upregulates OCT4 mRNA expression in KYSE30 cells. a – TWIST1 is overexpressed nine-fold in GFP-hTWIST1 compared to the control. This caused a statistically significant eight-fold increase in OCT4 mRNA expression. b – Phase contrast and fluorescent microscope images of GFP-hTWIST1 and control cells. The phase contrast images show the cells after 24 h and the fluorescent images are after 48 h
TWIST1 and OCT4 may be linked through the ETS2 gene
Having checked the probable interactions of TWIST1 and OCT4, we found that ETS2 can act as an intermediate communicator for these markers (Fig. 3a). TWIST1 can indirectly interact with OCT4 through ETS2 via protein–protein interaction. There is also predicted evidence for indirect interaction of these genes in different cellular processes, such as morphogenesis and regulation of gene transcription, through their DNA- or transcription factor-binding activities.
Computational relationship between genes based on the biograph database: http://www.biograph.be. a – Computational relationship between the TWIST1 and OCT4 genes. b – Computational relationship between the TWIST1 and ETS2 genes. c – Computational relationship between the OCT4 and ETS2 genes
Discussion
As a bHLH transcription factor, TWIST1 effectively plays a role in EMT, with a controlling function in the migration, invasion and metastasis of cancer cells [19]. Its overexpression has been reported in a variety of invasive tumors [20,21,22,23,24,25,26,27,28,29,30]. Such an elevated level of TWIST1 expression may have significant impact on the cellular transcription network and change cell behavior through deregulation of different cell signaling pathways [31].
In this study, TWIST1 was ectopically expressed in KYSE30 cells and its effect on the transcription of the stemness state marker OCT4 was investigated. Interestingly, TWIST1-transduced cells showed higher levels of OCT4 expression than the GFP control cells. This may both highlight the impact of TWIST1 on OCT4 expression and introduce a novel link between TWIST1 and the stemness state of cancer cells.
The crosstalk between TWIST1 and intermediaries of different cell signaling pathways has previously been demonstrated. It has been reported that TWIST1 has an inverse correlation with SNAIL in ESCC KYSE30 cells, with the suggestion that enforced expression of TWIST1 may negatively regulate SNAIL expression [16]. Furthermore, a new connection between TWIST1 and the testicular cancer antigen MAGEA4 was recently reported for KYSE30 cells. The performed functional study on ESCC cells showed an elevated level of MAGEA4 expression after TWIST1 ectopic expression, and confirmed indirect binding of TWIST1 to the MAGEA4 promoter region leading to increased expression of MAGEA4 at both the mRNA and protein levels [17].
In addition, a significant correlation between TWIST1 and MAML1, the main transcription factor of the Notch signaling pathway, was reported for ESCC patients through advanced stages of the disease, suggesting new crosstalk between these markers in ESCC invasion and metastasis [32].
This evidence clearly shows the great potential of TWIST1 in transcription regulation of a wide spectrum of genes involved in various cell signaling pathways. In line with this statement, the impact of TWIST1 ectopic expression on OCT4 gene expression was revealed in this study. The OCT4 upstream region consists of a proximal promoter located near the transcription start site and a number of distal enhancers. As shown in Fig. 2, there are seven different E-boxes in the OCT4 promoter region. TWIST1 may bind either directly or indirectly to these sequences and transcriptionally upregulate OCT4 gene expression. Confirming this correlation, our in silico analysis introduced ETS2 protein as an important linkage between TWIST1 and OCT4. As depicted in Fig. 3, TWIST1 and OCT4 can be associated with ETS2 through protein–protein interactions. Figure 3b shows some predicted paths of connection between TWIST1 and ETS2.
ETS is a downstream target of ERK1/2 and one the mitogen-activated protein kinase-dependent transcription factors, which can interact with coregulatory transcription factors, including bHLH transcription factors such as TWIST1. Based on its partner type, it can activate or repress the transcription of numerous target genes involved in cancer cell progression and invasion [33, 34].
The ability of TWIST1 to interact with ETS2 may be correlated to the capability of TWIST1 to repress Ras-mediated activation of p16INK4A in epithelial cancer cells [35]. An association between ETS2 and TWIST1 was also confirmed in Helicobacter pylori-infected gastric cancer cells (GCCs), where the two genes were found to enhance SIAH2 expression [36].
OCT4 also contributed significantly to the progression of ESCC. It has been reported to positively regulate survivin expression, promoting cancer cell proliferation and leading to a poor prognosis and poor survival of ESCC patients [15]. Furthermore, a possible role of OCT4 in identifying putative cancer stem cells in ESCC pathobiology has been determined [37].
It has been revealed that OCT4 and ETS2 may be linked together. OCT4 can prevent the tendency of pluripotent cells to differentiate through its ability to repress the ETS2 activity. A soluble complex containing these proteins is recognized in fully pluripotent ES cells [38]. Figure 3c depicts predicted linkages between OCT4 and ETS2.
Here, we showed the correlation between TWIST1 and OCT4 and hypothesized the transcriptional regulation of OCT4 by TWIST1. To confirm this statement, further experiments are required. For example, the generation of truncated mutants of the OCT4 promoter region and luciferase assays are needed to determine which E-box sites (Fig. 1) are essential to TWIST1-mediated regulation of OCT4 expression. Chromatin immunoprecipitation assays (CHiP) may help to elucidate the role of TWIST1 binding to the OCT4 promoter. In addition, an electrophoretic mobility shift assay (EMSA) would determine the direct or indirect binding of TWIST1 to the OCT4 promoter.
Conclusions
We showed that ectopic expression of TWIST1 in KYSE30 cells can upregulate OCT4 gene expression at the mRNA level. Based on the existence of different E-boxes in the OCT4 promoter region sequence, it may be hypothesized that TWIST1 binds either directly or indirectly to these and thus regulates OCT4 gene expression.
To the best of our knowledge, this is the first report on the connection between TWIST1 and OCT4 to introduce a correlation between TWIST1 and the stemness state in cells of the ESCC line KYSE30. Such linkage may expand our understanding of the biological role of TWIST1 in cancer cell progression and self-renewal.
Abbreviations
- bHLH:
-
Basic helix-loop-helix
- cDNA:
-
Complementary DNA
- CSCs:
-
Cancer stem cells
- DMEM:
-
Dulbecco’s Modified Eagle’s medium
- EMT:
-
Epithelial–mesenchymal transition
- ESCC:
-
Esophageal squamous cell carcinoma
- ETS2:
-
ETS proto-oncogene 2
- FBS:
-
Fetal bovine serum
- GAPDH:
-
Glyceraldehyde 3-phosphate dehydrogenase
- GFP:
-
Green fluorescent protein
- ICM:
-
Inner cell mass
- OCT4:
-
Octamer-binding transcription factor 4
References
Harirchi I, Kolahdoozan S, Hajizadeh S, Safari F, Sedighi Z, Nahvijou A, et al. Esophageal cancer in Iran; a population-based study regarding adequacy of cancer surgery and overall survival. Eur J Surg Oncol. 2014;40:352–7.
Schrump DS, Nguyen DM. Novel molecular targeted therapy for esophageal cancer. J Surg Oncol. 2005;92:257–61.
Wang L-S, Chow K-C, Chi K-H, Liu C-C, Li W-Y, Chiu J-H, et al. Prognosis of esophageal squamous cell carcinoma: analysis of clinicopathological and biological factors. Am J Gastroenterol. 1999;94:1933–40.
Ansieau S, Morel A, Hinkal G, Bastid J, Puisieux A. TWISTing an embryonic transcription factor into an oncoprotein. Oncogene. 2010;29:3173–84.
Li Q-Q, Xu J-D, Wang W-J, Cao X-X, Chen Q, Tang F, et al. Twist1-mediated adriamycin-induced epithelial-mesenchymal transition relates to multidrug resistance and invasive potential in breast cancer cells. Clin Cancer Res. 2009;15:2657–65.
Yang Z, Zhang X, Gang H, Li X, Li Z, Wang T, et al. Up-regulation of gastric cancer cell invasion by Twist is accompanied by N-cadherin and fibronectin expression. Biochem Biophys Res Commun. 2007;358:925–30.
Hoek K, Rimm DL, Williams KR, Zhao H, Ariyan S, Lin A, et al. Expression profiling reveals novel pathways in the transformation of melanocytes to melanomas. Cancer Res. 2004;64:5270–82.
Martin TA, Goyal A, Watkins G, Jiang WG. Expression of the transcription factors snail, slug, and twist and their clinical significance in human breast cancer. Ann Surg Oncol. 2005;12:488–96.
Lee K-W, Kim JH, Han S, Sung C-O, Do I-G, Ko Y-h, et al. Twist1 is an independent prognostic factor of esophageal squamous cell carcinoma and associated with its epithelial–mesenchymal transition. Ann Surg Oncol. 2012;19:326–35.
Li J, Zhou BP. Activation of β-catenin and Akt pathways by Twist are critical for the maintenance of EMT associated cancer stem cell-like characters. BMC Cancer. 2011;11:49.
Tysnes BB. Tumor-Initiating and-Propagating Cells: Cells That We Would to Identify and Control. Neoplasia. 2010;12:506–15.
Kellner S, Kikyo N. Transcriptional regulation of the Oct4 gene, a master gene for pluripotency. Histol Histopathol. 2010;25:405.
Ashok KA, Reddy K. Oct-4: more than a pluripotent marker. Yakhteh Med J. 2009;11:1–12.
Zhou X, Huang G-R, Hu P. Over-expression of Oct4 in human esophageal squamous cell carcinoma. Mol Cells. 2011;32:39–45.
Li C, Yan Y, Ji W, Bao L, Qian H, Chen L, et al. OCT4 positively regulates Survivin expression to promote cancer cell proliferation and leads to poor prognosis in esophageal squamous cell carcinoma. PloS One. 2012;7:e49693.
Forghanifard MM, Ardalan Khales S, Farshchian M, Rad A, Homayouni-Tabrizi M, Abbaszadegan MR. Negative Regulatory Role of TWIST1 on SNAIL Gene Expression. Pathol Oncol Res. 2016:1–6.
Forghanifard MM, Rad A, Farshchian M, Khaleghizadeh M, Gholamin M, Moghbeli M, et al. TWIST1 upregulates the MAGEA4 oncogene. Molecular Carcinogenesis. 2016;
Rubie C, Kempf K, Hans J, Su T, Tilton B, Georg T, et al. Housekeeping gene variability in normal and cancerous colorectal, pancreatic, esophageal, gastric and hepatic tissues. Mol Carcinog. 2005;19:101–9.
Qin Q, Xu Y, He T, Qin C, Xu J. Normal and disease-related biological functions of Twist1 and underlying molecular mechanisms. Cell Res. 2012;22:90–106.
Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–39.
Kwok WK, Ling M-T, Lee T-W, Lau TC, Zhou C, Zhang X, et al. Up-regulation of TWIST in prostate cancer and its implication as a therapeutic target. Cancer Res. 2005;65:5153–62.
Lee TK, Poon RT, Yuen AP, Ling MT, Kwok WK, Wang XH, et al. Twist overexpression correlates with hepatocellular carcinoma metastasis through induction of epithelial-mesenchymal transition. Clin Cancer Res. 2006;12:5369–76.
Yang MH, Chen CL, Chau GY, Chiou SH, Su CW, Chou TY, et al. Comprehensive analysis of the independent effect of twist and snail in promoting metastasis of hepatocellular carcinoma. Hepatol. 2009;50:1464–74.
Yuen HF, Chua CW, Chan YP, Wong YC, Wang X, Chan KW. Significance of TWIST and E-cadherin expression in the metastatic progression of prostatic cancer. Histopathol. 2007;50:648–58.
M-y F, Wang K, Song H-t, Yu H-w, Qin Y, Shi Q-t, et al. Metastasis-induction and apoptosis-protection by TWIST in gastric cancer cells. Clin Exp Metastasis. 2009;26:1013–23.
Yuen H-F, Chan Y-P, Wong ML-Y, Kwok W-K, Chan K-K, Lee P-Y, et al. Upregulation of Twist in oesophageal squamous cell carcinoma is associated with neoplastic transformation and distant metastasis. J Clin Pathol. 2007;60:510–4.
Sasaki K, Natsugoe S, Ishigami S, Matsumoto M, Okumura H, Setoyama T, et al. Significance of Twist expression and its association with E-cadherin in esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2009;28:1.
Zhang Z, Xie D, Li X, Wong Y-C, Xin D, Guan X-Y, et al. Significance of TWIST expression and its association with E-cadherin in bladder cancer. Hum Pathol. 2007;38:598–606.
Wallerand H, Robert G, Pasticier G, Ravaud A, Ballanger P, Reiter RE, et al. The epithelial-mesenchymal transition-inducing factor TWIST is an attractive target in advanced and/or metastatic bladder and prostate cancers. In: Urologic Oncology: Seminars and Original Investigations: Elsevier. Urol Oncol. 2010;28(5):473–9.
Satoh K, Hamada S, Kimura K, Kanno A, Hirota M, Umino J, et al. Up-regulation of MSX2 enhances the malignant phenotype and is associated with twist 1 expression in human pancreatic cancer cells. Am J Pathol. 2008;172:926–39.
Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818–29.
Forghanifard MM, Moaven O, Farshchian M, Montazer M, Raeisossadati R, Abdollahi A, et al. Expression analysis elucidates the roles of MAML1 and Twist1 in esophageal squamous cell carcinoma aggressiveness and metastasis. Ann Surg Oncol. 2012;19:743–9.
Petrovics G, Liu A, Shaheduzzaman S, Furasato B, Sun C, Chen Y, et al. Frequent overexpression of ETS-related gene-1 (ERG1) in prostate cancer transcriptome. Oncogene. 2005;24:3847–52.
Seth A, Watson DK. ETS transcription factors and their emerging roles in human cancer. Eur J Cancer. 2005;41:2462–78.
Ansieau S, Bastid J, Doreau A, Morel A-P, Bouchet BP, Thomas C, et al. Induction of EMT by twist proteins as a collateral effect of tumor-promoting inactivation of premature senescence. Cancer Cell. 2008;14:79–89.
Das L, Kokate SB, Rath S, Rout N, Singh SP, Crowe SE, et al. ETS2 and Twist1 promote invasiveness of Helicobacter pylori-infected gastric cancer cells by inducing Siah2. Biochem J. 2016;473:1629–40.
Vaiphei K, Sinha SK, Kochhar R. Comparative Analysis of Oct4 in Different Histological Subtypes of Esophageal Squamous Cell Carcinomas in Different Clinical Conditions. Asian Pac J Cancer Prev. 2014;15:3519–24.
Gupta R, Ezashi T, Roberts RM. Squelching of ETS2 transactivation by POU5F1 silences the human chorionic gonadotropin CGA subunit gene in human choriocarcinoma and embryonic stem cells. Mol Endocrinol. 2012;26:859–72.
Acknowledgments
The authors gratefully acknowledge their colleagues at the Division of Human Genetics of the Immunology Research Institute, MUMS, for their kind technical support.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial or not-for-profit sectors.
Availability of data and materials
Not applicable.
Author information
Authors and Affiliations
Contributions
MHI performed the majority of the work presented in this manuscript. MRA made a critical scientific revision of the manuscript. YF was involved in drafting and editing. MMF designed the concept, conducted the experiments, analyzed data, and drafted and edited the manuscript. All the authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable
Consent for publication
The authors read and approved the manuscript.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Izadpanah, M.H., Abbaszadegan, M.R., Fahim, Y. et al. Ectopic expression of TWIST1 upregulates the stemness marker OCT4 in the esophageal squamous cell carcinoma cell line KYSE30. Cell Mol Biol Lett 22, 33 (2017). https://doi.org/10.1186/s11658-017-0065-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s11658-017-0065-x