Abstract
The antigen-receptor genes of vertebrates are rearranged by a specialized somatic recombination mechanism in developing lymphocytes - and, unexpectedly, also in the germline of cartilaginous fishes. The recombination system that carries out these DNA rearrangements may thus be a significant evolutionary force, perhaps not limited to rearrangements at antigen-receptor loci.
Similar content being viewed by others
The vertebrate immune system employs a wide variety of antigen-specific receptors - the immunoglobulins and T-cell receptors - to recognize and neutralize foreign invaders. The receptor diversity necessary to recognize an almost limitless universe of potential pathogens is created by a site-specific DNA rearrangement process termed V(D)J recombination. This unique reaction assembles the receptor genes from separate V, D and J gene segments, a process ostensibly restricted to lymphocytes at a certain stage of their development. In this view, the prediction is that the germline DNA of a given organism should contain unrearranged receptor genes; rearranged versions of the genes should exist only in lymphocytes. This prediction was satisfied by examination of germline and lymphocyte DNA samples from a variety of familiar vertebrates, including mice, humans, birds, and common farm animals [1,2,3]. The fly in the ointment, however, appeared with an analysis of receptor genes in the germline of evolutionarily distant organisms - the cartilaginous fishes. In these primitive vertebrates, many of the immunoglobulin genes are actually found pre-rearranged in the germline (reviewed in [2,3]). This suggests that the V(D)J recombinase may actually be an evolutionary force, a notion that is strongly supported by recent studies in the nurse shark [4]. The evolutionary consequences of these site-specific germline gene rearrangements may reach far beyond the immune system.
The basics of V(D)J recombination
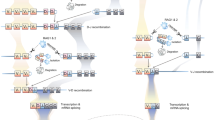
The recombination machinery recognizes DNA sequences called recombination signal sequences (RSSs) adjacent to each gene segment. Each RSS consists of conserved heptamer and nonamer motifs separated by 12- or 23-nucleotide 'spacer' sequences. The recombinase is made up of two proteins, RAG-1 and RAG-2, which, in conjunction with the non-specific DNA-bending proteins, HMG-1 or HMG-2, recognize the RSS and catalyze site-specific DNA cleavage (see [5,6] for review). Cleavage requires both RAG-1 and RAG-2, and co-expression of these proteins is thought to be limited to developing lymphocytes ([7], and reviewed in [6]). As illustrated in Figure 1, the RAG proteins introduce a double-strand DNA break precisely between the V, D or J coding sequence and the RSS, generating two types of DNA ends: blunt signal ends (which terminate in the RSS) and covalently sealed (hairpin) coding ends (which terminate in the V, D or J element). After cleavage, the two signal ends are joined, producing a signal joint. Prior to joining the coding ends, the hairpins must be opened; joining generates a coding joint that may have lost or gained nucleotides.
The precise details of the end-processing and joining reactions remain obscure, and are not important for this story; it is clear, however, that multiple, non-lymphoid-specific DNA repair proteins are involved. One detail that is important is that the opening of the hairpins frequently occurs 'off center', leading to palindromic single-stranded tails. Joining of these ends can give rise to a characteristic signature in the completed junction: a palindromic, or P nucleotide, insertion [8,9]. Another type of junctional insertion, N (non-templated) nucleotides, are added randomly by the enzyme terminal deoxynucleotidyl transferase. Thus, coding joints formed by V(D)J recombination have several distinguishing characteristics, including variable loss of nucleotides and the frequent presence of either N nucleotides, P nucleotides, or both [5].
V(D)J recombination bears many striking parallels to the movement of certain transposable elements, both in its general form and in important mechanistic details of the reaction (reviewed in [10,11]). In fact, recent biochemical experiments have shown that purified RAG proteins can catalyze transposition in the test tube, integrating a DNA fragment bearing signal ends into a target duplex (Figure 2) [12,13]. This reaction does not require specific DNA sequences in the target and, like many transposition reactions, creates a characteristic 'footprint': upon integration, three to five nucleotides of target DNA are duplicated on either side of the transposon. It should be noted that there is, as yet, no firm evidence that RAG-mediated transposition events can occur in living cells.
Surprises from sharks and skates
Early on, it was suggested that the V(D)J recombination system might have arisen by the fortuitous integration of a transposable element into an ancestral antigen-receptor gene [14]. This hypothesis was strengthened by the discovery that the RAG genes are tightly linked [7], and by the finding that the RAG proteins can act as a transposase. Thus, a plausible model for the acquisition of the V(D)J recombination system during vertebrate evolution is the integration of a transposable element carrying the linked RAG genes into a primordial antigen-receptor gene in an ancestral jawed vertebrate, approximately 450 million years ago (reviewed in [1,11]). Presumably, this initial integration event created the first rearranging antigen-receptor gene; subsequent gene duplication events then created the multiple immunoglobulin and T-cell receptor loci.
To learn more about the evolutionary origins of the combinatorial immune system, several laboratories have characterized antigen-receptor loci from a wide variety of species, including the cartilaginous fishes - the living jawed vertebrates most phylogenetically distant from mammals. The immunoglobulin loci of sharks and skates contain some fairly typical antibody genes, with multiple V, D and J elements. Surprisingly, however, many of the immunoglobulin genes are already partially or fully rearranged in the germline (see [2,3] for review). Recently, pre-rearranged immunoglobulin genes were also found in a teleost fish, the channel catfish [15].
How did these pre-rearranged genes arise? One possibility is that they are descendants of the ancestral antigen-receptor gene, before integration of the putative transposable element. A second possibility is that these genes arose from RAG-mediated DNA rearrangement events that occurred in the germline, an operation that violates the precept that the RAG recombinase is functional only in developing lymphocytes. A recent paper by Lee et al. [4] addresses these questions by examining immunoglobulin genes in the nurse shark. The nurse shark NS4 immunoglobulin light chain gene family provided very useful information, as there are several highly homologous genes present both in pre-rearranged and unrearranged forms. These features allowed the authors to evaluate sequences in sufficient detail to ascertain whether the genes bear characteristic features of V(D)J recombination or footprints of transposition. Their analysis revealed that the pre-rearranged genes did indeed contain tell-tale signs of coding joints formed by V(D)J recombination, including both N nucleotides and P nucleotides, which strongly suggest a hairpin intermediate. The presence of these features in several junctions suggests several independent germline V(D)J recombination events, although analysis of multiple unrelated individuals suggests that these events are not frequent. Importantly, analysis of the unrearranged NS4 genes failed to detect the target site duplications that are hallmarks of transposon insertions. Thus, while the pre-rearranged NS4 genes appear to have been derived from unrearranged genes by germline V(D)J recombination, there is as yet no evidence to support the hypothesis that the unrearranged genes were derived from the pre-rearranged genes by insertion of a transposable element; this is discussed in a recent review by Lewis and Wu [16].
The recent studies of the nurse shark NS4 genes strongly suggest that V(D)J recombination events can occur in the germline. Moreover, phylogenetic analysis indicates that these events occurred recently (at least from an evolutionary perspective), some time within the last 7 million years. What evolutionary benefit might there be in germline recombination events? Pre-rearranged immunoglobulin genes may confer certain advantages over genes that must be assembled by recombination in individual lymphocytes. For example, pre-rearranged genes may encode receptors capable of recognizing common pathogens likely to be encountered during the neonatal period, before the development of a full repertoire of rearranged antigen receptors [16]. Furthermore, germline joining could have contributed to evolution of gene segment clusters, and possibly the evolution of D segments [16].
Could the V(D)J recombinase aid the generation of evolutionary diversity on a genome-wide scale?
The results described above raise a number of intriguing questions. Are the RAG proteins normally expressed during germ cell development? To my knowledge, co-expression of both RAG proteins outside the lymphoid system has not been reported, but RNA species encoding RAG-1 and RAG-2 have been detected in zebrafish ovary and Xenopus oocytes, respectively [17,18]. Even if the recombinase is not normally expressed in these tissues, though, inappropriate expression might occur occasionally, perhaps as a result of improper reprogramming of tissue-specific gene expression during development. Such rare events could underlie the apparently infrequent germline rearrangements of the immunoglobulin loci.
If RAG expression does occur during the development of germ cells, another important question arises: how are loci chosen for rearrangement? During normal B-lymphocyte differentiation, immunoglobulin loci undergo a carefully orchestrated series of rearrangements. D to J rearrangements of the heavy-chain genes occur first, followed by V to DJ rearrangements, followed in turn by rearrangement of the light-chain genes. T-cell receptor gene rearrangements in developing T lymphocytes follow a similar pattern. The carefully ordered sequence of rearrangements is critical for proper lymphocyte differentiation and is thought to be controlled by accessibility of the loci, mediated by alterations in chromatin structure (reviewed in [5,19]). RSSs present in antigen-receptor loci that have not been 'targeted' for rearrangement are used rarely, if at all.
The careful control of locus accessibility seen in lymphocyte differentiation may not be recapitulated in the development of germ cells - after all, these cells are not supposed to be expressing RAG recombinase activity. Furthermore, the V(D)J recombinase is not particularly picky about the sequences it can target for rearrangement. Lewis and co-workers have found that 'cryptic' sites capable of supporting rearrangement occur at least once in every 600 base pairs of a commonly used plasmid sequence, and they have estimated that there are at least 10 million such sites in the mammalian genome [20]. In fact, there is evidence that cryptic sites in the mammalian genome can serve as targets for RAG-mediated rearrangement in lymphocytes (reviewed in [5]). Thus, it is possible that RAG expression during germ-cell development might cause rearrangements of regions of the genome far removed from the immune receptor loci. These considerations suggest that the V(D)J recombinase might have been (and could still remain) a significant force shaping vertebrate evolution, by catalyzing V(D)J-like rearrangements and, perhaps, transposition. Comparison of genome sequences from a variety of organisms may allow some aspects of this notion to be tested.
V(D)J recombination occurs in several steps. First, the RAG proteins bind to the RSSs (triangles) and bring them together into a synaptic complex. Cleavage ensues, generating a pair of blunt signal ends and a pair of DNA hairpin coding ends. Joining of these ends generates signal and coding joints, respectively. The boxes represent V, D or J coding elements.
Transposition catalyzed by the RAG proteins. A fragment of DNA generated by RAG-mediated cleavage, with RAG proteins bound to the signal ends (the donor), can capture another DNA duplex (the target). The RAG proteins bound to the signal ends catalyze integration into the target, generating a characteristic duplication of the target sequence at the integration site (arrowheads). Other symbols are as described in Figure 1.
References
Thompson CB: New insights into V(D)J recombination and its role in the evolution of the immune system. Immunity. 1995, 3: 531-539.
Litman GW, Anderson MK, Rast JP: Evolution of antigen binding receptors. Annu Rev Immunol. 1999, 17: 109-147. 10.1146/annurev.immunol.17.1.109.
Marchalonis JJ, Schluter SF, Bernstein RM, Shen S, Edmundson AB: Phylogenetic emergence and molecular evolution of the immunoglobulin family. Adv Immunol. 1998, 70: 417-506.
Lee SS, Fitch D, Flajnik MF, Hsu E: Rearrangement of immunoglobulin genes in shark germ cells,. J Exp Med. 2000, 191: 1637-1648. 10.1084/jem.191.10.1637.
Lewis SM: The mechanism of V(D)J joining: lessons from molecular, immunological and comparative analyses. Adv Immunol. 1994, 56: 27-150.
Fugmann SD, Lee Al, Shockett PE, Villey IJ, Schatz DG: The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu Rev Immunol. 2000, 18: 495-527. 10.1146/annurev.immunol.18.1.495.
Oettinger MA, Schatz DG, Gorka C, Baltimore D: RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990, 248: 1517-1523.
Lafaille JJ, DeCloux A, Bonneville M, Takagaki Y, Tonegawa S: Junctional sequences of T cell receptor γδ genes: implications for γδ T cell lineages and for a novel intermediate of V(D)J joining. Cell. 1989, 59: 859-870.
Lewis SM: P nucleotide insertions and the resolution of hairpin DNA structures in mammalian cells. Proc Natl Acad Sci USA. 1994, 91: 1332-1336.
Roth DB, Craig NL: VDJ recombination: a transposase goes to work. Cell. 1998, 94: 411-414.
Hansen JD, McBlane JF: Recombination-activating genes, transposition, and the lymphoid-specific combinatorial immune system: a common evolutionary connection. Curr Top Microbiol Immunol. 2000, 248: 111-135.
Agrawal A, Eastman QM, Schatz DG: Transposition mediated by RAGI and RAG2 and its implications for the evolution of the immune system. Nature. 1998, 394: 744-751. 10.1038/29457.
Hiom K, Melek M, Gellert M: DNA transposition by the RAGI and RAG2 proteins: a possible source of oncogenic translocations. Cell. 1998, 94: 463-470.
Sakano H, Huppi K, Heinrich G, Tonegawa S: Sequences at the somatic recombination sites of immunoglobulin light chain genes. Nature. 1979, 280: 288-294.
Ghaffari SH, Lobb CJ: Structure and genomic organization of a second cluster of immunoglobulin heavy chain gene segments in the channel catfish. J Immunol. 1999, 162: 1519-1529.
Lewis SM, Wu GE: The old and the restless. J Exp Med. 2000, 191: 1631-1636. 10.1084/jem.191.10.1631.
Greenhalgh P, Olesen CE, Steiner LA: Characterization and expression of recombination activating genes (RAG-1 and RAG-2) in Xenopus laevis. J Immunol. 1993, 151: 3100-3110.
Willett CE, Cherry JJ, Steiner LA: Characterization and expression of the recombination activating genes (rag I and rag2) of zebrafish. Immunogenetics. 1997, 45: 394-404. 10.1007/s002510050221.
Sleckman BP, Bassing CH, Bardon CG, Okada A, Khor B, Bories JC, Monroe R, Alt FW: Accessibility control of variable region gene assembly during T-cell development. Immunol Rev. 1998, 165: 121-130.
Lewis SM, Agard E, Suh S, Czyzyk L: Cryptic signals and the fidelity of V(D)J joining. Mol Cell Biol. 1997, 17: 3125-3136.
Acknowledgements
I thank Vicky Brandt for editorial assistance and Mark Landree for critical comments on the manuscript. Work in the author's laboratory is supported by the Howard Hughes Medical Institute and by grants from the National Institutes of Health, the Human Frontiers Science Program, and the American Cancer Society.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Roth, D.B. From lymphocytes to sharks: V(D)J recombinase moves to the germline. Genome Biol 1, reviews1014.1 (2000). https://doi.org/10.1186/gb-2000-1-2-reviews1014
Published:
DOI: https://doi.org/10.1186/gb-2000-1-2-reviews1014