Abstract
We present an archival format for electron back-scatter diffraction (EBSD) data based on the HDF5 scientific file format. We discuss the differences between archival and data work flow file formats, and present details of the archival file layout for the implementation of h5ebsd, a vendor-neutral EBSD-HDF5 format. Information on sample and external reference frames can be included in the archival file, so that the data is internally consistent and complete. We describe how the format can be extended to include additional experimental modalities, and present some thoughts on the interactions between working files and archival files. The complete file specification as well as an example h5ebsd formatted data set are made available to the reader.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
A significant amount of scientific research is carried out with financial support from government agencies, such as the Department of Education, the Department of Energy, the Department of Defense, the National Institutes of Health, and the National Science Foundation, to name just a few. Since this financial support can be traced back to taxpayer dollars, it stands to reason that taxpayers should have access to the results from government-sponsored scientific research. Such access can take on many forms, from free access to scientific journal articles to access to the actual data produced by the research projects, an issue addressed by a recent response from the White House Office of Science and Technology to a petition regarding such free access [1]. While free scientific journal access will require a dialogue between funding agencies and large publishing companies, access to actual research data presents a more complex issue, in part due to the myriad of file formats used across the scientific community; many of these formats are proprietary, often created by the manufacturers of scientific equipment, but there are several public domain formats that could potentially be suitable for open data access. In this contribution, we discuss the nature of such an open archival data format, and provide an example of an open source format suitable for archiving the results from electron back-scatter diffraction data, a technique that is in widespread use, both in the materials community and the geological community, as well as in the materials industry.
The massive amounts of experimental and simulation data produced by modern characterization instruments and computational platforms also introduce many challenges in terms of scalability, data storage, complexity, high dimensionality, interpretation, and retrieval. This makes it imperative to employ advanced methods for efficient data storage, retrieval, and analysis, thereby providing important opportunities and scope for high performance computing and data mining. Our colleagues in the biological and medical communities long ago realized that a standardized approach to data storage, retrieval, tagging, and visualization is an essential component of a successful research field; they have established an impressive array of searchable and linked databases (from genes, proteins, nucleotides, etc., to cells and organisms), all accessible through a single portal, the National Center for Biotechnology Information[2]. To date, there is no equivalent structures database in the materials community, although the recent report from the National Research Council on Integrated Computational Materials Engineering (ICME) [3] makes a strong case for establishing such a materials-centric database network. The ONR/DARPA-sponsored Materials Atlas, hosted by TMS and Iowa State University [4], is a step in the right direction, but has not received much attention thus far.
Modern materials characterization instruments, often equipped with multiple detector systems, now routinely produce large amounts of data in relatively short time periods. For instance, it is not uncommon for time-dependent synchrotron x-ray tomography data sets to consist of several hundreds of gigabytes of raw data. Similarly, advanced multimodal serial sectioning tools can produce large amounts of raw data, especially when stitching techniques are used to combine multiple fields-of-view into single large area observations. Repeating such measurements on parallel slices can again generate hundreds of gigabytes of raw data. While initial storage of such data sets often employs the proprietary data formats that are part of the instrument’s data acquisition modules, long term archival storage in an open access format has not yet become a standard modus operandi in the materials community. With the present contribution, we attempt to carry out two important tasks: provide an example of a data file format that is uniquely suited for the storage of EBSD data in a vendor-transparent way, and extend this file format to include multi-modal data.
One of the main motivations behind the drive to create a flexible open data file format is the fact that archival data storage is, currently, to a large extent an afterthought. In the materials characterization field, much of the experimental data acquisition is carried out by graduate students, post-doctoral researchers and facilities staff members, and, recently, also by robotic setups. Once an experiment has been completed, the task of data analysis begins. Ideally, however, the first task should be to archive the raw data, so that one can always retrieve it, along with all the meta-data that is relevant to the experiment. Since there is no universal standard for file naming or data organization, it is not uncommon for the raw data to end up in a vulnerable state, i.e., one or more parts of the data could easily get lost or misplaced, potentially rendering the entire data set unusable. Individual data files could end up scattered over multiple storage devices, with no clear pathway to retrieving them. In addition, researchers may not always take sufficient notes for future researchers to be able to decipher the structure of a large data set, including how to read particular file formats; often, the notes regarding a particular experiment do not even end up with the corresponding data files, further increasing the risk of total data loss.
Every materials characterization experiment has a real cost associated with it, and this cost can be quite significant, in particular when personnel time, facilities, utilities, etc. are included. Consider the case of Rowenhorst et al. [5, 6] wherein a set of optical serial sections combined with a subset of EBSD sections of a beta titanium alloy were collected, processed and analyzed. The dataset is comprised of 200 optical micrograph montages and 20 EBSD maps. The total time to collect the dataset was on the order of 200 man-hours plus 260 hours of machine time; image segmentation of the optical images and fusing the EBSD data took ≈600 man-hours, and development and validation of the data analysis tools required the investment of the better part of three man years. All told, this one dataset represents nearly $1M of investment. While much of this cost would not necessarily be repeated in future datasets due to reuse of data analysis codes and improvements in automation, future datasets that utilize higher levels of automation in the collection process will incur higher equipment costs.
With the advent of both faster computing systems and increased storage size at a relatively low cost, the ability to obtain 3D volumes of multi-modal data sets is within financial reach of most laboratories. This wealth of information acquired from the instruments can be instrumental in allowing the materials researcher to obtain new insights into the properties of any particular material system. For instance, in a typical multi-modal setup on a scanning electron microscope (SEM), there are detectors for EBSD (Electron Back-Scatter Diffraction), EDS (Energy Dispersive Spectroscopy), BEI (Back-Scatter Electron Imaging) and SEI (Secondary Electron Imaging). Each modality is acquired by a different detector driven by acquisition software from different vendors, with vastly different storage organizations ranging from simple text and XML files to raw binary files and proprietary file formats. In addition to these issues, each data acquisition system has its own coordinate system to consider and mapping from one system to another may not be immediately obvious. The open data file format introduced in this contribution aims to resolve these data storage issues by storing enough meta-data alongside the raw acquired data; in addition, it should be possible to include the proper coordinate transformations to allow all data sets to be aligned with respect to the same reference frame.
In the remainder of this paper, we introduce the h5ebsd data format for the storage of EBSD data sets. We will initially focus on only a single data modality, and describe how one can define an archival format based on the HDF5 (hierarchical data format [7]) open source standard. We will illustrate this format by means of schematic data layouts. Small data sets will be made available to the reader as Additional files 1 and 2.
Methods
Archival file formats vs. dataflow file formats
At this point it is necessary to clarify our viewpoint on the difference between an archival format and a working data file that would be used within a data processing workflow. The differences between these two scenarios is subtle, but nonetheless significant. The role of the archival data format is to store the information from an experimental measurement exactly as it is represented by the machine, with no processing that alters this information. In addition, the archival file format should strive to include enough metadata describing the experimental set up to make it possible to recollect the same data at a future time (assuming a similar sample is available). In the case of EBSD data collection, this would include acquisition software versions, geometrical calibrations, stage position, electron beam conditions, EBSD camera settings (exposure time, gain, binning, image processing parameters,...), as well as sample information and labeling. In addition, the archival file format should include a description of the relationship between the spatial descriptions within the data and some reference spatial coordinate system. It should be noted, that while this external information is collected, in an archival format the external data should not be allowed to act on the measured information within the dataset. Thus future users of the data have confidence that the data exists in an unaltered state from the point of collection, and has a description of the conditions that provided the data. In this way, the archival data format is very similar to the various RAW imaging formats used in digital photography. A concrete implementation of the H5EBSD archival format can be found in DREAM3D software package (http://dream3d.bluequartz.net) and the accompanying source code at http://www.github.com/DREAM3D/dream3d.git.
In contrast to the archival data format, a working data file format records the result of data analysis operations on the data, with no guarantee that the integrity of the original data is maintained in the final result. At the simplest level this could include the application of an image segmentation method that identifies separate regions of interest within the images. Another example might include, for the ease of further data processing, rearranging the measured data so that it is no longer arranged as separate slices, but rather as a single 3D volume of data described by a single reference frame. Following the analogy from digital photography, these files would be analogous to a TIFF file that has been produced by applying filters to the original RAW image file. Ideally, the parameters of the entire processing pipeline would also be included in this file, including a reference to the original archival data file, thus providing a direct pedigree for the processed data and the ability to recreate that processing pipeline if necessary. An example of this type of file is the.DREAM3D file format currently used in the DREAM3D analysis package; details of how to design such a working data format will be described in a separate article [8].
Basic HDF5 file layout
The Hierarchical Data Format Version 5 (HDF5) is an open-source library developed and maintained by “The HDFGroup” (http://www.hdfgroup.org) that implements a file format designed to be flexible, scalable, performance oriented and portable. HDF5 is superbly flexible in that each application can organize its data in a hierarchy that makes sense for the application. Virtually any type of data, from simple scalar values to complex data structures, can be stored in an HDF5 file. The HDF5 format can handle computing environments ranging from embedded systems to desktop computers to High Performance Computers (HPC) systems. Because scalability has been a design consideration from the outset, HDF5 can handle data objects of almost any size or dimensionality. The library has also been designed to be quick and efficient at reading and writing data objects, including utilizing parallel I/O when needed. In addition to being fast and efficient, the library offers the option to employ one of several types of compression when storing the data objects, always using a lossless compression scheme so that the actual data is never modified or lost. Applications also have the opportunity to implement domain specific compression schemes if needed.
One of the most important aspects of HDF5 is its portability across all the major computing operating systems. By using HDF5 to store data, this data then becomes accessible to any researcher running anything from a consumer level operating system to a huge cluster operating system. Because HDF5 is developed by an entire group of people, application programmers have access to this flexible and scalable storage library for little to no cost. The cost that is incurred is in the developer time to learn the HDF5 functions. From a developer or software engineering point of view, HDF5 has support for “C”, “C++”, Fortran and Java as its native implementations. Many other higher level programming languages also have direct support for HDF5, including IDL (Interactive Data Language), MATLAB and python. This truly makes data stored in an HDF5 file portable and cross platform. The HDFGroup also makes available a free Java based program that allows the user to open, view and export data from within any HDF5 file; therefore, anyone with this viewer program can examine any HDF5 file, regardless of how its contents are organized.
Description of typical EBSD data sets
In a typical EBSD data acquisition run, one scans the electron beam across a region of interest; such a scan employs a preset step size and beam dwell time, which determines the total duration of the run. At each beam position, an electron back-scatter pattern (EBSP) is recorded by the camera; after background subtraction, the pattern geometry is analyzed, usually amounting to the determination of a set of Kikuchi bands by means of a Hough transform. This band information is then matched against a precomputed set of interplanar/interzonal angles, and the most likely indexing of the bands is stored and converted into an Euler angle triplet, expressing the orientation of the crystal reference frame with respect to the sample reference frame. Along with the Euler angles, the indexing program usually produces a confidence index and a pattern quality index; those numbers, combined with the coordinates of the sampling point, are then stored in the experiment’s output file. Depending on the vendor, this file may be a simple ASCII text file, or a binary file with a proprietary data format. Although the original EBSP is usually discarded, the user does have the option to also store each pattern. This increases the size of the data set significantly, but allows for further analysis of the raw data by alternative indexing algorithms, or extraction of other data from the patterns. Once again, the storage format for the individual patterns is vendor-specific, from the standard BMP format to proprietary binary formats.
Tables 1 and 2 show the various outputs reported by two of the major EBSD vendors, EDAX Inc. and Oxford Instruments. Both companies use an ASCII text file to store the indexed data. While there are some minor differences in the actual data items being stored, it is important to realize that this is not the whole story. For instance, to compare data generated on each of these EBSD systems, one must fully understand how each company defines its reference frames, both for the indexed data and for the array of scan points on the sample. In the following section we will describe how to deal with multiple reference frames within the context of HDF5 archival storage.
Results and discussion
Mapping EBSD data onto an HDF5 file
During an EBSD scan, numerous pieces of data are collected at each scan point, including the 2D Kikuchi pattern, the coordinates of the scan point, and the phase identifier at the scan point. Using this information, the acquisition system computes several additional quantities, including the Euler angles (φ1, Φ, φ2) and several diagnostic parameters that provide an indication of the quality of the data point. Working with the various ASCII files can be cumbersome and error prone due to the encoding of the textual data and numerical data, i.e., the differences between European and American decimal formats. In addition to these issues, converting data from an ASCII representation to a binary representation is relatively slow in computer terms and generally the textual representations require more storage space than a compact binary representation. Lastly, with the advent of 3D EBSD data sets, some vendors store each slice of data as a separate file on the file system, making data management and archiving more difficult.
It is in these latter areas that the flexibility of HDF5 excels. During the conversion of EBSD data, from a set of either.ang or.ctf files, the ASCII representations are converted into a standard binary representation where each column of data is stored as a complete block of data, and all slices of a 3D data set are stored within the same HDF5 file. Because of differences in reference frames, some additional meta-data is also gathered from the user at the time of conversion; this includes the z-ordering of the data, the sample and Euler transformation axes/angles, the manufacturer’s name and other meta-data values. For each slice of data from a data set, the header is parsed into values that are stored as individual data sets within the HDF5 file (for easy processing later on); they are also stored as a complete block of text in the HDF5 file in case the original file needs to be reconstructed or parsed values validated. One should note that during the conversion process, no data is ever transformed or converted except to cast the data into a binary representation. The advantage of storing all the data into a single HDF5 file is immediately obvious from a data management point of view. Fewer files on the file system means less chance of a file being lost, which would compromise the entire data set. Another benefit is that during data processing, the time required to read the data from the local storage medium is greatly reduced.
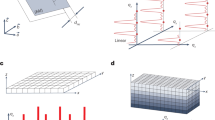
The essential aspect of EBSD data is the recording at a specific spatial location of the orientation of a crystal as related to a crystal reference coordinate system. Due to differing experimental setups as well as tradition, typically the spatial coordinate frame is not coincident with the crystal reference frame. For illustration, Figure 1 shows the typical experimental set-up for EDAX EBSD data collection. Figure 1a shows the view of the sample as it is mounted in the SEM chamber, including the crystal reference frame, RD-TD-ND. In addition, we have included a user defined global reference frame, Xg - Yg - Zg. The choice of the global reference frame is arbitrary but should be clearly defined. Here the global reference frame is defined so that as the observer looks at the sectioning surface, the bottom left-hand corner of the sample will be the origin, with Xg increasing to the right, Yg increasing as one moves up along the surface, and Zg is normal to the surface of the sample, pointing towards the observer. Figure 1b illustrates how the data set would appear within data collection software using the typical SEM and EBSD defaults. Note that the default configuration has the shortest working distance (labeled on the figure as “Top”) at the bottom of the image. Figure 1b also shows the orientation of the scanning (or spatial) reference frame (Xs - Ys - Zs), the crystal reference frame, and the global reference frame within the data collection software. The relationships between these reference frames are critical for the further analysis of the data. Thus, within the h5ebsd file, the orientations of the crystal reference frame and spatial reference frame are recorded as unit vectors which are combined in the global reference frame into a 3×3 array. In this case, Xs is thus equivalent to [-1 0 0] in Xg - Yg - Zg, Ys is [0 1 0] and Zs is [0 0 -1]. Thus the “Scan Reference Directions” within the.h5ebsd file would be recorded as:
Similarly, the “Crystal Reference Directions” are recorded as:
Note that what is represented here is simply the transformation matrix that describes how to transform the coordinate systems into the global reference frame. Thus an alternative, more compact notation is also provided that represents the transformations as angle-axis pairs in “SampleTransformation Angle”/“SampleTransformation Axis” and “EulerTransformation Angle”/“EulerTransformation Axis” respectively.
Schematic of EBSD data collection. (a) Schematic of typical EBSD data collection showing the orientation of the crystal reference frame, RD-TD-ND, as well as a user defined global reference frame, Xg - Yg - Zg. (b) Schematic of how the EBSD data appears within the data collection software with all relevant coordinate frames, including the scanning reference frame, Xs - Ys - Zs.
Another ambiguity that exists within EBSD data is the meaning of the Euler angles recorded for each point. The Euler angles describe the three rotations needed to bring the crystal reference frame into coincidence with the local crystal orientation. However, there are a number of conventions that could be used to describe these rotations. The most common convention used in EBSD data collection is the Bunge notation, where in φ1 represents a rotation of the reference frame about the sample reference Z-axis, Φ is a rotation about the new coordinate frame X-axis, and φ2 is a rotation of the coordinate frame about the new Z-axis. Note that all of these rotations are rotations of the coordinate reference frames, thus they are passive rotations (rotations of the coordinate frame) and not active rotations (rotations of the vectors). The.h5ebsd file records this convention with the attribute “Euler Angle Definition” with the notation “ZXZ+”, indicating the rotations around each of the axes, with the “+” indicating that they describe passive rotations from the crystal reference frame to the local crystal orientation. Alternatively the Bunge Euler angles could describe the passive rotations from the local crystal orientation to the reference crystal frame, in which case this would be recorded within the.h5ebsd file as “ZXZ-”.
Figure 2 shows schematically how the.h5ebsd file format is constructed; red lines correspond to individual data sets, whereas blue lines indicate data groups, which eventually consist of individual data sets. At the top level of the file (left-most box, labeled MY_EBSD_experiment.h5ebsd in this example), one stores information that is of a general nature and pertains to the entire experiment; this includes version numbers, vendor information, and the reference frame information described in the preceding paragraphs. If the data set has multiple slices (as is the case, for instance, in a serial sectioning experiment), then one creates a data group for each slice; this can be compared with the creation of individual folders in a file system. For each slice, we introduce two data groups with the names Header and Data. In the header data group, we store all relevant experimental information about the acquisition of this particular data set, including a description of the individual phases that might be present in the sample; such crystal structure information would be essential for a potential later re-indexing of the individual Kikuchi patterns, which may also be stored in the archive. In the Data group, we store all experimental data, including information extracted from the Kikuchi patterns by the data acquisition program; this includes Euler angles, image quality, phase identifications, a secondary or back-scatter image of the region of interest, and, potentially, all the individual Kikuchi patterns. An example h5ebsd file is made available to the reader as Additional files 1 and 2, and can be analyzed using the public domain Java reader program available from the HDF5 web site.
Extending h5ebsd to multimodal data sets
Extending the archival h5ebsd file to accommodate multimodal data sets is a straightforward process in principle because of the flexibility of the HDF5 format. Each data modality can contain its own reference frame information and modality-specific metadata, and these data objects can be easily appended to the HDF5 file. Note that the data types could have different spatial resolutions, or even correspond to an irregular grid. The archival version of the data format described here does not require any consistency with respect to the sampling densities of different modes.
One should note that ultimately relating each of the individual modalities to a common reference frame requires additional information that may or may not be determined during the data collection process. For example, co-registering two different data modalities usually requires correcting for non-linear spatial distortions within each modality, as well as determining an additional affine transformation matrix to register the modalities to a common reference frame. The serial section data set of Rowenhorst serves as an illustrative example of the type of post-collection spatial registration information that could be included in the archival multimodal format. As mentioned previously, this data set consists of both optical micrograph montages (comprised of multiple image tiles) along with EBSD maps. The EBSD maps were collected on a sparser spatial grid compared to the optical micrograph data. Co-registering all of this data to a common spatial reference frame required removal of non-linear spatial distortions in the EBSD maps, determining translation matrices to create the optical micrograph montages from the individual tiles, calculating the affine transformation to align the EBSD maps to the optical micrograph montages, and determining the serial sectioning removal rate. Ideally, these transformation processes and associated coefficients would be saved to the archival file. Storing this additional information allows for each of the optical micrograph tile and the EBSD maps to be treated as individual (sub-) data sets.
As mentioned, the transformations often must be calculated after the data is collected and initially archived. In some instances, certain transformations may be known ahead of time, i.e. a well characterized distortion field from an optical microscope or the translation matrix of a single tile in a montage. These transformations and any computed after the initial archiving should be stored in the archival file. Each (sub-) data set could potentially have multiple non-linear and affine transformations associated with it. For example, one could imagine computing distortions in an image using multiple methods and in this case each transformation should be stored with the (sub-) data set in the file along with details of how the transformation was obtained. Storing the various transformations separately and with multiple options allows the user to interact with the raw data in a very transparent and flexible manner without ever compromising the data itself.
Thoughts on creating a working file from an archival file
Under the paradigm of the working vs. archival file discussed previously, working with the proposed file format requires establishing a new file. The benefits of this approach are that (1) the raw data remains raw and archived and (2) the working file can take on any sampling strategy desired by the user. This second point is worth discussing in more detail as it illustrates the method by which the user obtains data from the archival file. The user (or a software tool such as DREAM.3D [8]) generates a set of sample points on which they would like to have data. That set of points, which exists in the global reference frame, can be transformed into the reference frames of the individual (sub-) data sets using the transformation matrices in the archival file. Note that if multiple matrices exist to describe either or both the non-linear and affine transformations, the user could easily select the mapping path between the sampling points and the raw data. The transformed sampling points will inevitably fall between points in the raw data images/scans. At that point, multiple methods for inferring the data can be employed. The data on the nearest data point could be used or some form of interpolation (e.g., bilinear) could be used. By transforming the points into the reference frames of the (sub-) data sets and interpolating there, the fewest number of interpolations are applied to the data. Criteria could also be set in case no raw data point of a certain mode is within a specified distance of the transformed sampling point. Note that this process is currently not employed when working with the h5ebsd file, because with a single mode (and currently no example of storage of the non-linear distortions) the raw data grid is treated directly as the grid in the working file.
A powerful aspect of this paradigm is that there is not inherently a resolution associated with a data set. The user can specify any point sampling and the data can be generated quickly from the raw data, not sampled from an already processed or interpolated data set. Uncertainty or error analyses could then be coupled to the choice of sampling points, transformations used and the interpolation technique. Furthermore, the user can specify which modes they are interested in, so down-selecting a multimodal data set to a single mode becomes easy as well. It is important to point out, however, that such operations are only possible if the raw data has been stored in a carefully structured archival data file format.
Conclusions
In this paper, we have described a dedicated archival file format, based on the Hierarchical Data Format HDF5, for the storage of raw data from electron back-scatter diffraction experiments. While the detailed requirements for other experimental modalities will be different from those for the EBSD implementation, we believe that the h5ebsd file format reported on in this article serves as a representative example for how one can adapt the broad HDF5 environment to the specific needs of an experimental modality. Extension to multimodal data sets, for which multiple acquisition modalities would be stored in a single archival file, can proceed along similar lines, and efforts to include back-scattered and secondary electron images and/or optical microscopy images along with EBSD data are currently underway.
It is our hope that the h5ebsd implementation will serve as an example for both the materials community and the instrument vendors of how one can employ a public domain format to provide open access to large data sets in a way that does not require proprietary writers/readers. The most straightforward way in which vendors should be able to accommodate this type of open format would be via new “Save As...” and “Import From...” options in the file menu of their data acquisition programs. While there is definitely a need and a use for the vendor’s original and proprietary file formats, the increasing demands placed on the research community by funding agencies to make experimental data available to the general public will require the use of open data formats. Members of the materials community should work with vendor representatives to ensure that open archival file formats are incorporated into commercial platforms. There are plenty of precedents for such inclusions; for instance, the TIFF file format is a public domain specification and many data acquisition programs have a “Save As TIFF” option in their File menu. We believe it will be in the long-term general interest of the materials community for vendors to start adding “Save As HDF5” and “Import From HDF5” options to their commercial data acquisition and instrument control programs, or, alternatively, allowing for such additions via user scripts.
References
Holdren J: Increasing Public Access to the Results of Scientific Research. 2013. https: //petitions.whitehouse.gov/response/increasing-public-access-results-scientific-research
National Institutes for Health: National Center for Biotechnology Information http://www.ncbi.nlm.nih.gov/
Committee on IntegratedComputationalMaterialsEngineering: Integrated Computational Materials Engineering: A transformational discipline for improved competitiveness and national security. Washington, DC: National Research Council, The National Academies Press; 2008.
Materials Atlas T 2009.https://cosmicweb.mse.iastate.edu/wiki/display/home/Materials+Atlas+Home
Rowenhorst DJ, Lewis AC, Spanos G: Three-dimensional analysis of grain topology and interface curvature in a β -titanium alloy. Acta Materialia 2010, 58(16):5511–5519. 10.1016/j.actamat.2010.06.030
Rowenhorst DJ, Lewis AC: Image processing and analysis of 3-D microscopy data. JOM 2011, 63(3):53–57. 10.1007/s11837-011-0046-x
HDF5 2013.http://www.hdfgroup.org/HDF5/
Jackson M, Groeber M: DREAM.3D: A Digital Representation Environment for the Analysis of Microstructure in 3D. Integrating Mater Manuf Innov 2014, 3: 5. 10.1186/2193-9772-3-5
Acknowledgements
The authors would like to acknowledge stimulating interactions with A.D. Rollet and G.S. Rohrer.
MDG would like to acknowledge financial support from the Air Force Office of Scientific Research, MURI contract # FA9550-12-1-0458. MAJ would like to acknowledge financial support from the Air Force Research Laboratories contract # FA8650-07-D-5800. DRJ would like to acknowledge financial support from the U.S. Naval Research Laboratory under the auspices of the Office of Naval Research and the Office of Naval Research under the Alloys and Joining program.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
The h5ebsd file format is the result of multiple interactions and discussions between all of the co-authors. MJ and MG implemented the h5ebsd file format as a part of the DREAM.3D software environment; DR contributed the section on the various reference frames of importance to EBSD data sets; MU and MG contributed the sections on multi-modal extensions and thoughts on creating a working file from an archival file. MDG contributed the introduction and took charge of the editing of the final version of this paper. All authors read and approved the final manuscript.
Electronic supplementary material
40192_2013_8_MOESM2_ESM.pdf
Additional file 2: Condensed version of Figure 2.(PDF 31 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Jackson, M.A., Groeber, M.A., Uchic, M.D. et al. h5ebsd: an archival data format for electron back-scatter diffraction data sets. Integr Mater Manuf Innov 3, 44–55 (2014). https://doi.org/10.1186/2193-9772-3-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1186/2193-9772-3-4