Abstract
Myxozoan spores were observed in yellowfin goby Acanthogobius flavimanus collected from Suisun Marsh, San Francisco Estuary (SFE). Although histopathological changes associated with the parasite were not observed, the spores formed plasmodia that partially blocked the gastric and intestinal mucosa and gut lumen and may affect the perfomance and survival of the yellowfin goby. Morphological features of the spores resembled Henneguya sp. and molecular analysis of the 18S ribosomal DNA (Domain III) confirmed close similarity to H. rhinogobii and H. pseudorhinogobii isolated from the Japanese freshwater goby. The yellowfin goby myxozoan however, is likely an undescribed species based on phylogenetic analysis and morphologic features. Detailed description of vegetative and spore stages are currently lacking for proposal to a new species of Henneguya. A specific PCR test was developed, which confirmed a 100% prevalence of the parasite among randomly collected gobies in group 1 (N = 30) and group 2 (N = 15) at termination of the study at one month in captivity. The myxozoan was also detected from 18 gobies (12%) that died in the first group within two weeks in captivity. Apparently healthy gobies that served as controls did not reveal the presence of the myxozoan by PCR. This study documents the occurrence of a potentially new species of myxozoan in the yellowfin goby and underscores the detection of a parasitic infection in an introduced fish in the SFE. Although the pathogenesis of the myxozoan was not assessed and the prevalence as reported here is restricted to a comparatively small collection site in Suisun slough, the reemergence, identification, and ecological relevance of the parasite on goby populations in the SFE may be investigated in the future using the specific diagnostic tool developed in this study.
Similar content being viewed by others
Background
The San Francisco Estuary (hereafter SFE) is the largest estuary on the U.S. Pacific Coast. It provides drinking water to 25 million California residents, irrigation water to one of the most productive agricultural economies worldwide, and an open−water habitat to hundreds of native plants and aquatic organisms including 212 exotic and 123 cryptic species of unknown origin (Cohen and Carlton 1998Service RF 2007). Although the SFE is one of the most invaded (Cohen and Carlton 1998) and perturbed (Nichols et al. 1986) water bodies worldwide, determining the occurrence of pathogens and diseases on native and introduced fishes in this estuarine ecosystem has not been a goal of the various fish monitoring surveys of the Sacramento San Joaquin Delta. The yellowfin goby (hereafter goby) Acanthogobius flavimanus is native to northern Asia and Japan (Akihito et al. 2002) and has been an abundant bottom fish (Moyle 2002Feyrer and Healey 2003) following its introduction in the SFE via ballast water and first detection in 1963 (Brittan et al. 1963Dill and Cordone 1997).
This study describes the occurrence and identification of a myxozoan parasite among juvenile yellowfin gobies collected during a monitoring program that assessed the distribution and abundance of various fish species in Suisun Marsh, a critical rearing habitat for juvenile fishes in the SFE. Our goals are two-fold: 1) identify the myxozoan using a molecular-based approach and 2) evaluate the prevalence of infections among collected gobies by designing and using a PCR test specific to the myxozoan. The development and application of this molecular tool is an important first step for specific identification and assessment of the parasite among gobies and other fish species of economic and ecologic importance in the SFE that may be infected with the myxozoan.
Results
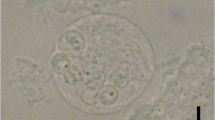
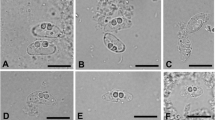
Eighteen of 151 gobies (average size = 1.5 gm, 50 mm) in the first group died within two weeks of captivity, whereas no mortalities occurred in the second group (N = 55, average size: 10.0 gm, 100 mm) until termination of the study after one month in captivity. Although none of the fish died in captivity from the second group, the 15 fish examined for histopathology showed variable presence of the parasite and all were positive by PCR including the 10 fish sampled from group 1. Spores were formed within plasmodia in the gastric and intestinal mucosa that extended into the gut lumen and partially blocked the intestinal cavities (Figure 1) however, microscopic changes associated with the parasite were not observed. Stained sections (hematoxylin and eosin) showed variable presence of myxozoan spores from stomach and intestines but difficult to confirm for vegetative stages. Moribund fish (N = 18) that succumbed to infection in the first group (12%) revealed the presence of spores in the intestine and stomach but gross clinical signs of infection were not observed. In addition, these moribund fish did not show the presence of the myxozoan in other tissues such as the liver, kidney, and gills. Examination for ectoparasites and culture of kidney and spleen tissues on general isolation medium (e.g. blood agar plate) did not reveal any bacterial growth as potential causes of mortality.
Myxozoan spores in the yellowfin goby. A) Spore plasmodia (arrows) partially blocking the intestinal cavity, Scale bar = 333 μm, B) Closer view of spores showing a thin line outside the plasmodium indicating the original intestinal lining (arrow), Scale bar = 67 μm, C) Spores (arrow) in the stomach, Scale bar = 333 μm, and D) Closer view of spores (arrows) in the stomach. Scale bar = 22 μm.
Initial validation of the PCR assay did not show any cross-reaction with Myxobolus sp. and M. cerebralis (Kent et al. 2001). Other myxozoans need to be tested in the future to further confirm the specificity of the PCR method. Using the specific PCR assay, the prevalence of infections with the myxozoan was assessed from randomly collected fish remaining in both groups at the end of the study at one month in captivity. All of the fish examined from group 1 (N = 30 of 123) and group 2 (N = 15 of 40) were positive for the myxozoan by PCR (Table 1). Genomic DNA extracted from the stomach and intestine of healthy goby and used as negative control did not show any amplification using the PCR assay specific to the myxozoan. Freshly dead or moribund gobies from group 1 and fish sampled for histopathology from both groups were not included for estimating the parasite prevalence since these fish were specifically collected and used for spore extraction and identification, and histopathological evaluations.
The spores obtained from the stomach and intestine of moribund gobies were almost spindle-shaped with mildly pointed anterior end. One sporoplasm was present in the spore body with a caudal appendage. Two almost equal polar capsules occupied most half of the spore cavity. The mean dimension of the spores (N = 10): spore body = 5.1 × 2.4 μm, tail = 9.9 μm, and polar capsule = 2.0 × 1.0 μm. Compared to other species of Henneguya found in goby species with body length ranging from 14.2 – 17.8 μm (Kageyama et al. 2009), the yellowfin goby myxozoan is significantly smaller.
The 18 rDNA sequence of the myxozoan (1,987 bp) showed the highest similarity to Henneguya rhinogobii and H. pseudorhinogobii found in fresh water goby Rhinogobius sp. in Japan (Kageyama et al. 2009). The sequence data were deposited in GenBank (Accession No. JN566045). The goby myxozoan also showed the highest similarity with H. pseudorhinogobii and H. rhinogobii by pairwise comparison of the 18S rDNA Domain III (97.9%) and the long region covering Domain I through III (83–84%), which includes three variable regions (Table 2). The percentage similarity scores, especially for Domain III, further support that the goby parasite is closely related to the two myxozoans H. pseudorhinogobii and H. rhinogobii from Japanese gobies (Kageyama et al. 2009). However, the phylogenetic tree showed that the yellowfin goby myxozoan clearly branched separately as supported by high posterior probability value (1.00) suggesting a different species (Figure 2).
Phylogenetic tree of the yellowfin goby myxozoan and closely related myxozoans using the 18S ribosomal DNA. The tree was generated by MrBayes program ver. 3.1.2 based on the 18S rDNA long sequences (ca. 1.3 kb) covering Domain I through III. Posterior probabilities are listed at each node. The branch lengths, indicated by bars, are based on the number of inferred substitutions. Myxobolus albi was chosen as the out-group.
Discussion
The goby myxozoan is consistent with the description of the family Myxobolidae and the genus Henneguya based on morphological criteria of the spore stages (Lom and Arthur 1989Lom and Dykova 19922006). The taxonomic identification of the goby myxozoan was confirmed by molecular analyses showing close similarity to H. rhinogobii and H. pseudorhinogobii previously reported from the Japanese fresh water goby (Kageyama et al. 2009). Confirmatory identification of the goby myxozoan in the genus Henneguya is based on the pairwise comparison of the 18S rDNA conserved region (Domain III), which is 97.9% similar with H. rhinogobii and H. pseudorhinogobii and from phylogenetic analysis by forming a clade with both species of Henneguya from the Japanese goby. While high similarity values were observed among the domain III of the Japanese goby and the yellowfin goby myxozoans, the sequence similarity of the long region (Domain I through III, 83–84%) and the phylogenetic tree suggests the yellowfin goby myxozoan is a different species. The morphologic features (small spore size) further indicate the yellowfin goby myxozoan is different from the freshwater goby myxozoan (Kageyama et al. 2009). An in-depth morphological description of the spores and vegetative stages is currently lacking to support the proposal to a new species of Henneguya. The goby myxozoan will therefore be referred to as Henneguya sp. until additional morphological data are available to support its classification to a new species.
Because myxozoan spores were present in dead, moribund, and in some apparently healthy gobies that were examined, infections were contracted prior to their collection from the field. In this context, gobies may provide reservoirs of infection at Suisun Marsh although the exact mechanism and the pathogenicity of the goby myxozoan have yet to be determined. Considered the second largest genus within Myxosporea, Henneguya is one of the most important groups of pathogens affecting both freshwater and marine fishes (Lom and Dykova 1992). Certain myxozoan species are known agents of serious diseases in fish (Kent et al. 2001). Henneguya infections occur mainly on the gills rendering respiratory failure and mortality due to asphyxia (Lom and Dykova 1992El-Mansy 2002). The Henneguya sp. was not observed in the gill tissues of gobies examined in this study. The only Henneguya species found in intestinal tissues of goby is H. rhinogobii found in the goby Rhinogobius giurinus from China (Kageyama et al. 2009). While the myxozoan was prevalent from gobies in both groups, mortalities (N = 18) only occurred in the first group with smaller size fish. Histopathological changes associated with the parasite were not observed among moribund gobies in the first group. Exposure experiments are warranted to determine if the mortalities are directly attributed to the myxozoan and if certain life stages of the gobies are more susceptible to infections. For these reasons, the significance of the myxozoan as a potential pathogen is currently unknown.
In their native habitats, gobies feed on small fish, benthic crustaceans, and worms (Kikuchi and Yamashita 1992Hironouchi and Sano 2000Workman and Merz 2007). Fish and oligochaetes are the fundamental hosts in the myxozoan life cycle (Kent et al. 2001). When these hosts die, they liberate spores into the water column that are infectious to the other host (Hedrick et al. 1998Kent et al. 2001). At present, the mode of transmission and geographic source of the parasite as contracted by the goby in Suisun Marsh have yet to be determined. Horizontal transmission has been shown only in the marine myxozoan Enteromyxum leei (Diamant 1997). Except for E. leei, parasite transmission precluding an obligate alternate host has not been demonstrated for other myxozoans.
Whether this pathogen was introduced by the goby from its native origin in Japan or whether the goby contracted it from its current environment (e.g. water column, sediment, infected preys such as worms being a part of their natural diet) is unknown. However, based on the presence of similar parasites from gobies in Japan, it seems likely that the parasite invaded along with its host. Furthermore, the similarity score in the 18S rDNA Domain III is very high (97.9%) between the gobiid myxozoans from the two distant locations.
Of interest in the myxozoan is their relevance in the abundance of the yellowfin goby being an introduced species in the SFE. It is important to note that monitoring for pathogens and diseases has not been a focus of the various fish surveys in the Sacramento San Joaquin Delta. Interestingly, monitoring programs in Suisun Marsh conducted from 1979 to 2007 demonstrates the peak of goby abundance from 1992 to 2001 but significantly declined from 2002 (Meng et al. 1994Matern et al. 2002;O’Rear and Moyle , O’Rear and Moyle O’Rear and Moyle 2010). Invasion ecology suggests that disease can either reduce (if it affects the invader) or increase (if initiated by a relatively immune invader) the impact of introduced species (Simberloff and Gibbons 2004). While many pathogens are known as disease agents in captive fish populations, the effects of diseases on wild populations are notoriously difficult to assess due to the complex interactions of many variables in the aquatic environment (Hedrick 1998). In a stressed ecosystem as the SFE, invasive species harboring exotic pathogens may affect the abundance of the host population by carrying pathogens deemed more pathogenic to naïve hosts (Lafferty et al. 2004Riley et al. 2008). Effects on hosts may be broad if the disease can infect new species that have no means of avoiding or reducing infection (Lafferty et al. 2005). Empirical evidence on the potential impacts of invasive species harboring pathogens and diseases in natural systems is unknown. Environmental factors however, may help create unique conditions for alien organisms to dominate and out-compete native species (Brook 2008Bradley et al. 2010).
Conclusion
This study documents the occurrence of a potentially new species of myxozoan in the yellowfin goby and underscores the detection of a parasitic infection in an introduced fish in the San Francisco Estuary. Although the significance of this parasite as a potential pathogen is unknown, the myxozoan may alter the performance and survival of yellowfin gobies by blocking the linings of the intestine and stomach. The PCR assay that we developed will provide a specific and rapid diagnostic tool to identify carriers of the myxozoan. PCR screening of species that may harbor the parasite and histopathological assessment on the severity of infections will provide a better understanding of the parasite impact on the long-term health of gobies and the potential transmission of infections to other susceptible fish species. While the prevalence of the myxozoan as reported here is restricted to a comparatively small collection site, the reemergence, identification, and ecological relevance of the parasite on goby populations in the San Francisco Estuary may be investigated in the future using the specific diagnostic tool developed in this study.
Methods
Two groups of juvenile gobies were collected in June and July 2005 in the northern part of Suisun slough at Suisun Marsh (Figure 3) as part of a monitoring survey of juvenile fishes. The gobies were transported live to the Center for Aquatic Biology and Aquaculture (CABA) at UC Davis in aerated tanks containing ambient water (22 ± 1°C) from the marsh. Gobies in the first group (N = 151, average size: 1.5 gm, 50 mm) were collected in June 2005 at Suisun slough by beach seining. The second group (N = 55, average size: 10.0 gm, 100 mm) were sampled in the same location in July 2005 using otter trawl and beach seine. The two groups were held separately in aerated 130-L tanks receiving (22–23°C) non-recirculating well water at CABA, UC Davis for one month.
After collection from the estuary, moribund gobies (N = 5) were observed from the first group and were therefore collected to determine if an agent is likely associated with the morbidity. Various organs such as the gill, spleen, and kidney were initially examined from these fish by wet mounts and did not reveal any parasitic stages. However, the stomach and intestinal tissues showed the presence of spores by light microscopy and were therefore extracted and purified by Percoll method (Hamilton and Canning 1988). The purified spores were used for analysis of morphologic features (Lom and Arthur 1989Lom and Dykova 2006) and for DNA extraction for identification by molecular approaches and to develop a specific PCR. Moribund gobies were euthanized with ethyl-p-aminobenzoate (500 μg/L, Benzocaine, Sigma) prior to dissection for observation of spores from stomach and intestinal tissues. Spores were measured for dimensions and features following the guidelines for myxozoan species (Lom and Arthur 1989). Gobies were also collected (group 1: N = 10, group 2: N = 15) and processed for histopathology (Humason 1979) and stained with hematoxylin and eosin. These samples were also PCR tested using the primers and conditions as described below. Gobies were euthanized with benzocaine as above prior to processing for PCR assays and fixing in buffered formalin for histopathology.
Genomic DNA (gDNA) was extracted from spores using a QIAamp DNA Mini kit (Qiagen). The 18S ribosomal gene (rDNA) region was PCR-amplified using a universal primer set (18e–18 g’) targeting ca. 1,900 bp fragment (Hillis and Dixon 1991Andree et al. 1999). The PCR products were purified (QIAEX II, Qiagen), and ligated into pGEM-T Easy vector (Promega) for transformation (Invitrogen). The plasmids containing the PCR amplified DNA fragment were sequenced using an ABI 377 automated DNA sequencer (Applied Biosciences).
The entire 18S rDNA sequence of the myxozoan obtained in this study (1,987 bp) was used for similarity search using the BLASTN program (http://www.ncbi.nlm.nih.gov/). The conserved (Domain III, ca. 540 bp) and long regions (Domain I through III including variable regions, 1.3 kb) of the sequence were selected for further analyses. The multiple sequence alignments were generated for each of the region for the goby myxozoan and closely-related myxozoans listed in Table 3 using MUSCLE ver. 3.8.31 (Edgar 2004). This was followed by pairwise comparison using Geneious ver. 6.1 Drummond et al. 2011). The phylogenetic tree was generated by MrBayes program (ver. 3.1.2) for Domain I − III (1.3 kb) using Markov chain Monte Carlo method with the following settings: Ngen = 10000000, Nchain = 4, Temp = 0.5, Stopval = 0.01, Samplefreq = 50, Printfreq = 1000 (Ronquist and Huelsenbeck 2003). The general time-reversible (GTR) model with gamma-distributed rates was used for the analysis as chosen by jModelTest ver. 2.1.4 (Darriba et al. 2012). Myxobolus albi, a closely related myxozoan that does not belong to Henneguya spp., was used as an out-group for the phylogenetic tree analysis. The phylogenetic trees were depicted by FigTree ver 1.3.1 (http://tree.bio.ed.ac.uk/software/figtree/).
The goby myxozoan parasite was detected by developing a specific PCR assay targeting a unique region of the 18S rDNA. A primer pair, Goby Myx 2 F (5’-ATG CTT CCG GGT ACT GTA GG-3’) and Goby Myx 2R (5’-CAC GCT CGT GAG AAC GAT TC-3’) generated a 150 bp PCR product. The PCR cocktail (Invitrogen) for a 50 μl reaction contained 200 μM of dNTPs, 1.5 mM of MgCl2, 40 pmol of each primer, 1 unit Platinum® Taq DNA polymerase, and 10x buffer at 1/10 the volume. The PCR cycle profile was performed consisting of an initial denaturation step of 95°C for 5 min, 40 cycles of 95°C for 30 s, 55°C for 1 min, 72°C for 30 s, and a final extension step at 72°C for 5 min. The specificity of the PCR test was verified by demonstrating the inability to amplify 18S rDNA fragment from other closely related myxozoans including Myxobolus sp. and M. cerebralis.
The prevalence of the myxozoan among the collected gobies was assessed at termination of the study after one month in captivity by randomly collecting gobies from the first group (30 from a total of 123 remaining fish) and from the second group (15 from a total of 40 remaining fish). Gobies that were moribund or dead and fish used for histopathological analysis were not included for estimating the myxozoan prevalence in the two groups. Apparently healthy gobies were also processed as negative controls. The sampled gobies from both groups and the controls were euthanized with 500 μg/L benzocaine and were processed for diagnostic PCR testing using the primers and conditions described above. Genomic DNA was extracted from pooled stomach and intestinal tissues of each fish and subjected to the reaction following the optimized condition. Amplified DNA fragments were randomly chosen and processed for direct sequencing at Davis Sequencing Service to confirm the parasite DNA sequence.
Authors’ information
DVB, TK, SJT are members of the Aquatic Health Program at UC Davis, School of Veterinary Medicine: http://www.vetmed.ucdavis.edu/aquatic_health/index.cfm.
References
Akihito K, Sakamoto S, Ikeda Y, Sugiyama K: Gobiidae. In Fishes of Japan with pictorial keys to the species. Edited by: Nakabo T. Tokyo, Japan: Tokai University Press; 2002:1154-1259.
Andree KB, Székely C, Molnár K, Gresoviac SJ, Hedrick RP: Relationships among members of the genus Myxobolus (Myxozoa: Bivalvidae) based on small subunit ribosomal DNA sequences. J Parasitol 1999, 85: 68-74. 10.2307/3285702
Bahri S, Andree KB, Hedrick RP: Morphological and phylogenetic studies of marine Myxobolus spp. from mullet in Ichkeul Lake, Tunisia. J Eukaryot Microbiol 2003, 50: 463-470. 10.1111/j.1550-7408.2003.tb00272.x
Bradley BA, Blumenthal DM, Wilcove DS, Ziska LH: Predicting plant invasions in an era of global change. Trend Ecol Evol 2010, 25: 310-318. 10.1016/j.tree.2009.12.003
Brittan MR, Albrecht AB, Hopkirk JB: An oriental goby collected in the San Joaquin River Delta near Stockton, California. California Fish and Game 1963, 49: 302-304.
Brook BW: Synergies between climate change, extinctions and invasive vertebrates. Wildlife Res 2008, 35: 249-252. 10.1071/WR07116
Cohen AN, Carlton JT: Accelerating invasion rate in a highly invaded estuary. Science 1998, 279: 555-558. 10.1126/science.279.5350.555
Darriba D, Taboada GL, Doallo R, Posada D: jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 2012, 9(8):772.
Diamant A: Fish-to-fish transmission of a marine myxosporean. Dis Aquat Org 1997, 30: 99-105.
Dill AW, Cordone AJ: History and status of introduced fishes in California. California Department of Fish and Game. Sacramento, California: Fishery Bulletin; 1997. 178: 1871–1996
Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, Duran C, Field M, Heled J, Kearse M, Markowitz S, Moir R, Stones-Havas S, Sturrock S, Thierer T, Wilson A: Geneious v5.4. 2011. Available from Accessed 8/10/2011 http://www.geneious.com/ Available from Accessed 8/10/2011
Edgar RC: MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32: 1792-1797. 10.1093/nar/gkh340
El-Mansy A: Immature stages and re-description of Henneguya suprabranchiae (Myxosporea: Myxobolidae), an intestinal parasite of the catfish Clarias gariepinus in the River Nile, Egypt. Dis Aquat Org 2002, 51: 179-186.
Feyrer F, Healey MP: Fish community structure and environmental correlates in the highly altered southern Sacramento-San Joaquin Delta. Environ Biol Fish 2003, 66: 123-132. 10.1023/A:1023670404997
Fiala I: The phylogeny of Myxosporea (Myxozoa) based on small subunit ribosomal RNA gene analysis. Int J Parasitol 2006, 36: 1521-1534. 10.1016/j.ijpara.2006.06.016
Hamilton AJ, Canning EU: The production of anti- Myxosoma cerebralis antiserum from Percoll-purified spores and its use in immunofluorescent labelling of historesin-embedded cartilage derived from infected rainbow trout, Salmo gairdneri Richardson. J Fish Dis 1988, 11: 185-190. 10.1111/j.1365-2761.1988.tb00538.x
Hanson LA, Lin D, Pote LM, Shivaji R: Small subunit rRNA gene comparison of four actinosporean species to establish a polymerase chain reaction test for the causative agent of proliferative gill disease in channel catfish. J Aquat Anim Health 2001, 13: 117-123. 10.1577/1548-8667(2001)013<0117:SSRGCO>2.0.CO;2
Hedrick RP: Relationships of the host, pathogen and environment: implications for diseases of cultured and wild fish populations. J Aquat Anim Health 1998, 10: 107-111. 10.1577/1548-8667(1998)010<0107:ROTHPA>2.0.CO;2
Hedrick RP, El-Matbouli M, Adkison MA, MacConnell E: Whirling disease: re-emergence among wild trout. Immunol Rev 1998, 166: 365-376. 10.1111/j.1600-065X.1998.tb01276.x
Hillis DM, Dixon T: Ribosomal DNA: molecular evolution and phylogenetic inference. Q Rev Biol 1991, 66: 411-453. 10.1086/417338
Hironouchi M, Sano M: Food habits of fishes in a Zostera marina bed at Aburatsubo, central Japan. Ichthyol Res 2000, 47: 163-173. 10.1007/BF02684237
Humason GL: Animal tissue techniques. San Francisco: WH Freeman Co; 1979. 569 p
Kageyama T, YanagidaT OK, Yokoyama H: Henneguya pseudorhinogobii n. sp. (Myxozoa: Myxosporea) parasitizing the gills of the freshwater goby Rhinogobius sp. OR from the Nagara River and redescription of Henneguya rhinogobii . Fish Sci 2009, 75: 657-663. 10.1007/s12562-009-0096-y
Kent ML, Andree KB, Bartholomew JL, El-Matbouli M, Desser S, Xiao C, Devlin RH, Hedrick RP, Khattra L, Plaenzuela O, Siddall M: Recent advances in our knowledge of the Myxozoa. J Eukaryot Microbiol 2001, 48: 395-413. 10.1111/j.1550-7408.2001.tb00173.x
Kikuchi T, Yamashita Y: Seasonal occurrence of gobiid fish and their food habit in a small mud flat in Amakusa. Publ Amakusa Mar Lab 1992, 11: 73-93.
Lafferty KD, Porter J, Ford SE: Are diseases increasing in the oceans? Annu Rev Ecol Evol Syst 2004, 35: 31-54. 10.1146/annurev.ecolsys.35.021103.105704
Lafferty K, Smith K, Torchin M, Dobson A, Kuris A, Sax D, Stachowicz J, Gaines SD: The role of infectious disease in natural communities: what introduced species tell us. In Species Invasions: Insights into Ecology, Evolution, and Biogeography. Edited by: Sax DF, Stachowicz JJ, Gaines SD. Sunderland: Sinauer; 2005:111-134. Mass, pp
Lom J, Arthur JR: A guideline for the preparation of species descriptions in Myxosporea. J Fish Dis 1989, 12: 151-156. 10.1111/j.1365-2761.1989.tb00287.x
Lom J, Dykova I: Protozoan parasites of fishes. New York: Elsevier Science Publishers; 1992:159-227.
Lom J, Dykova I: Myxozoan genera: definition and notes on taxonomy, life-cycle terminology and pathogenic species. Folia Parasitol 2006, 53: 1-36.
Matern SA, Moyle PB, Pierce L: Native and alien fishes in a California estuarine marsh: Twenty-one years of changing assemblages. Trans Am Fish Soc 2002, 131: 797-816. 10.1577/1548-8659(2002)131<0797:NAAFIA>2.0.CO;2
Meng L, Moyle PB, Herbold B: Changes in abundance and distribution of native and introduced fishes of Suisun Marsh. Trans Am Fish Soc 1994, 123: 498-507. 10.1577/1548-8659(1994)123<0498:CIAADO>2.3.CO;2
Moyle PB: Inland fishes of California, revised and expanded. Berkeley, CA: University of California Press; 2002. 502 p
Nichols FH, Cloern JE, Luoma SN, Peterson DH: The modification of an estuary. Science 1986, 231: 567-573. 10.1126/science.231.4738.567
O’Rear TA, Moyle PB: Long term and recent trends of fishes and invertebrates in Suisun Marsh. Interagency Ecological Program for the San Francisco Estuary (IEP). Newsletter 2010, 23: 26-49.
Picon-Camacho SM, Astrid A, Holzer S, Mark B, Freeman A, David A, Morrisa J, Shinn AP: Myxobolus albi n. sp. (Myxozoa) from the gills of the common goby Pomatoschistus microps Krøyer (Teleostei: Gobiidae). J Eukaryot Microbiol 2009, 56: 421-427. 10.1111/j.1550-7408.2009.00419.x
Riley SC, Munkittirick KR, Evans AN, Krueger CC: Understanding the ecology of disease in Great Lakes fish populations. Aquat Ecosyst Health Manag 2008, 11: 321-334. 10.1080/14634980802301638
Ronquist F, Huelsenbeck JP: MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19: 1572-1574. 10.1093/bioinformatics/btg180
Service RF: Delta blues, California style. Science 2007, 317: 442-445. 10.1126/science.317.5837.442
Siddall ME, Martin DS, Bridge D, Desser SS, Cone DK: The demise of a phylum of protists: phylogeny of Myxozoa and other parasitic cnidaria. J Parasitol 1995, 81: 961-967. 10.2307/3284049
Simberloff D, Gibbons L: Now you see them, now you don’t! – population crashes of established introduced species. Biol Invasions 2004, 6: 161-172.
Smothers JF, Von Dohlen CD, Smith LH Jr, Spall RD: Molecular evidence that the myxozoan protists are metazoans. Science 1994, 265: 1719-1721. 10.1126/science.8085160
Workman ML, Merz JE: Introduced yellowfin goby, Acanthogobius flavimanus : diet and habitat use in the lower Mokelumne River, California. San Francisco Estuary and Watershed Science 2007, 5: 1-13.
Yokoyama H, Itoh N, Tanaka S: Henneguya pagri n. sp. (Myxozoa: Myxosporea) causing cardiac henneguyosis in red sea bream, Pagrus major (Temminck & Schlegel). J Fish Dis 2005, 28: 479-487. 10.1111/j.1365-2761.2005.00655.x
Acknowledgements
Ms. Terry McDowell, Ms. Susan Yun, and Ms. Kavery Mukkatira are gratefully acknowledged for their assistance on spore extraction and purification by Percoll method and other laboratory techniques used in the study. We thank Dr. Thomas Waltzek for his support on phylogenetic analysis and Dr. Alireza Javidmehr for verifying the validity of random sampling. We also thank Dr. Moyle’s staff at the Wildlife, Fish, and Conservation Biology program at UC Davis for collection of the yellowfin gobies at Suisun Marsh. Partial funding support for this study was provided by the Ecosystem Restoration Program, UC Davis Agreement # E1183004.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
RPH, AS, and DVB designed the lab experiments. PBM directed the field sampling and identification of gobies. AS and MC isolated and processed the spores for 18S rDNA sequencing and designed the PCR primers. TK generated the sequence alignments, pairwise comparison of 18S rDNA sequences, and phylogenetic trees. DVB conducted the PCR analyses of samples. RPH and SJT performed histopathology and photomicrography. DVB interpreted the findings and wrote the paper. All co-authors contributed, read, and approved the manuscript revisions.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Baxa, D.V., Stover, A., Clifford, M. et al. Henneguya sp. in yellowfin goby Acanthogobius flavimanus from the San Francisco Estuary. SpringerPlus 2, 420 (2013). https://doi.org/10.1186/2193-1801-2-420
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/2193-1801-2-420