Abstract
Background
Skeletal muscle fibres represent one of the most abundant cell types in mammals. Their highly specialised contractile and metabolic functions depend on a large number of membrane-associated proteins with very high molecular masses, proteins with extensive posttranslational modifications and components that exist in highly complex supramolecular structures. This makes it extremely difficult to perform conventional biochemical studies of potential changes in protein clusters during physiological adaptations or pathological processes.
Results
Skeletal muscle proteomics attempts to establish the global identification and biochemical characterisation of all members of the muscle-associated protein complement. A considerable number of proteomic studies have employed large-scale separation techniques, such as high-resolution two-dimensional gel electrophoresis or liquid chromatography, and combined them with mass spectrometry as the method of choice for high-throughput protein identification. Muscle proteomics has been applied to the comprehensive biochemical profiling of developing, maturing and aging muscle, as well as the analysis of contractile tissues undergoing physiological adaptations seen in disuse atrophy, physical exercise and chronic muscle transformation. Biomedical investigations into proteome-wide alterations in skeletal muscle tissues were also used to establish novel biomarker signatures of neuromuscular disorders. Importantly, mass spectrometric studies have confirmed the enormous complexity of posttranslational modifications in skeletal muscle proteins.
Conclusions
This review critically examines the scientific impact of modern muscle proteomics and discusses its successful application for a better understanding of muscle biology, but also outlines its technical limitations and emerging techniques to establish new biomarker candidates.
Similar content being viewed by others
Introduction
Proteomics is an unbiased and technology-driven approach for the comprehensive cataloging of entire protein complements and represents an ideal analytical tool for the high-throughput discovery of protein alterations in health and disease [1]. Mass spectrometry-based proteomics is concerned with the global analysis of protein composition, posttranslational modifications and the dynamic nature of expression levels [2–4]. The generation of large data sets on protein expression levels makes proteomics a preeminent hypothesis-generating approach in modern biology [5]. Proteomics has now been accepted as a key technology in biochemistry, cell biology, systems biology and drug discovery [6–9]. In this respect, proteomics suggests itself as a thorough approach for the detailed biochemical analysis of heterogeneous and plastic types of tissue, such as muscles. Skeletal muscle proteomics aims at the global identification, detailed cataloguing and biochemical characterisation of the entire protein complement of voluntary contractile tissues in normal and pathological specimens [10–12]. Although mass spectrometry-based proteomics is a relatively new analytical approach in the general field of muscle biology, large-scale proteomic studies have already provided a plethora of new information on global changes during myogenesis, fibre maturation, muscle transformation and natural muscle aging [12–14]. High-throughput surveys of common neuromuscular diseases, such as x-linked muscular dystrophy [15], have revealed many new proteome-wide changes and adaptations on the molecular and cellular level [13]. Proteomics routinely employs high-resolution separation techniques, such as two-dimensional gel electrophoresis and/or liquid chromatography, in combination with advanced mass spectrometric methods for the unequivocal identification of peptides and proteins of interest [16–19]. The independent verification of proteomic data is usually accomplished by using immunoblotting surveys, activity assays and immunofluorescence microscopic analysis [12]. Over the past few years, technical advances in mass spectrometry [20–22] and the development of vastly improved bioinformatic analysis tools [23–25] have driven the remarkable progress of proteomic science. This review outlines the findings from recent applications of mass spectrometry-based proteomics for studying physiological adaptations and pathological alterations in skeletal muscle tissues and critically examines novel analytical strategies to establish muscle-specific biomarker signatures.
The complex biochemistry of skeletal muscle tissues
Contractile fibres of skeletal muscle tissues constitute the cellular units that provide coordinated excitation-contraction-relaxation cycles for voluntary movements and postural control [26]. In addition, skeletal muscles play a central physiological role in heat homeostasis and present a crucial metabolic tissue that integrates various biochemical pathways. For example, skeletal muscle fibres have the highest capacity for insulin-mediated uptake of glucose in the body, making muscle tissues a critical organ in carbohydrate metabolism [27]. The complex cellular tasks of muscles are performed by a large number of proteins with specialised functions, structures and interactions. Skeletal muscles contain a considerable amount of integral membrane proteins and high molecular mass complexes. Some of the largest protein species present in mammalian tissues are expressed in skeletal muscle, such as nebulin of 600-800 kDa and titin with a molecular mass exceeding 1,200 kDa [28, 29]. Although supramolecular membrane assemblies are also found in many other types of tissue, notably the nervous system, they represent a major biochemical feature of contractile fibres. This includes the nicotinic acetylcholine receptor of the postsynaptic muscle membrane [30], the acetylcholinesterase of the basal lamina [31], the voltage-sensing dihydropyridine receptor of the transverse tubules [32], the ryanodine receptor Ca2+-release channel of the triad junctions [33], the dystrophin-glycoprotein complex of the sarcolemma [34], the respiratory chain of muscle mitochondria [35] and the abundant actomyosin machinery with its regulatory troponin-tropomyosin system [36]. The large number of membrane-associated proteins, the exceptionally high molecular mass of many muscle components, extensive posttranslational modifications in various muscle proteins and their organisation in highly complex supramolecular structures make it extremely difficult to carry out conventional biochemical studies of potential changes in protein clusters during physiological adaptations or pathological processes. In this respect, proteomics and subcellular proteomics attempt to isolate, separate and identify the entire protein constellation of a given muscle tissue or fibre population. Most initial proteomic studies have focused on total extracts of mostly soluble muscle proteins, and more recent investigations have made an effort to also encompass integral proteins and high molecular mass components in their analysis.
Dynamics and heterogeneity of the skeletal muscle proteome
The biological hierarchy from genome to transcriptome to proteome is diagrammatically shown in Figure 1. In contrast to the stable neuromuscular genome, the muscle transcriptome is highly dynamic and the muscle proteome is constantly changing and adapting to altered functional demands, making its comprehensive analysis very challenging. The estimated 25,000 protein-coding genes in the human genome probably translate into several hundred thousand different protein species [37]. Although alternative splicing and posttranslational modifications depend on the exact definition of the term 'protein isoform', they result in the production of a very large number of distinct subspecies of proteins. In mammals, the majority of genes appear to be differentially spliced, so that a single gene often codes for multiple proteins, which in turn may undergo different types of posttranslational modifications. A small number of muscle-specific genes can thus generate a large number of protein species encoded by the muscle genome. An excellent example of this phenomenon is the family of sarcoendoplasmic reticulum Ca2+-ATPase (SERCA)-type Ca2+ pumps from slow and fast muscles. Although the SERCA proteins are encoded by three genes, of which the SERCA1 and SERCA2 genes are expressed in voluntary fibres, the isoform diversity of this abundant ion pump is drastically increased by alternative splicing of the transcripts and various posttranslational modifications [38], producing more than 10 different SERCA isoforms [39]. These cellular processes significantly increase muscle protein diversity.
Biological hierarchy of the neuromuscular system. Shown are the organisation of the genome, transcriptome and proteome of motor neurons and skeletal muscles. The histological image illustrates neuromuscular junctions on individual muscle fibres labelled for the presence of the enzyme acetylcholinesterase.
The wide and dynamic expression range of proteins within a specific tissue makes it impossible to separate and detect all protein species with currently available biochemical techniques. In addition, most tissue types are heterogeneous in composition. Aside from the main contractile cells of differing contractile properties, such as slow oxidative (type I), moderately fast oxidative glycolytic (type IIa) and fast glycolytic (type IIb) fibres, muscle contains extended layers of connective tissues, capillaries and nerve cells [40–42]. Thus the starting material for almost all invasive biochemical studies, that is, homogenised muscle tissues, contains a certain degree of cell types that have originated from the tendon, epimysium, endomysium, perimysium, muscle spindles, satellite cells, blood vessels and motor neurons. This analytical fact has to be taken into account when one interprets proteomic findings from total tissue extracts. In the past, microassay systems have been developed to study single-fibre preparations biochemically [43], so it might be feasible to investigate such cellular preparations by using miniaturised separation protocols and proteomic techniques with enhanced sensitivity in the future. Concentration differences between highly abundant muscle proteins, such as glycolytic enzymes, and low-abundance muscle proteins, such as surface signaling receptors, have been estimated to be several orders of magnitude. Standard gel electrophoretic or liquid chromatographic methods are not capable of separating this vast range of muscle proteins with differing densities.
Technical limitations of two-dimensional gel electrophoresis
Besides the biological fact that it is extremely difficult to accurately define a fixed protein complement in highly adaptable and dynamic fibre populations as 'the skeletal muscle proteome', the most obvious technical obstacle to the study of entire muscle proteomes is the limited availability of all-encompassing protein analytical capabilities. Currently no one set of biochemical techniques exists that can efficiently separate and consistently detect the total protein complement of a given cell type. Hence, with respect to interpreting findings from analytical gel electrophoresis, it is important to take into account various technical limitations. Two-dimensional gel electrophoresis of crude tissue extracts usually underestimates the presence of low abundance elements, the amount of integral membrane proteins and components with very high molecular masses in complex tissues [19, 44–46]. In addition, proteins with extreme pI values are often not properly resolved at the edge of large gel systems with a wide pI range. This problem can be partially addressed by using narrow-range pI gels in the first dimension or by employing overlapping gel systems covering several pI ranges [44]. Another important issue in two-dimensional gel electrophoresis is the distortion of protein spots due to abundant protein species or the presence of isforms with extensive posttranslational modifications [45]. For example, high levels of heterogeneous glycosylation patterns can result in broad or overlapping spot patterns, which are difficult to pick for in-gel digestion procedures. Importantly, the presence of muscle proteins with a high density, such as myosin heavy chains, myosin light chains, troponins, tropomyosins and actins, can distort certain zones within the two-dimensional separation pattern and thus potentially contaminate other protein spots. In such a case, the densitometric analysis of a specific spot and the identification of the most abundant protein species present in this gel region might not perfectly correlate. Proteolytic degradation products of high molecular mass proteins may also complicate the analysis of the gel image.
However, despite these technical limitations, gel electrophoresis-based proteomics results in excellent coverage of soluble and abundant muscle proteins involved in the regulation and execution of the contraction-relaxation cycle, energy metabolism and the cellular stress response [13]. The use of crude tissue extracts as starting material has the advantage of representing the entire soluble protein complement without the potential danger of protein desorption or artefactual entrapment by complex separation steps. On the other hand, organelle and membrane proteomics reduces sample complexity by focusing on distinct subsets of protein populations, as outlined below. Ideally, proteomic studies of skeletal muscle tissues should first use both crude extracts and distinct subcellular fractions as starting material and second employ gel electrophoresis and liquid chromatography in parallel for the separation of as many different classes of muscle proteins as possible.
Mass spectrometry-based muscle proteomics
The long-term goal of proteomic profiling studies is the cataloguing of all expressed protein species and the establishment of comprehensive biomarker signatures that characterise physiological processes during development and natural aging, as well as disease progression in common pathologies. The flowchart in Figure 2 outlines the main steps of proteomic profiling studies. The various steps involved in routine muscle proteomics involve (1) the extraction of the target protein complement, resulting in total crude preparations of mostly soluble components, distinct subcellular fractions including membrane-associated elements or isolated complexes; (2) the separation of proteins by one-dimensional gel electrophoresis, high-resolution two-dimensional gel electrophoresis and/or liquid chromatography; (3) in the case of gel electrophoretic approaches, the determination of altered expression levels in protein maps by densitometric surveys; (4) the mass spectrometric identification of distinct protein species, usually by analysing trypsin-generated peptide mixtures of proteins of interest; and (5) the validation of proteomic data by routine assay systems, such as immunoblotting, enzyme assays, confocal microscopy, binding assays and physiological tests.
Proteomic profiling of skeletal muscle. The flowchart outlines the various preparative and analytical steps involved in the routine mass spectrometry-based proteomic investigation of contractile tissues. Protein separation is usually carried out by one-dimensional polyacrylamide gel electrophoresis (1-D PAGE), 2-D gel electrophoresis with isoelectric focusing in the first dimension and PAGE in the second slab gel dimension (2-D IEF-PAGE) and/or liquid chromatography.
Over the past few years, mass spectrometry-based proteomics has successfully catalogued several hundred of the most abundant and soluble muscle-associated protein species and identified several thousand distinct protein isoforms present in skeletal muscle tissues [47–50]. Muscle proteomics has been applied to the comprehensive biochemical profiling of developing, maturing and aging muscle [51–56], as well as the analysis of contractile tissues undergoing physiological adaptations seen in disuse atrophy, physical exercise and chronic muscle transformation [57–63]. Biomedical investigations into proteome-wide alterations in skeletal muscle tissues were also used to establish novel biomarker signatures of neuromuscular pathologies. Disease-specific markers were determined for muscle-associated diseases such as dystrophinopathy [64–66], dysferlinopathy [67], traumatic denervation [68], obesity [69], diabetes-related contractile weakness [70], sepsis [71], hypokalemic myopathy [72], inclusion body myositis [73] and reducing body myopathy [74]. Since skeletal muscle biology is highly relevant to the meat industry, muscle proteomics has been widely applied to cataloguing and studying protein complements in livestock [75–77]. These studies have especially focused on the proteomic evaluation of hypertrophy in chicken [78, 79], sheep [80], pig [81–83] and cow [84] muscles. Table 1 lists key findings from recent proteomic studies focusing on the biochemical characterisation of skeletal muscle tissues. Biomarker signatures are listed only when more than one study has been conducted on a specific cell biological, physiological or pathological topic.
Besides bulk skeletal muscle, refined studies of muscle subtypes with an unusual histology such as extraocular muscles have been conducted [66, 85], including the proteomic profiling of sarcomere-associated elements [86]. In addition, posttranslational changes were studied in skeletal muscle preparations by proteomics, focusing especially on protein nitration [87], carbonylation [88], glycosylation [89–91] and phosphorylation [92, 93]. Comprehensive reviews have covered the critical examination of these proteomic studies [10–15, 94, 95]. Thus, instead of recapitulating the considerable impact of these early studies of muscle proteomics, this review instead outlines the more recent application of fluorescent gel electrophoresis, organelle proteomics and membrane proteomics for studying skeletal muscle tissues.
Fluorescence gel electrophoresis of the muscle proteome
Labeling of proteins with fluorescent dyes has been extensively applied in proteomic investigations [96–99]. However, the one technique that stands out for its potential to directly compare two different sets of protein complements is fluorescence difference in-gel electrophoresis, usually abbreviated as DIGE [100]. This advanced gel electrophoretic method represents one of the most powerful analytical tools for conducting comparative protein biochemical investigations [101]. The fluorescence DIGE technique covers the same type of proteins as conventional two-dimensional gel electrophoretic approaches. If proper labeling protocols are followed, the fluorescence tagging procedure does not significantly interfere with the chemical properties of proteins with respect to their gel electrophoretic mobility. Minden et al. [102] first described this two-dimensional gel electrophoretic technique and the first extensive evaluation of its two-dimensional software analysis was conducted by Tonge et al. [103]. The DIGE technique is an ideal method for comparing entire soluble proteomes in one swift analytical approach [100] if one accepts that proteins with extreme pI values, proteins with a very low density, certain classes of high molecular mass proteins, extremely hydrophobic proteins and elements with certain posttranslational modifications may be underrepresented in two-dimensional gel systems [19, 44–46]. DIGE greatly reduces gel-to-gel variations and thereby greatly improves the evaluation of trends in changed protein expression patterns [104].
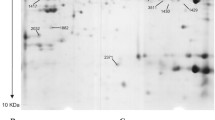
The DIGE method has also captured a lot of attention in the field of skeletal muscle proteomics over the past few years [55–57, 59, 63–66]. If the experimental design of fluorescent studies maximises the sensitivity for detecting changes in protein expression levels and takes into account statistical variations in dye binding, labelling artefacts of soluble protein species can be kept to a minimum [105–107]. Therefore, reverse DIGE labelling controls are not routinely employed. Dye-to-dye variability is usually minimal, and the findings from expression analyses with different dye combinations do not differ to a large extent. Analytical DIGE systems can be employed with two-dye or three-dye systems, depending on specific applications. Advanced DIGE, using an internal pooled standard, is a highly accurate quantitative method that enables multiple protein samples to be separated on the same two-dimensional gel. For example, a recent study conducted at our laboratory of the effects of experimental exon skipping on the dystrophic diaphragm from the x-linked muscular dystrophy (mdx) model of Duchenne muscular dystrophy combined the proteomic profiling of normal versus mdx versus treated mdx specimens [65]. In one set of six DIGE gels, 12 individual biological replicates of muscle samples were differentially labelled with different CyDyes Fluors (GE Healthcare Amersham Biosciences UK, Little Chalfont, Buckinghamshire, UK) for direct comparison on two-dimensional gels. A pool of all protein samples was also prepared and labelled with another CyDye to be employed as a standard on all gels. A pooled standard greatly aids image matching and cross-gel statistical analysis. Ideally, samples are evenly distributed between both CyDye fluors and analytical gels [65]. Figure 3 outlines the use of differential dye labelling for the analysis of two different muscle specimens. To illustrate the sensitivity of the fluorescent method, an enlarged DIGE image of distinct changes in the isoform expression pattern of myosin light chain during fast-to-slow muscle transitions is shown. Our proteomic profiling of fast muscle following chronic low-frequency stimulation has been studied by using DIGE analysis, and it clearly revealed a switch to slower isoforms of myosin light and heavy chains, as well as increased levels of oxidative enzymes [63].
Overview of the fluorescence difference in-gel electrophoretic (DIGE) method. Shown is a diagram of the differential labeling of muscle specimens with the fluorescent CyDyes Cy2, Cy3 and Cy5, as well as an example of a DIGE analysis of myosin light chain isoforms during fast-to-slow transitions of electrostimulated skeletal muscle.
Subcellular proteomics of skeletal muscle
Large-scale gel electrophoretic and liquid chromatographic methods have limitations with respect to analysing highly complex protein mixtures and protein populations with an extensive dynamic expression range. Thus, to reduce sample complexity, organelle proteomic studies have been initiated that focus on the protein complement of distinct subcellular fractions. Organelle proteomic approaches include sample prefractionation and the use of narrow pH ranges for the isoelectric focusing of low copy number proteins in two-dimensional gels as well as in one-dimensional gels for studying hydrophobic and high molecular mass proteins [108–110]. Reference maps of subcellular fractions from skeletal muscle include microsomes, sarcolemma, cytosol, contractile apparatus and mitochondria [111–120]. Since mitochondria are involved in various diseases and cellular aging, many subcellular proteomic studies have focused on this crucial organelle [121–123]. Proteomic profiling of the mitochondria-enriched fraction from senescent rat muscle has revealed a shift to more aerobic oxidative metabolism in a slower-twitching fibre population during age-related muscle degeneration [118]. This organelle proteomic study has identified many new potential biomarkers of sarcopenia of old age [14]. Maughan et al. [111] showed that the 10 glycolytic enzymes represent the most abundant proteins in the diffusible fraction of the rabbit skeletal muscle proteome. This makes the key metabolic proteins that mediate the core glycolytic pathway ideal candidates to be studied by using mass spectrometry-based proteomics [95]. In contrast, organelle-associated proteins are usually more difficult to identify and characterise. In this respect, Figure 4 shows a novel approach for studying subcellular fractions by on-membrane digestion of electrophoretically transferred proteins.
An inefficient trypsination of certain target proteins often hampers in-gel digestion procedures. To address this technical problem in the proteomic identification of proteins, on-membrane digestion has been developed [124–127]. The advantage of on-membrane digestion is superior protein sequence coverage [124]. In addition, on-membrane digestion is more efficient and faster compared with conventional in-gel digestion methods [127]. This technical fact reduces complications due to trypsin autolysis and makes the on-membrane digestion technique especially suitable for the mass spectrometric identification of low copy number proteins and large hydrophobic proteins. This technique has recently been applied to the biochemical analysis of the large-membrane cytoskeletal protein dystrophin from rabbit skeletal muscle [120]. Standard two-dimensional gels are usually unable to separate muscle proteins with a molecular mass greater than 200 kDa [56]. In contrast, one-dimensional gradient gels are capable of presenting very large membrane proteins up to 2,000 kDa [128]. The mass spectrometric analysis of muscle membranes using one-dimensional gels identified membrane-associated muscle proteins as well as the Dp427 isoform of dystrophin, including the dihydropyridine receptor of the transverse tubules and the 565-kDa ryanodine receptor Ca2+-release channel of the junctional sarcoplasmic reticulum [120]. On-membrane digestion was demonstrated to be highly suitable for studying high molecular mass proteins that would otherwise not be properly recognised by gel electrophoresis-based proteomic studies. Muscle proteins adsorbed onto nitrocellulose sheets appear to be more accessible to proteases, increasing digestion efficiency [120]. This makes this novel technique the method of choice for studying large membrane proteins.
Since membrane proteins play a crucial role in cellular signaling, regulation, homeostasis and metabolism, the large-scale identification and characterisation of peripheral and integral membrane proteomics is an important aspect of modern proteomics [129–131]. Recently, detergent phase extraction was applied to the proteomic analysis of the membrane-associated fraction from skeletal muscle [132]. This method is based on the principle of temperature-dependent phase extraction with Triton X-114. Since phase separation with this nonionic detergent occurs at temperatures above 22°C, skeletal muscle protein fractionation was carried out at 37°C [132]. As outlined in the flowchart in Figure 5, the separation step resulted in an aqueous phase enriched in hydrophilic proteins and a detergent phase with predominantly hydrophobic proteins. However, phase transition approaches always result in a certain degree of cross-contamination between soluble and membrane-associated proteins. The partitioning and cross-contamination of hydrophobic versus hydrophilic protein populations can be conveniently monitored by immunoblotting (Figure 5).
Detergent phase extraction method for muscle proteomics. The flowchart outlines the application of detergent phase extraction for the mass spectrometric identification of membrane-associated proteins from skeletal muscle tissues. The separation of protein species into an aqueous (AQ) fraction versus a detergent (DT) fraction can be conveniently verified by immunoblotting with antibodies to soluble versus integral markers.
Emerging technologies for muscle proteomics
Mass spectrometry-based proteomics is a fast-moving field with a variety of emerging technologies that have not yet been fully exploited in the field of skeletal muscle proteomics. Although several studies on muscle cells have included quantification methods involving the incorporation of stable isotopes through metabolic or chemical labelling [53, 61, 82, 133–135], as well as through label-free shotgun proteomics [50, 51, 85, 136, 137], future applications of a variety of advanced gel-free techniques promise an even greater impact of proteomics on muscle biology and biomarker discovery. The advantages and technical challenges of label-free proteomics versus the use of stable isotopes for metabolic or chemical labelling have been extensively outlined in several excellent reviews [4, 138–140]. Numerous stable isotope-labelling techniques have been employed in quantitative shotgun proteomics, including isobaric tags for relative and absolute quantification, or iTRAQ; isotope-coded affinity tag, or ICAT; and stable isotope labelling by amino acids in cell culture, or SILAC. In skeletal muscle proteomics, the SILAC method was successfully applied to the quantitative profiling of differential protein expression between myoblasts and myotubes [53] and the identification of proteins that interact with the glucose transporter GLUT4 in an insulin-regulated manner [133]. The ICAT labelling approach has been used for the proteomic profiling of the cytosolic fraction from atrophying mouse tibialis anterior muscle [135]. The iTRAQ technique was employed for the analysis of changes in the protein complement of human vastus lateralis muscle in response to interval exercise training [61], the cataloguing of the skeletal muscle proteome from pigs [82] and the identification of carbonylated proteins from rat muscle mitochondria [134].
In future proteomic shotgun experiments, protein populations from skeletal muscle tissues could be profiled by label-free quantification approaches such as peptide spectral counts. The term spectral counts refers to the number of mass spectrometry (MS)/MS identifications per protein species. Since abundant peptide species are detectable across a wide period of retention time and are therefore repeatedly sampled, a linear relationship exists between protein concentration and spectral counts [141]. This makes identification results from MS/MS spectra an attractive option for quantitative shotgun proteomics. With respect to the validation of protein biomarkers, stable isotope dilution (SID)-MS might be a useful addition to the analytical repertoire of muscle proteomics. This technique is especially useful for the characterisation of low-abundance biomarkers. Multiple reaction monitoring (MRM) coupled with SID-MS represents an ideal method for the quantitative measurement of low-abundance biomarkers [142]. In SID-MRM-MS investigations, the quantification of a biomarker protein of interest is achieved by selecting signature peptides of the digested target protein [143]. Unique signature peptides are employed as quantitative substitutes of the marker protein. The concentration of the biomarker protein can be determined by comparing the signals from the exogenous SID-labelled signature peptide and the endogenous unlabelled peptide population [144].
To tackle the technical challenges associated with analysing membrane proteins, an exciting universal sample preparation method has been developed: filter-aided sample preparation (FASP). On the basis of the fact that membrane proteins can be depleted of detergent by gel filtration in 8 M urea [145], Wisniewski et al. [146] established a proteomic sample preparation technique based on the complete solubilisation of a protein complement in 4% sodium dodecyl sulphate and subsequent exchange of detergent by urea on spin filter devices. The FASP method depletes the suspension of low molecular mass components in urea-containing buffer, digests a representative cohort of proteins including hydrophobic elements and elutes pure peptide mixtures [146]. Thus, FASP is an ideal tool to be applied to the proteomic identification and characterisation of high molecular mass membrane proteins from skeletal muscle, because peptide mixtures eluted after digestion on the spin filter were highly suitable for mass spectrometric analysis [146]. Since a large number of hydrophobic proteins are involved in physiological adaptations and pathological changes in contractile fibres, this new universal sample preparation tool promises to be extremely useful for future proteomic investigations of muscle organelles.
Conclusions
Over the past few years, skeletal muscle proteomics has successfully catalogued the majority of abundant and soluble fibre-associated proteins. The refined proteomic analysis of isoform expression patterns and biochemical studies of dynamic posttranslational modifications has identified thousands of distinct muscle protein species. Myogenesis, muscle maturation, muscle transformation and aging-related muscle wasting have been intensively investigated by using proteomic methods and has resulted in the establishment of a comprehensive biomarker signature for major physiological adaptation processes in contractile tissues. The proteomic characterisation of common neuromuscular disorders has revealed novel disease-specific marker proteins of disuse atrophy, muscular dystrophy, obesity, type 2 diabetes, sepsis, hypokalemic myopathy, inclusion body myositis and reducing body myopathy. Thus, MS-based proteomics has decisively improved our general understanding of physiological and pathophysiological mechanisms in muscle tissues. New biomarker candidates can now be used for improving diagnostic methods, the identification of novel therapeutic targets, better comprehension of the molecular pathogenesis of muscular disorders, improved monitoring of disease progression and the judging of potential side effects of experimental drugs. Importantly, if the proteomic workflow could be successfully miniaturised, then single-cell proteomics of different fibre populations would lead to more comprehensive coverage of the skeletal muscle proteome. This depends on technical developments in the field of MS [20–22]. In the future, organelle and membrane proteomics will probably play a more prominent role in muscle biochemistry to study less abundant and more hydrophobic proteins. Filter-aided sample preparation and on-membrane digestion may be preferred for the proteomic analysis of high molecular mass membrane proteins. Once the majority of large and integral muscle proteins have been catalogued by proteomics, it will be crucial to correlate these findings with genomic, transcriptomic and metabolomic databanks [147] and establish the global relationship of biomolecules in striated voluntary muscle tissues.
References
Hochstrasser DF, Sanchez JC, Appel RD: Proteomics and its trends facing nature's complexity. Proteomics 2002, 2: 807-812. 10.1002/1615-9861(200207)2:7<807::AID-PROT807>3.0.CO;2-4
Aebersold R, Mann M: Mass spectrometry-based proteomics. Nature 2003, 422: 198-207. 10.1038/nature01511
Guerrera IC, Kleiner O: Application of mass spectrometry in proteomics. Biosci Rep 2005, 25: 71-93. 10.1007/s10540-005-2849-x
Yates JR, Ruse CI, Nakorchevsky A: Proteomics by mass spectrometry: approaches, advances, and applications. Annu Rev Biomed Eng 2009, 11: 49-79. 10.1146/annurev-bioeng-061008-124934
Cravatt BF, Simon GM, Yates JR: The biological impact of mass-spectrometry-based proteomics. Nature 2007, 450: 991-1000. 10.1038/nature06525
Zhu H, Bilgin M, Snyder M: Proteomics. Annu Rev Biochem 2003, 72: 783-812. 10.1146/annurev.biochem.72.121801.161511
Gstaiger M, Aebersold R: Applying mass spectrometry-based proteomics to genetics, genomics and network biology. Nat Rev Genet 2009, 10: 617-627. 10.1038/nrg2633
Walther TC, Mann M: Mass spectrometry-based proteomics in cell biology. J Cell Biol 2010, 190: 491-500. 10.1083/jcb.201004052
Chakravarti B, Mallik B, Chakravarti DN: Proteomics and systems biology: application in drug discovery and development. Methods Mol Biol 2010, 662: 3-28. full_text
Isfort RJ: Proteomic analysis of striated muscle. J Chromatogr B Analyt Technol Biomed Life Sci 2002, 771: 155-165. 10.1016/S1570-0232(02)00056-9
Hittel DS, Hathout Y, Hoffman EP: Proteomics and systems biology in exercise and sport sciences research. Exerc Sport Sci Rev 2007, 35: 5-11. 10.1097/jes.0b013e31802d744a
Doran P, Donoghue P, O'Connell K, Gannon J, Ohlendieck K: Proteomic profiling of pathological and aged skeletal muscle fibres by peptide mass fingerprinting. Int J Mol Med 2007, 19: 547-564.
Ohlendieck K: Proteomics of skeletal muscle differentiation, neuromuscular disorders and fiber aging. Expert Rev Proteomics 2010, 7: 283-296. 10.1586/epr.10.2
Doran P, Donoghue P, O'Connell K, Gannon J, Ohlendieck K: Proteomics of skeletal muscle aging. Proteomics 2009, 9: 989-1003. 10.1002/pmic.200800365
Lewis C, Carberry S, Ohlendieck K: Proteomic profiling of x-linked muscular dystrophy. J Muscle Res Cell Motil 2009, 30: 267-9. 10.1007/s10974-009-9197-6
Ali-Khan N, Zuo X, Speicher DW: Overview of proteome analysis. Curr Protoc Protein Sci 2003., 22: Unit 22.1
Baggerman G, Vierstraete E, De Loof A, Schoofs L: Gel-based versus gel-free proteomics: a review. Comb Chem High Throughput Screen 2005, 8: 669-677. 10.2174/138620705774962490
Falk R, Ramström M, Stahl S, Hober S: Approaches for systematic proteome exploration. Biomol Eng 2007, 24: 155-168. 10.1016/j.bioeng.2007.01.001
Rabilloud T, Chevallet M, Luche S, Lelong C: Two-dimensional gel electrophoresis in proteomics: past, present and future. J Proteomics 2010, 73: 2064-2077. 10.1016/j.jprot.2010.05.016
Canas B, Lopez-Ferrer D, Ramos-Fernandez A, Camafeita E, Calvo E: Mass spectrometry technologies for proteomics. Brief Funct Genomic Proteomic 2006, 4: 295-320. 10.1093/bfgp/eli002
Han X, Aslanian A, Yates JR: Mass spectrometry for proteomics. Curr Opin Chem Biol 2008, 12: 483-490. 10.1016/j.cbpa.2008.07.024
Domon B, Aebersold R: Mass spectrometry and protein analysis. Science 2006, 312: 212-217. 10.1126/science.1124619
Mueller LN, Brusniak MY, Mani DR, Aebersold R: An assessment of software solutions for the analysis of mass spectrometry based quantitative proteomics data. J Proteome Res 2008, 7: 51-61. 10.1021/pr700758r
Kumar C, Mann M: Bioinformatics analysis of mass spectrometry-based proteomics data sets. FEBS Lett 2009, 583: 1703-1712. 10.1016/j.febslet.2009.03.035
Jacob RJ: Bioinformatics for LC-MS/MS-based proteomics. Methods Mol Biol 2010, 658: 61-91. full_text
Gordon AM, Homsher E, Regnier M: Regulation of contraction in striated muscle. Physiol Rev 2000, 80: 853-924.
Abdul-Ghani MA, DeFronzo RA: Pathogenesis of insulin resistance in skeletal muscle. J Biomed Biotechnol 2010, 2010: 476279.
Ottenheijm CA, Granzier H: New insights into the structural roles of nebulin in skeletal muscle. J Biomed Biotechnol 2010, 2010: 968139.
Ottenheijm CA, Granzier H: Role of titin in skeletal muscle function and disease. Adv Exp Med Biol 2010, 682: 105-122. full_text
Fagerlund MJ, Eriksson LI: Current concepts in neuromuscular transmission. Br J Anaesth 2009, 103: 108-114. 10.1093/bja/aep150
Aldunate R, Casar JC, Brandan E, Inestrosa NC: Structural and functional organization of synaptic acetylcholinesterase. Brain Res Brain Res Rev 2004, 47: 96-104. 10.1016/j.brainresrev.2004.07.019
Arikkath J, Campbell KP: Auxiliary subunits: essential components of the voltage-gated calcium channel complex. Curr Opin Neurobiol 2003, 13: 298-307. 10.1016/S0959-4388(03)00066-7
Franzini-Armstrong C: Architecture and regulation of the Ca2+delivery system in muscle cells. Appl Physiol Nutr Metab 2009, 34: 323-327. 10.1139/H09-017
Michele DE, Campbell KP: Dystrophin-glycoprotein complex: post-translational processing and dystroglycan function. J Biol Chem 2003, 278: 15457-15460. 10.1074/jbc.R200031200
Hood DA: Mechanisms of exercise-induced mitochondrial biogenesis in skeletal muscle. Appl Physiol Nutr Metab 2009, 34: 465-472. 10.1139/H09-045
Huxley HE: Fifty years of muscle and the sliding filament hypothesis. Eur J Biochem 2004, 271: 1403-1415. 10.1111/j.1432-1033.2004.04044.x
Southan C: Has the yo-yo stopped? an assessment of human protein-coding gene number. Proteomics 2004, 4: 1712-1726. 10.1002/pmic.200300700
Brini M, Carafoli E: Calcium pumps in health and disease. Physiol Rev 2009, 89: 1341-1378. 10.1152/physrev.00032.2008
Periasamy M, Kalyanasundaram A: SERCA pump isoforms: their role in calcium transport and disease. Muscle Nerve 2007, 35: 430-442. 10.1002/mus.20745
Punkt K: Fibre types in skeletal muscles. Adv Anat Embryol Cell Biol 2002, 162: 1-109.
Spangenburg EE, Booth FW: Molecular regulation of individual skeletal muscle fibre types. Acta Physiol Scand 2003, 178: 413-424. 10.1046/j.1365-201X.2003.01158.x
Pette D, Staron RS: Cellular and molecular diversities of mammalian skeletal muscle fibers. Rev Physiol Biochem Pharmacol 1990, 116: 1-76.
Pette D, Peuker H, Staron RS: The impact of biochemical methods for single muscle fibre analysis. Acta Physiol Scand 1999, 166: 261-277. 10.1046/j.1365-201x.1999.00573.x
Gorg A, Weiss W, Dunn MJ: Current two-dimensional electrophoresis technology for proteomics. Proteomics 2004, 4: 3665-3685. 10.1002/pmic.200401031
Wittmann-Liebold B, Graack HR, Pohl T: Two-dimensional gel electrophoresis as tool for proteomics studies in combination with protein identification by mass spectrometry. Proteomics 2006, 6: 4688-4703. 10.1002/pmic.200500874
Weiss W, Gorg A: High-resolution two-dimensional electrophoresis. Methods Mol Biol 2009, 564: 13-32. full_text
Capitanio D, Vigano A, Ricci E, Cerretelli P, Wait R, Gelfi C: Comparison of protein expression in human deltoideus and vastus lateralis muscles using two-dimensional gel electrophoresis. Proteomics 2005, 5: 2577-2586. 10.1002/pmic.200401183
Hojlund K, Yi Z, Hwang H, Bowen B, Lefort N, Flynn CR, Langlais P, Weintraub ST, Mandarino LJ: Characterization of the human skeletal muscle proteome by one-dimensional gel electrophoresis and HPLC-ESI-MS/MS. Mol Cell Proteomics 2008, 7: 257-267.
Raddatz K, Albrecht D, Hochgraefe F, Hecker M, Gotthardt M: A proteome map of murine heart and skeletal muscle. Proteomics 2008, 8: 885-1897. 10.1002/pmic.200700902
Parker KC, Walsh RJ, Salajegheh M, Amato AA, Krastins B, Sarracino DA, Greenberg SA: Characterization of human skeletal muscle biopsy samples using shotgun proteomics. J Proteome Res 2009, 8: 3265-3277. 10.1021/pr800873q
Kislinger T, Gramolini AO, Pan Y, Rahman K, MacLennan DH, Emili A: Proteome dynamics during C2C12 myoblast differentiation. Mol Cell Proteomics 2005, 24: 887-901.
Gonnet F, Bouazza B, Millot GA, Ziaei S, Garcia L, Butler-Browne GS, Mouly V, Tortajada J, Danos O, Svinartchouk F: Proteome analysis of differentiating human myoblasts by dialysis-assisted two-dimensional gel electrophoresis (DAGE). Proteomics 2008, 8: 264-278. 10.1002/pmic.200700261
Cui Z, Chen X, Lu B, Park SK, Xu T, Xie Z, Xue P, Hou J, Hang H, Yates JR, Yang F: Preliminary quantitative profile of differential protein expression between rat L6 myoblasts and myotubes by stable isotope labeling with amino acids in cell culture. Proteomics 2009, 9: 1274-1292. 10.1002/pmic.200800354
Sun H, Zhu T, Ding F, Hu N, Gu X: Proteomic studies of rat tibialis anterior muscle during postnatal growth and development. Mol Cell Biochem 2009, 332: 161-171. 10.1007/s11010-009-0186-2
Gelfi C, Vigano A, Ripamonti M, Pontoglio A, Begum S, Pellegrino MA, Grassi B, Bottinelli R, Wait R, Cerretelli P: The human muscle proteome in aging. J Proteome Res 2006, 5: 1344-1353. 10.1021/pr050414x
Doran P, O'Connell K, Gannon J, Kavanagh M, Ohlendieck K: Opposite pathobiochemical fate of pyruvate kinase and adenylate kinase in aged rat skeletal muscle as revealed by proteomic DIGE analysis. Proteomics 2008, 8: 364-377. 10.1002/pmic.200700475
Moriggi M, Cassano P, Vasso M, Capitanio D, Fania C, Musicco C, Pesce V, Gadaleta MN, Gelfi CA: DIGE approach for the assessment of rat soleus muscle changes during unloading: effect of acetyl-L-carnitine supplementation. Proteomics 2008, 8: 3588-3604. 10.1002/pmic.200701176
Ferreira R, Vitorino R, Neuparth MJ, Appell HJ, Duarte JA, Amado F: Proteolysis activation and proteome alterations in murine skeletal muscle submitted to 1 week of hindlimb suspension. Eur J Appl Physiol 2009, 107: 553-563. 10.1007/s00421-009-1151-1
Moriggi M, Vasso M, Fania C, Capitanio D, Bonifacio G, Salanova M, Blottner D, Rittweger J, Felsenberg D, Cerretelli P, Gelfi C: Long term bed rest with and without vibration exercise countermeasures: effects on human muscle protein dysregulation. Proteomics 2010, 10: 3756-3774. 10.1002/pmic.200900817
Burniston JG: Changes in the rat skeletal muscle proteome induced by moderate-intensity endurance exercise. Biochim Biophys Acta 2008, 1784: 1077-1086.
Holloway KV, O'Gorman M, Woods P, Morton JP, Evans L, Cable NT, Goldspink DF, Burniston JG: Proteomic investigation of changes in human vastus lateralis muscle in response to interval-exercise training. Proteomics 2009, 9: 5155-5174. 10.1002/pmic.200900068
Donoghue P, Doran P, Dowling P, Ohlendieck K: Differential expression of the fast skeletal muscle proteome following chronic low-frequency stimulation. Biochim Biophys Acta 2005, 1752: 166-176.
Donoghue P, Doran P, Wynne K, Pedersen K, Dunn MJ, Ohlendieck K: Proteomic profiling of chronic low-frequency stimulated fast muscle. Proteomics 2007, 7: 3417-3430. 10.1002/pmic.200700262
Doran P, Martin G, Dowling P, Jockusch H, Ohlendieck K: Proteome analysis of the dystrophin-deficient MDX diaphragm reveals a drastic increase in the heat shock protein cvHSP. Proteomics 2006, 6: 4610-4621. 10.1002/pmic.200600082
Doran P, Wilton SD, Fletcher S, Ohlendieck K: Proteomic profiling of antisense-induced exon skipping reveals reversal of pathobiochemical abnormalities in dystrophic mdx diaphragm. Proteomics 2009, 9: 671-685. 10.1002/pmic.200800441
Lewis C, Ohlendieck K: Proteomic profiling of naturally protected extraocular muscles from the dystrophin-deficient mdx mouse. Biochem Biophys Res Commun 2010, 396: 1024-1029. 10.1016/j.bbrc.2010.05.052
De Palma S, Morandi L, Mariani E, Begum S, Cerretelli P, Wait R, Gelfi C: Proteomic investigation of the molecular pathophysiology of dysferlinopathy. Proteomics 2006, 6: 379-385. 10.1002/pmic.200500098
Sun H, Liu J, Ding F, Wang X, Liu M, Gu X: Investigation of differentially expressed proteins in rat gastrocnemius muscle during denervation-reinnervation. J Muscle Res Cell Motil 2006, 27: 241-250. 10.1007/s10974-006-9067-4
Hittel DS, Hathout Y, Hoffman EP, Houmard JA: Proteome analysis of skeletal muscle from obese and morbidly obese women. Diabetes 2005, 54: 1283-1288. 10.2337/diabetes.54.5.1283
Mullen E, Ohlendieck K: Proteomic profiling of non-obese type 2 diabetic skeletal muscle. Int J Mol Med 2010, 25: 445-458.
Duan X, Berthiaume F, Yarmush D, Yarmush ML: Proteomic analysis of altered protein expression in skeletal muscle of rats in a hypermetabolic state induced by burn sepsis. Biochem J 2006, 397: 149-158. 10.1042/BJ20051710
Thongboonkerd V, Kanlaya R, Sinchaikul S, Parichatikanond P, Chen ST, Malasit P: Proteomic identification of altered proteins in skeletal muscle during chronic potassium depletion: Implications for hypokalemic myopathy. J Proteome Res 2006, 5: 3326-3335. 10.1021/pr060136h
Li J, Yin C, Okamoto H, Jaffe H, Oldfield EH, Zhuang Z, Vortmeyer AO, Rushing EJ: Proteomic analysis of inclusion body myositis. J Neuropathol Exp Neurol 2006, 65: 826-833. 10.1097/01.jnen.0000228204.19915.69
Schessl J, Zou Y, McGrath MJ, Cowling BS, Maiti B, Chin SS, Sewry C, Battini R, Hu Y, Cottle DL, Rosenblatt M, Spruce L, Ganguly A, Kirschner J, Judkins AR, Golden JA, Goebel HH, Muntoni F, Flanigan KM, Mitchell CA, Bonnemann CG: Proteomic identification of FHL1 as the protein mutated in human reducing body myopathy. J Clin Invest 2008, 118: 904-912.
Doherty MK, McLean L, Beynon RJ: Avian proteomics: advances, challenges and new technologies. Cytogenet Genome Res 2007, 117: 358-369. 10.1159/000103199
D'Ambrosio C, Arena S, Talamo F, Ledda L, Renzone G, Ferrara L, Scaloni A: Comparative proteomic analysis of mammalian animal tissues and body fluids: bovine proteome database. J Chromatogr B Analyt Technol Biomed Life Sci 2005, 815: 157-168.
Picard B, Berri C, Lefaucheur L, Molette C, Sayd T, Terlouw C: Skeletal muscle proteomics in livestock production. Brief Funct Genomics 2010, 9: 259-278.
Doherty MK, McLean L, Hayter JR, Pratt JM, Robertson DH, El-Shafei A, Gaskell SJ, Beynon RJ: The proteome of chicken skeletal muscle: changes in soluble protein expression during growth in a layer strain. Proteomics 2004, 4: 2082-2093. 10.1002/pmic.200300716
Teltathum T, Mekchay S: Proteome changes in Thai indigenous chicken muscle during growth period. Int J Biol Sci 2009, 5: 679-688.
Hamelin M, Sayd T, Chambon C, Bouix J, Bibé B, Milenkovic D, Leveziel H, Georges M, Clop A, Marinova P, Laville E: Proteomic analysis of ovine muscle hypertrophy. J Anim Sci 2006, 84: 3266-3276. 10.2527/jas.2006-162
Lametsch R, Kristensen L, Larsen MR, Therkildsen M, Oksbjerg N, Ertbjerg P: Changes in the muscle proteome after compensatory growth in pigs. J Anim Sci 2006, 84: 918-924.
Hakimov HA, Walters S, Wright TC, Meidinger RG, Verschoor CP, Gadish M, Chiu DK, Stromvik MV, Forsberg CW, Golovan SP: Application of iTRAQ to catalogue the skeletal muscle proteome in pigs and assessment of effects of gender and diet dephytinization. Proteomics 2009, 9: 4000-4016. 10.1002/pmic.200900049
Mach N, Keuning E, Kruijt L, Hortós M, Arnau J, Te Pas MF: Comparative proteomic profiling of 2 muscles from 5 different pure pig breeds using surface-enhanced laser desorption/ionization time-of-flight proteomics technology. J Anim Sci 2010, 88: 1522-1534. 10.2527/jas.2009-2256
Bouley J, Meunier B, Chambon C, De Smet S, Hocquette JF, Picard B: Proteomic analysis of bovine skeletal muscle hypertrophy. Proteomics 2005, 5: 490-500. 10.1002/pmic.200400925
Fraterman S, Zeiger U, Khurana TS, Rubinstein NA, Wilm M: Combination of peptide OFFGEL fractionation and label-free quantitation facilitated proteomics profiling of extraocular muscle. Proteomics 2007, 7: 3404-3416. 10.1002/pmic.200700382
Fraterman S, Zeiger U, Khurana TS, Wilm M, Rubinstein NA: Quantitative proteomics profiling of sarcomere associated proteins in limb and extraocular muscle allotypes. Mol Cell Proteomics 2007, 6: 728-737. 10.1074/mcp.M600345-MCP200
Kanski J, Hong SJ, Schoneich C: Proteomic analysis of protein nitration in aging skeletal muscle and identification of nitrotyrosine-containing sequences in vivo by nanoelectrospray ionization tandem mass spectrometry. J Biol Chem 2005, 280: 24261-24266. 10.1074/jbc.M501773200
Feng J, Xie H, Meany DL, Thompson LV, Arriaga EA, Griffin TJ: Quantitative proteomic profiling of muscle type-dependent and age-dependent protein carbonylation in rat skeletal muscle mitochondria. J Gerontol A Biol Sci Med Sci 2008, 63: 1137-1152.
Cieniewski-Bernard C, Bastide B, Lefebvre T, Lemoine J, Mounier Y, Michalski JC: Identification of O -linked N -acetylglucosamine proteins in rat skeletal muscle using two-dimensional gel electrophoresis and mass spectrometry. Mol Cell Proteomics 2004, 3: 577-585. 10.1074/mcp.M400024-MCP200
O'Connell K, Doran P, Gannon J, Ohlendieck K: Lectin-based proteomic profiling of aged skeletal muscle: decreased pyruvate kinase isozyme M1 exhibits drastically increased levels of N -glycosylation. Eur J Cell Biol 2008, 87: 793-805.
Hedou J, Bastide B, Page A, Michalski JC, Morelle W: Mapping of O -linked β- N -acetylglucosamine modification sites in key contractile proteins of rat skeletal muscle. Proteomics 2009, 9: 2139-2148. 10.1002/pmic.200800617
Gannon J, Staunton L, O'Connell K, Doran P, Ohlendieck K: Phosphoproteomic analysis of aged skeletal muscle. Int J Mol Med 2008, 22: 33-42.
Hojlund K, Bowen BP, Hwang H, Flynn CR, Madireddy L, Geetha T, Langlais P, Meyer C, Mandarino LJ, Yi Z: In vivo phosphoproteome of human skeletal muscle revealed by phosphopeptide enrichment and HPLC-ESI-MS/MS. J Proteome Res 2009, 8: 4954-4965. 10.1021/pr9007267
Doran P, Gannon J, O'Connell K, Ohlendieck K: Proteomic profiling of animal models mimicking skeletal muscle disorders. Proteomics Clin Appl 2007, 1: 1169-1184. 10.1002/prca.200700042
Ohlendieck K: Proteomics of skeletal muscle glycolysis. Biochim Biophys Acta 2010, 1804: 2089-2101.
Westermeier R, Marouga R: Protein detection methods in proteomics research. Biosci Rep 2005, 25: 19-32. 10.1007/s10540-005-2845-1
Waggoner A: Fluorescent labels for proteomics and genomics. Curr Opin Chem Biol 2006, 10: 62-66. 10.1016/j.cbpa.2006.01.005
Riederer BM: Non-covalent and covalent protein labeling in two-dimensional gel electrophoresis. J Proteomics 2008, 71: 231-44. 10.1016/j.jprot.2008.05.001
Sasse J, Gallagher SR: Staining proteins in gels. Curr Protoc Mol Biol 2009., Chapter 10: Unit 10.6
Minden JS, Dowd SR, Meyer HE, Stuehler K: Difference gel electrophoresis. Electrophoresis 2009, 30: S156-S161. 10.1002/elps.200900098
Marouga R, David S, Hawkins E: The development of the DIGE system: 2D fluorescence difference gel analysis technology. Anal Bioanal Chem 2005, 382: 669-678. 10.1007/s00216-005-3126-3
Unlu M, Morgan ME, Minden JS: Difference gel electrophoresis: a single gel method for detecting changes in protein extracts. Electrophoresis 1997, 18: 2071-2077. 10.1002/elps.1150181133
Tonge R, Shaw J, Middleton B, Rowlinson R, Rayner S, Young J, Pognan F, Hawkins E, Currie I, Davison M: Validation and development of fluorescence two-dimensional differential gel electrophoresis proteomics technology. Proteomics 2001, 1: 377-396. 10.1002/1615-9861(200103)1:3<377::AID-PROT377>3.0.CO;2-6
Van den Bergh G, Arckens L: Fluorescent two-dimensional difference gel electrophoresis unveils the potential of gel-based proteomics. Curr Opin Biotechnol 2004, 15: 38-43. 10.1016/j.copbio.2003.12.001
Karp NA, Lilley KS: Maximising sensitivity for detecting changes in protein expression: experimental design using minimal CyDyes. Proteomics 2005, 5: 3105-3115. 10.1002/pmic.200500083
Corzett TH, Fodor IK, Choi MW, Walsworth VL, Chromy BA, Turteltaub KW, McCutchen-Maloney SL: Statistical analysis of the experimental variation in the proteomic characterization of human plasma by two-dimensional difference gel electrophoresis. J Proteome Res 2006, 5: 2611-2619. 10.1021/pr060100p
Karp NA, McCormick PS, Russell MR, Lilley KS: Experimental and statistical considerations to avoid false conclusions in proteomics studies using differential in-gel electrophoresis. Mol Cell Proteomics 2007, 6: 1354-1364. 10.1074/mcp.M600274-MCP200
Cordwell SJ, Nouwens AS, Verrills NM, Basseal DJ, Walsh BJ: Subproteomics based upon protein cellular location and relative solubilities in conjunction with composite two-dimensional electrophoresis gels. Electrophoresis 2000, 21: 1094-1103. 10.1002/(SICI)1522-2683(20000401)21:6<1094::AID-ELPS1094>3.0.CO;2-0
Zuo X, Echan L, Hembach P, Tang HY, Speicher KD, Santoli D, Speicher DW: Towards global analysis of mammalian proteomes using sample prefractionation prior to narrow pH range two-dimensional gels and using one-dimensional gels for insoluble and large proteins. Electrophoresis 2001, 22: 1603-1615. 10.1002/1522-2683(200105)22:9<1603::AID-ELPS1603>3.0.CO;2-I
Elschenbroich S, Kim Y, Medin JA, Kislinger T: Isolation of cell surface proteins for mass spectrometry-based proteomics. Expert Rev Proteomics 2010, 7: 141-154. 10.1586/epr.09.97
Maughan DW, Henkin JA, Vigoreaux JO: Concentrations of glycolytic enzymes and other cytosolic proteins in the diffusible fraction of a vertebrate muscle proteome. Mol Cell Proteomics 2005, 4: 1541-1549. 10.1074/mcp.M500053-MCP200
Forner F, Foster LJ, Campanaro S, Valle G, Mann M: Quantitative proteomic comparison of rat mitochondria from muscle, heart, and liver. Mol Cell Proteomics 2006, 5: 608-619.
Reifschneider NH, Goto S, Nakamoto H, Takahashi R, Sugawa M, Dencher NA, Krause F: Defining the mitochondrial proteomes from five rat organs in a physiologically significant context using 2D blue-native/SDS-PAGE. J Proteome Res 2006, 5: 1117-1132. 10.1021/pr0504440
Hamelin M, Sayd T, Chambon C, Bouix J, Bibé B, Milenkovic D, Leveziel H, Georges M, Clop A, Marinova P, Laville E: Differential expression of sarcoplasmic proteins in four heterogeneous ovine skeletal muscles. Proteomics 2007, 7: 271-280. 10.1002/pmic.200600309
Vitorino R, Ferreira R, Neuparth M, Guedes S, Williams J, Tomer KB, Domingues PM, Appell HJ, Duarte JA, Amado FM: Subcellular proteomics of mice gastrocnemius and soleus muscles. Anal Biochem 2007, 366: 156-169. 10.1016/j.ab.2007.04.009
Gannon J, Doran P, Kirwan A, Ohlendieck K: Drastic increase of myosin light chain MLC-2 in senescent skeletal muscle indicates fast-to-slow fibre transition in sarcopenia of old age. Eur J Cell Biol 2009, 88: 685-700. 10.1016/j.ejcb.2009.06.004
Lefort N, Yi Z, Bowen B, Glancy B, De Filippis EA, Mapes R, Hwang H, Flynn CR, Willis WT, Civitarese A, Højlund K, Mandarino LJ: Proteome profile of functional mitochondria from human skeletal muscle using one-dimensional gel electrophoresis and HPLC-ESI-MS/MS. J Proteomics 2009, 72: 1046-1060. 10.1016/j.jprot.2009.06.011
O'Connell K, Ohlendieck K: Proteomic DIGE analysis of the mitochondria-enriched fraction from aged rat skeletal muscle. Proteomics 2009, 9: 5509-5524.
Ferreira R, Vitorino R, Alves RM, Appell HJ, Powers SK, Duarte JA, Amado F: Subsarcolemmal and intermyofibrillar mitochondria proteome differences disclose functional specializations in skeletal muscle. Proteomics 2010, 10: 3142-3154. 10.1002/pmic.201000173
Lewis C, Ohlendieck K: Mass spectrometric identification of dystrophin isoform Dp427 by on-membrane digestion of sarcolemma from skeletal muscle. Anal Biochem 2010, 404: 197-203. 10.1016/j.ab.2010.05.017
Ruiz-Romero C, Blanco FJ: Mitochondrial proteomics and its application in biomedical research. Mol Biosyst 2009, 5: 1130-1142. 10.1039/b906296n
Schmidt O, Pfanner N, Meisinger C: Mitochondrial protein import: from proteomics to functional mechanisms. Nat Rev Mol Cell Biol 2010, 11: 655-667. 10.1038/nrm2959
Chen X, Li J, Hou J, Xie Z, Yang F: Mammalian mitochondrial proteomics: insights into mitochondrial functions and mitochondria-related diseases. Expert Rev Proteomics 2010, 7: 333-345. 10.1586/epr.10.22
Aebersold RH, Leavitt J, Saavedra RA, Hood LE, Kent SB: Internal amino acid sequence analysis of proteins separated by one- or two-dimensional gel electrophoresis after in situ protease digestion on nitrocellulose. Proc Natl Acad Sci USA 1987, 84: 6970-6697. 10.1073/pnas.84.20.6970
Luque-Garcia JL, Zhou G, Sun TT, Neubert TA: Use of nitrocellulose membranes for protein characterization by matrix-assisted laser desorption/ionization mass spectrometry. Anal Chem 2006, 78: 5102-5108. 10.1021/ac060344t
Luque-Garcia JL, Zhou G, Spellman DS, Tung-Tien S, Neubert TA: Analysis of electroblotted proteins by mass spectrometry: protein identification after Western blotting. Mol Cell Proteomics 2008, 7: 308-314.
Luque-Garcia JL, Neubert TA: On-membrane tryptic digestion of proteins for mass spectrometry analysis. Methods Mol Biol 2009, 536: 331-341. full_text
Froemming GR, Murray BE, Ohlendieck K: Self-aggregation of triadin in the sarcoplasmic reticulum of rabbit skeletal muscle. Biochim Biophys Acta 1999, 1418: 197-205. 10.1016/S0005-2736(99)00024-3
Tan S, Tan HT, Chung MC: Membrane proteins and membrane proteomics. Proteomics 2008, 8: 3924-3932. 10.1002/pmic.200800597
Sadowski PG, Groen AJ, Dupree P, Lilley KS: Subcellular localization of membrane proteins. Proteomics 2008, 8: 3991-4011. 10.1002/pmic.200800217
Zheng YZ, Foster LJ: Biochemical and proteomic approaches for the study of membrane microdomains. J Proteomics 2009, 72: 12-22. 10.1016/j.jprot.2008.09.003
Donoghue P, Staunton L, Mullen E, Manning G, Ohlendieck K: DIGE analysis of rat skeletal muscle proteins using nonionic detergent phase extraction of young adult versus aged gastrocnemius tissue. J Proteomics 2010, 73: 1441-1453. 10.1016/j.jprot.2010.01.014
Foster LJ, Rudich A, Talior I, Patel N, Huang X, Furtado LM, Bilan PJ, Mann M, Klip A: Insulin-dependent interactions of proteins with GLUT4 revealed through stable isotope labeling by amino acids in cell culture (SILAC). J Proteome Res 2006, 5: 64-75. 10.1021/pr0502626
Meany DL, Xie H, Thompson LV, Arriaga EA, Griffin TJ: Identification of carbonylated proteins from enriched rat skeletal muscle mitochondria using affinity chromatography-stable isotope labeling and tandem mass spectrometry. Proteomics 2007, 7: 1150-1163. 10.1002/pmic.200600450
Toigo M, Donohoe S, Sperrazzo G, Jarrold B, Wang F, Hinkle R, Dolan E, Isfort RJ, Aebersold R: ICAT-MS-MS time course analysis of atrophying mouse skeletal muscle cytosolic subproteome. Mol Biosyst 2005, 1: 229-241. 10.1039/b507839c
Kislinger T, Gramolini AO, Pan Y, Rahman K, MacLennan DH, Emili A: Proteome dynamics during C2C12 myoblast differentiation. Mol Cell Proteomics 2005, 4: 887-901. 10.1074/mcp.M400182-MCP200
Elschenbroich S, Ignatchenko V, Sharma P, Schmitt-Ulms G, Gramolini AO, Kislinger T: Peptide separations by on-line MudPIT compared to isoelectric focusing in an off-gel format: application to a membrane-enriched fraction from C2C12 mouse skeletal muscle cells. J Proteome Res 2009, 8: 4860-4869. 10.1021/pr900318k
Brewis IA, Brennan P: Proteomics technologies for the global identification and quantification of proteins. Adv Protein Chem Struct Biol 2010, 80: 1-44. full_text
Gilmore JM, Washburn MP: Advances in shotgun proteomics and the analysis of membrane proteomes. J Proteomics 2010, 73: 2078-2091. 10.1016/j.jprot.2010.08.005
Zhu W, Smith JW, Huang CM: Mass spectrometry-based label-free quantitative proteomics. J Biomed Biotechnol 2010, 2010: 840518.
Podwojski K, Eisenacher M, Kohl M, Turewicz M, Meyer HE, Rahnenfuhrer J, Stephan C: Peek a peak: a glance at statistics for quantitative label-free proteomics. Expert Rev Proteomics 2010, 7: 249-261. 10.1586/epr.09.107
Keshishian H, Addona T, Burgess M, Kuhn E, Carr SA: Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol Cell Proteomics 2007, 6: 2212-2229. 10.1074/mcp.M700354-MCP200
Carr SA, Anderson L: Protein quantitation through targeted mass spectrometry: the way out of biomarker purgatory? Clin Chem 2008, 54: 1749-1752. 10.1373/clinchem.2008.114686
Gerszten RE, Carr SA, Sabatine M: Integration of proteomic-based tools for improved biomarkers of myocardial injury. Clin Chem 2010, 56: 194-201. 10.1373/clinchem.2009.127878
Nagaraj N, Lu A, Mann M, Wisniewski JR: Detergent-based but gel-free method allows identification of several hundred membrane proteins in single LC-MS runs. J Proteome Res 2008, 7: 5028-5032. 10.1021/pr800412j
Wisniewski JR, Zougman A, Nagaraj N, Mann M: Universal sample preparation method for proteome analysis. Nat Methods 2009, 6: 359-362. 10.1038/nmeth.1322
Yi Z, Bowen BP, Hwang H, Jenkinson CP, Coletta DK, Lefort N, Bajaj M, Kashyap S, Berria R, De Filippis EA, Mandarino LJ: Global relationship between the proteome and transcriptome of human skeletal muscle. J Proteome Res 2008, 7: 3230-3241. 10.1021/pr800064s
Acknowledgements
Research in the author's laboratory was supported by grants from the Irish Health Research Board, the Irish Higher Education Authority, Muscular Dystrophy Ireland, Science Foundation Ireland and the European Commission. The author thanks all members of the NUIM Muscle Biology Laboratory and our many international collaborators for their help and encouragement over the past few years.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The author declares that they have no competing interests.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Ohlendieck, K. Skeletal muscle proteomics: current approaches, technical challenges and emerging techniques. Skeletal Muscle 1, 6 (2011). https://doi.org/10.1186/2044-5040-1-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/2044-5040-1-6