Abstract
Background
In the marine brown macroalgae, the morphological characters are highly similar between two widely distributed genera, Homoeostrichus and Zonaria (Dictyotaceae), thereby resulting in the difficulty of exploring their hidden biodiversity. Owing to the help of the molecular tools, it is now easy for scientists to objectively describe a new species in nature. In this study, we make a description on the Homoeostrichus formosana sp. nov. from Taiwan, Indo-Pacific Ocean based on the morphological evidence and molecular data.
Results
Our morphological observations revealed that this species has marginal row of apical cells responsible for thallus growth and the thallus with four layers of cells except the marginal regions. The cortical cell lies upon each medullary cell in transverse section, and two cortical cells upon each medullary cell in longitudinal section. Tetrasporangium is developed from cortical cell with stalk cell and singly scattered over the thallus surface, and has no indusia and paraphyses. Molecularly, the phylogenetic trees based on SSU, psaA, psbA, and rbcL gene sequences supported that Homoeostrichus species are closely related to Exallosorus species and clearly separated from each others in addition to Zonaria species.
Conclusions
Homoeostrichus formosana sp. nov. can now be clearly distinguished from E. harveyanus and Japanese H. flabellatus.
Similar content being viewed by others
Background
Phillips The three genera, Exallosorus1997 J. Agardh , Homoeostrichus 1894 C. Agardh and Zonaria1817 were established based on the characteristics of their reproductive structures, which used as key characters in the taxonomy of Dictyotaceae (Papenfuss 1944; Womersley 1987; Phillips and Clayton 199319941997; Phillips et al. 1994; Phillips 1997). Genus Homoeostrichus was established to include Zonaria canaliculata J. Agardh, Z. multifida Harvey ex J. Agardh, Z. sinclairii Hooker et Harvey and Z. stuposa R. Brown ex J. Agardh (J. Agardh 1894). Genus Zonaria had included five sections with ca. 50 species (C. Agardh 1817see Silva 1952), of which several species were transferred to Dictyota Lamouroux and Padina Adanson. Ten species of Zonaria are currently recognized (Phillips 1997; Phillips and Nelson 1998), and most of them are endemic to Australia (Womersley 1987; Phillips 1997; Phillips and Nelson 1998), whereas Z. diesingiana J. Agardh and Z. tournefortii (Lamouroux) Montagne are widely distributed from subtropical to temperate waters (Børgesen 1926; Taylor 1960; Gayral 1966; Allender and Kraft 1983; Seagarief 1984; Yoshida et al. 1985; Silva et al. 19871996; Womersley 1987; Farrant and King 1989; Ribera et al. 1992; Phillips et al. 1994; Phillips 1997; Phillips and Clayton 1997; Yoshida 1998).
Papenfuss (1944) suggested that Homoeostrichus and Zonaria shared characteristics in vegetative morphology and subsumed Homoeostrichus in Zonaria. However, Womersley (1987) argued that species of Zonaria had octosporangia and paraphyses whereas species of Homoeostrichus had only tetrasporangia and no paraphyses. He kept distinguishing Homoeostrichus from Zonaria and recognized three species of Homoeostrichus (H. canaliculatus J. Agardh, H. olsenii Womersley and H. sinclairii (Hooker et Harvey) J. Agardh). Phillips (1997) established Exallosorus based on two Australian species, Zonaria harveyana (Pappe ex Kützing) Areschoug (as Homoeostrichus multifidus J. Agardh) and Homoeostrichus olsenii Womersley [as E. harveyanus (Pappe ex Kützing) Phillips and E. olsenii (Womersley) Phillips]. She suggested that these species of Exallosorus have tetraspornagia with a stalk cell and within the indusiate sori which lack paraphyses and mucilage. The plants of genus Homoeostrichus commonly distributed in southeastern Australia and currently are recognized as two species: H. canaliculatus (Womersley and H. sinclairii1987; Phillips 1997).
A species of brown alga with external morphology similar to Exallosorus and Zonaria was collected from several collecting sites (Figure 1) in southern Taiwan. The plants of Homoeostrichus formosana Wang, Lin, Lee et Liu sp. nov. have been identified as Z. diesingiana or Z. harveyana in Taiwan, due to short information of their reproductive structures and morphological characteristics, especially no gametangia. It is the first time to describe the characteristics of sporangia of H. formosana sp. nov. in this study. We also described the morphological and phenological characteristics of this species, and determined its phylogeny among the related species based on nuclear-encoded SSU rRNA and plastid encoded rbcL, psaA, and psbA gene sequences.
Methods
Survey on morphological characteristics
Collections were made by SCUBA or snorkeling in southern Taiwan (Figure 1) from 1999 to 2002. Voucher specimens were fixed with 10% formalin/sea water or pressed on herbarium sheets and deposited in the Herbarium of the Department of Biology, National Chunghua University of Education, Taiwan. Microscopic sections were made using a freezing microtome (Leica CM1850), then stained with 0.1% Toluidine Blue O (TBO) and mounted in 50% Karo syrup. Microphotographs were taken on a Pixera digital camera attached to a Carl Zeiss Axioskop 2 microscope with differential interference contrast (DIC) optics.
Other specimens deposited in the following institutions were also examined: the Institute of Oceanography, National Taiwan University, Taipei (IONTU), the National Museum of Natural Science, Taichung, Taiwan (NMNS) and the National Museum of Marine Biology and Aquarium, Hengchun, Taiwan (NMMBA).
Gene sequence analyses
Collections for gene sequencing were made by SCUBA or snorkeling at Kenting, in southern Taiwan on 23 April 2004. Nuclear-encoded SSU rRNA and plastid encoded rbcL gene were selected for elucidating the phylogenetic relationship of Homoeostrichus formosana sp. nov. with other Dictyotaceae. Genomic DNA was extracted from 0.01 g of powder ground in liquid nitrogen using Dneasy Plant Mini Kit™ (Qiagen, Hilden in Germany), according to the manufacturer’s instructions. The partial rbcL gene and rbcS, except for short 3′-terminal of rbcL and 5′-terminal region of the rbcS, were amplified and sequenced as two fragments using the primers sets, DRL1F-DRL2R and DRL2F-RU2 (Hwang et al. 2005). The psaA gene sequences were also amplified and sequenced by two 130 F-970R and 870 F-1760R primers sets, psbA gene by one fragment with psbA F- psbA R primers set (Yoon et al. 2002). The partial 18S rRNA gene (SSU) was amplified and sequenced using primers set, SR1-SR7 and SR4-SR12. The amplified DNA was purified using High Pure PCR Product Purification Kit™ (Roche, Indianapolis,USA), in accordance with the manufacturer’s instructions. The forward and the reverse sequences were determined for all samples using an ABI PRISM 377 DNA sequencer. The sequences were aligned using PHYDIT (Chun 1995) with final visual confirmation and then submitted to GenBank under the accession numbers (Table 1). The alignment of each coding gene sequence was based on the alignment of inferred amino acid sequences, and reconfirmed by eye. The Padina species were selected as the outgroup species in the phylogenetic analyses.
Phylogenetic analysis was conducted using the software MEGA with a maximum likelihood method (, Tamura et al.2011). Prior to the phylogenetic analysis, the best fit of nucleotide evolutionary model for each gene was selected based on maximum-likelihood model fitting in the software MEGA. The chosen model is TN93+G model for SSU [lnL = -4717.63, rates of nucleotide changes (AT: 0.05, AC: 0.04, AG: 0.08, TA: 0.05, TC: 0.20, TG: 0.06, CA: 0.05, CT: 0.25, CG: 0.06, GA: 0.07, GT: 0.05, GC: 0.04), G = 0.08, and nucleotide frequencies (A: 0.24, T: 0.26, C: 0.22, G: 0.28)], GTR+G model for rbc L [lnL = -8507.61, rates of nucleotide changes (AT: 0.12, AC: 0.02, AG: 0.09, TA: 0.11, TC: 0.13, TG: 0.03, CA: 0.04, CT: 0.27, CG: 0.02, GA: 0.12, GT: 0.04, GC: 0.02), G = 0.22, and nucleotide frequencies (A: 0.30, T: 0.32, C: 0.16, G: 0.22)], TN93+G+I model for psaA [lnL = -10500.23, rates of nucleotide changes (AT: 0.06, AC: 0.03, AG: 0.07, TA: 0.05, TC: 0.15, TG: 0.03, CA: 0.05, CT: 0.33, CG: 0.03, GA: 0.12, GT: 0.06, GC: 0.03), I = 0.47, G = 0.53, and nucleotide frequencies (A: 0.30, T: 0.36, C: 0.16, G: 0.19)], and GTR+G model for psbA [lnL = -4454.15, rates of nucleotide changes (AT: 0.15, AC: 0.01, AG: 0.06, TA: 0.11, TC: 0.17, TG: 0.02, CA: 0.01, CT: 0.37, CG: 0.004, GA: 0.07, GT: 0.03, GC: 0.003), G = 0.17, and nucleotide frequencies (A: 0.26, T: 0.36, C: 0.17, G: 0.21)]. The ML bootstrap analyses were conducted with 500 replicates because of high computational demands.
Results
Species description
Homoeostrichus formosana
W.-L. Wang, C.-S. Lin, W.-J. Lee & S.-L. Liu sp. nov. (Figures 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12 and 13)
Huius plantae thallus, qui mensuratur 5–23 cm altitudine ac (1-)3-7(-10) cm latitudine, est fuscus, planus ac flabellatus; in ramos dividitur quorum axes inferiors angusti, superiors vero segmentati atque flabellate sunt. Folia in basi sunt erecta aut plana atque exsurgunt e stipite manifeste rhizoidali. Thallus autem componitur ex duo aut quattuor cellularum ordinibus, crassitudine 88–100 μm. In transversali sectione, medullares cellulae, 80–157 μm altitudine ac 15–25 μm latitudine, conteguntur a singulari cellula corticali, cui mensuratio est 25–50 μm altitudine ac 15–25 μm crassitudine. In sectione autem longitudinali, duo tresve corticales cellulae contegunt singulariam cellulam medularem. Tetrasporangia sphaerica, dispersa supra superficies, marginibus exceptis, mensuram habent 80–100 μm altitudine ac 85–95 μm in diametro, cum singularia cellula basilari quae se protrudit ultra thalli superficiem, sed sine ullis excrescentiis sori, indusii aut paraphysis.
Thalli are 5–23 cm in height and (1-)3-7(-10) cm in width, dark brown in color, complanate, flabellate, split to form branches with narrow lower axes and upper flabellate segment, and prostrate at the base arising from a conspicuously rhizoid holdfast to upright blades. Thallus composed of two to four layers of cells throughout, 88–100 μm in thickness. In transverse section medullary cells, 80–157 μm in height, 15–25 μm in width, are overlain by a single cortical cell, 25–50 μm in height, 15–25 μm in width, and then in longitudinal section, two to three cortical cells over lay each medullary cell. Tetrasporangia are spherical, scattered over the both sides of thallus except the margins, 80–100 μ m in height by 85–95 μ m in diameter, with one basal stalk cell projecting out from the thallus surface, without forming a sorus, indusia and paraphyses absent.
Holotype
The holotype is deposited at Department of Biology, National Changhua University of Education, Changhua (NCUE-CAF91072101) (Figure 2).
Type locality
Chuan-Fan-Shih, Southern Taiwan (21˚56′01″N, 120˚49′21″E).
Etymology
“formosana” refers to Taiwan, where the alga was collected.
Distribution
Known only from southern Taiwan (Figure 1).
Habitat and phenology
Absence of perennial stipes indicates that this species may be annual. Plants were found all year round, mainly at 2–5 m depth, where they were abundant on coral reefs or on reef rocks.
Specimens examined and localities
Pingtung County, southern Taiwan: Chu-Shui-Kou, 5–7 m, coll. C-S Lin, CAF91041401, 14 April 2002; Chuan-Fan-Shih, 1–4 m, coll. C-C Peng, 840013(NTU), 25 Oct.1995; coll. W-L Wang, CAF85053101, 31 Mar.1996; coll. C-S Lin, CAF90030301, sporophyte, 03 Mar.2001; coll. C-S Lin, CAF90050501, 05 May 2001; coll. S-M Lin, CAF90102601, 26 Oct. 2001; coll. S-M Lin, CAF90112801, sporophyte, 28 Nov. 2001; coll. C-S Lin, CAF91011301, sporophyte, 13 Jan. 2002; coll. C-S Lin, CAF91020601, 06 Feb. 2002; coll. C-S Lin, CAF91030101, 01 Mar. 2002; coll. S-M Lin, CAF91031401, 14 Mar. 2002; coll. C-S Lin, CAF91041301, 13 April 2002; coll. C-S Lin, CAF91061501, 15 June 2002; coll. C-S Lin, CAF91072101, sporophyte, 21 July 2002; coll. C-S Lin (Holotype), CAF91100201, 02 Oct. 2002; coll. S-L Lau, CAF91103001, sporophyte, 30 Oct. 2002; Hsiao-Wan, 1–3 m, coll. S-M Lin, CAF82063001, 30 June 1993; coll. W-L Wang, CAF86042601, 26.iv.1997; coll. C-S Lin, CAF91051101, 11.v.2002; coll. C-S Lin, CAF91072001, sporophyte, 20 July 2002; Hsiang-Chiao-Wan, 1–3 m coll. S-M Lin, CAF91032901, 29 May 2002; Feng-Chui-Sha, 1–5 m, coll. C-S Lin, CAF91051102, 11 May 2002; Chiu-Peng, 2–3 m, coll. G-L Lin, CAF82071101, 11 July 1993; coll. G-L Lin, CAF82102901, 29 Oct.1993.
Habitat and anatomical structures
Thalli are yellow or dark brown in color, composed of upright blades (Figures 2 and 3), and which basal portions are creeping with a conspicuously rhizoid holdfast. They are 5–23 cm in height and (1-)3-7(-10) cm in width (Figures 2 and 3). Thallii are fan-shaped when young and splitting into numerous bladelets when old. The surfaces of thallus are covered with hyaline hairs that are arranged in interrupted concentric bands (Figure 4), and with the blanketing brown rhizoidal filament at the base (Figure 5). Thallus growth is by a row of marginal meristem cells, which are dark in color (Figure 6). The apical cell is 120–240 μ m in length and 70–78 μ m in width (Figure 12). The blades are polystromatic, two or four cell layers, with 88–100 μ m in thickness. Cortical cells are 25–50 μm in height and 15–25 μ m in width. Those cells occurred on either side of two-cell layers of medullary cells, which measure 80–157 μ m in height and 15–25 μ m in width (Figures 7 and 8). In longitudinal section of thallus, two or three cortical cells overlay a single medullary cell (Figure 8), whereas a single cortical cell overlays each medullary cell in transverse section (Figures 11 and 13).
Reproductive structures
Sporangia are scattered over the surface on both sides of the blade (Figures 9 and 10). Tetrasporangia are roughly spherical and projected above the surface of the thallus, 80–100 μ m in height and 85–95 μ m in diameter, with a basal stalk cell which measured 12–26 μ m in height by 17–25 μ m in diameter, and lacked indusium and paraphyses (Figures 11 and 13). Gametophytes were not observed.
Characteristics of gene sequences
The SSU sequences determined and aligned in this study were 1,814 nucleotides long. The 20 aligned SSU sequences had 106 (5.8%) variable bases and 176 (9.7%) parsimoniously informative sites and 49.4% G+C contents. Transitions occurred more than transversions (Ts/Tv=1.16). The average of uncorrected pairwise distances (p-distances) was 0.059 from the aligned data set (Figure 14). The uncorrected pair wise distance (p-distances) between Zonaria species and Homoeostrichus species ranged from 0.057 to 0.077, and between Exallosorus species and Homoeostrichus species from 0.009 to 0.014. We could find 5 nucleotide base pairs differences in the aligned 1,723 nucleotide base pairs sequences between H. formosana sp. nov. and H. flabellatus from Japan (~0.3%), and 11 nucleotide base pairs differences between H. formosana sp. nov. and H. sinclairii.
We determined and aligned 1,351 nucleotides long rbcL sequences in this study. The 28 aligned rbcL sequences had 82 (6.07%) variable bases and 420 (31.08%) parsimoniously informative sites. The G+C content was 38.2% in the aligned sequence data set. Transitions were almost less than transversions (Ts/Tv=0.89). The average of p-distances was 0.122 from the aligned data set (Figure 14). The “p-distance” between Zonaria species and Homoeostrichus species ranged from 0.118 to 0.125, and between Exallosorus species and Homoeostrichus species from 0.098 to 0.119. Sixteen nucleotide differences were found between H. formosana sp. nov. and H. flabellatus in 1,305 nucleotide base aligned sequences (~1.2%).
The psaA sequences determined and aligned in this study were 1,395 nucleotides long. The aligned 24 psaA sequences had 87 (4.49%) variable bases and 496 (35.55%) parsimoniously informative sites and had 35.1% G+C content, and ratio of 0.82 transitions to transversions (Ts/Tv). The “p-distances” was 0.154 from the aligned psaA sequences data set (Figure 14). The “p-distances” between Zonaria species and Homoeostrichus species ranged from 0.143 to 0.152, and between Exallosorus species and Homoeostrichus species from 0.132 to 0.137. We found 182 nucleotide differences between H. formosana sp. nov. and H. sinclairii in aligned sequences of 1,394 base pairs.
The total 845 base pairs of psbA sequences were determined and aligned in this study. The aligned 25 psbA sequences had 45 (5.33%) variable bases and 213 (25.21%) parsimoniously informative sites with 37.8% G+C content. Transitions occurred more frequently than transversions (Ts/Tv=1.22) and p-distance ranged from 0.030 to 0.134 with average of 0.089 in aligned psbA sequences data set (Figure 14). The “p-distances” between Zonaria species and Homoeostrichus species ranged from 0.072 to 0.102, and between Exallosorus species and Homoeostrichus species from 0.084 to 0.098.
Overall, the sequence divergence is smallest in SSU, followed by psbA (Figure 14). In contrast, the sequence divergence is much larger in rbc L and psaA (Figure 14). Our observations suggest that SSU is more suitable to resolve the phylogenetic relationship of higher taxonomic level and other plastid genes used in this study are more suitable to tackle the phylogenetic relationship for the lower taxonomic level.
The phylogeny based on gene sequences
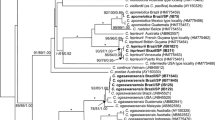
The phylogenetic tree based on SSU sequences showed that genera of tribe Zonarieae made four clades with no phylogenetic resolution among them in the ML analyses (Figure 15). The clade of Homoeostrichus and Exallosorus species is separated from that of Zonaria and Lobophora species. Three Homoeostrichus species made a subclade distinguished from Exallosorus species except for H. canaliculatus. Especially H. formosana sp. nov. made a clade with H. flabellatus with very low supporting value in three analyses.
The topology of phylogenetic tree based on rbcL sequences also show that four clades are distinguished (Figure 15). The clade comprising Homeostrichus and Exallosorus species figured out as basal sister group in this phylogeny although the results showed pale phylogenetic resolution. Homoeostrichus formosana sp. nov. made a clade with H. flabellatus with very high supporting value, and a sister group of H. sinclairii with low supporting value in three analyses. Exallosorum olsenii also made a sister group to three Homoeostrichus species clade and closely related to H. cnaliculatus. The clade of Zonaria and Lobophora species made a concrete clade with high supporting value distinguished from others.
The aligned psaA gene sequences data set made the phylogenetic tree with five clades, which have a basal clade of Stypopodium species although with pale phylogenetic resolution (Figure 15). As in the former trees, the clade of Homeostrichus and Exallosorus species is distinguished as basal sister group in this phylogeny although having pale phylogenetic resolution. Homoeostrichus formosana sp. nov. made a clade with H. flabellatus with very high supporting value, with a sister group of H. sinclairii with low supporting value in three analyses. Exallosorum olsenii also made a sister calde with three Homoeostrichus species and closely related to H. cnaliculatus as in the phylogeny of rbcL. The clade of Zonaria and Lobophora species made a concrete clade with high supporting value distinguished from Homoeostrichus and Exallosorus species.
The phylogenic tree based on psbA gene also show that H. formosana sp. nov. is involved in a clade with H. sinclairii and H. canaliculatus. This phylogenic tree is composed of five clades with very pale phylogenic resolution (Figure 15). Exallosorus species are closely related to Homoeostrichus species as in the other phylogenic trees.
Discussion
The taxonomy of the Dictyotales is largely based on the comparison of vegetative and reproductive growth and organization (Phillips 1997). Homoeostrichus formosana sp. nov. is mainly characterized by blades composing of two to four layers of cells, single tetrasporangia scattered over both thallus surfaces, sporangia borne on a stalk cell, and lacking indusium and paraphyses. In the erect to recumbent fan-like fronds of Lobophora, unusual large medullary cell and indusiate sporangial sorus, and Padina, rolling margin and concentric arrangement of reproductive structures, which both are conspicuously differed from Homoeostrichus, whatever the habit, texture, anatomical and reproductive structures. Homoeostrichus has been very easily confused with Exallosorus and Zonaria, based on the vegetative and reproductive characters used for separating among them summarized in Table 2. The phylogenic trees based on SSU, psaA, psbA, and rbcL gene sequences supported that Homoeostrichus species are closely related to Exallosorus species but clearly separated from each others in addition to Zonaria species.
The genus Exallosorus is separated from Zonaria and Homoeostrichus in having regularly arranged cells in transverse section, densely placed basally stalked sporangia within sori that possess brown paraphyses and indusium (Phillips 1997) (Table 2). Sporangia of Zonaria lacked basal stalk cells, are surrounded by whitish paraphyses (except in Z. angustata) in the indusiate sori, and released eight spores (Womersley 1987; Phillips et al. 1994; Phillips 1997) (Table 2). Sporangia of Homoeostrichus are distributed among brown paraphyses in non-indusiate sori, and released four spores (Womersley 1987; Phillips et al. 1994; Phillips 1997) (Table 2). In this study, we also found the sporangia in H. formosana sp. nov. are singly scattered over the surfaces of the thallus without forming a sorus and lacking indusium and paraphyses (Table 3). Classifying the genera of tribe Zonariae based on these morphological and anatomical characteristics is basically agreed to five clades in phylogenic analyses based on gene sequences.
Homoeostrichus formosana sp. nov. is superficially similar to Zonaria diesingiana found from Taiwan in external morphology. However, H. formosana sp. nov. can be distinguished vegetatively and reproductively from Z. diesingiana, especially it makes four cell layers. The thallus of Z. diesingiana is composed of 4–8 layers of cells, in which the one medullary cell is flanked by 2 cortical cells in transverse section, the octosporangia are borne on no stalk cell, and white paraphyses are present in indusiate sori. However, the tetrasporangium of H. formosana sp. nov. is borne on a basal stalk cell and lacks paraphyses and indusium.
Homoeostrichus formosana sp. nov. was previously misidentified as E. harveyanus (as Z. harveyana, H. multifidus) in Taiwan (Yamada 1925; Okamura 1936; Shen and Fan 1950; Chiang 1960; Lewis and Norris 1987). The thallus of E. harveyanus is composed of 6 layers of cells, which measured 120–170 μ m in thickness, and the sporangia are formed in a dark brown band of an indusiate sorus, whereas the sporangia in H. formosana are singly scattered over the surfaces of the thallus without forming a sorus (see Table 3). Although Yamada (1925) and Okamura (1936) had documented the thallus of “Homoeostrichus multifidus” (as H. formosana sp. nov. in this study) as being composed of four layers of cells, they did not observe reproductive structures, moreover, it is now known that E. harveyanus (as H. multifidus) is only distributed in southern Africa, the type locality (Silva et al. 1996; Phillips 1997). All molecular data also supported that H. formonosa sp. nov. is clearly distinguished from E. harveyanus in the psbA sequences molecular analyses in this study. Another Exallosorus species, E. olsenii, comprised of six cell layers, has sporangia assembled in indusiate sori that are connected with hairs and paraphyses, and with the reproductive structures only occurring on one thallus surface (Womersley 1987, as H. olsenii; Phillips et al. 1994; Phillips 1997), which is not agreed with H. formosana sp. nov. (Table 3).
Homoeostrichus formosana sp. nov can possibly be confused with other species of Homoeostrichus: H. canaliculatus (Womersley and H. sinclairii1987; Phillips 1997). However, H. formosana sp. nov. can be distinguished from the other species of Homoeostrichus by its 2–4 layers of cells thallus and sporangial stalk cells opposed to a 6–7 cell layer thallus and by multicellular stalk cells which are found in Homoeostrichus (see Table 3). The phylogentic tree especially based on psbA gene sequences showed that H. canaliculatus is distinguished from other Homeostrichus species and from Exallosorus species. Moreover, H. flabellatus Okamura, another Dictyotaceae species from Taiwan, might also be confused with H. formosana sp. nov. (Taniguti 1976; Lewis and Norris 1987; Wang and Chiang 2001). Okamura (1936) reported the thallus of H. flabellatus was composed of three layers of cells but he did not observe reproductive structures. Womersley (1987) speculated that Japanese H. flabellatus did not belong to the genus Homoeostrichus, and Papenfuss (1944) transferred H. flabellatus to Zonaria flabellata (Okamura) C. However, this combination is not recognized by some phycologists (see Phillips 1997; Phillips and Nelson 1998). The molecular characteristics of SSU show Japanese H. flabellatus is more related to H. formosana sp. nov. in this study. These show that the status of this taxon should be required further study especially examining voucher specimens of H. flabellatus. Furthermore, it is noted that an undescribed Zonaria sp. was recently reported from Chaojing, Keelung, northern Taiwan by Kitayama and Lin (2012). Though they only showed single photo of the thallus of this alga without any anatomical observations, this alga is highly similar to H. formasana in appearance. Considering that H. flabellatus (as Zonaria flabellatus) in Okinawa is biogeographically close to northern Taiwan (Figure 15), it will be interesting to examine the phylogenetic affinity of this undescribed Zonaria sp. from the northern Taiwan to test whether this alga is phylogenetically close to H. flabellatus or H. formasana.
Conclusions
We describes a new species, Homoeostrichus formosana Wang, Lin, Lee et Liu, collected from Taiwan. This species has marginal row of apical cells responsible for thallus growth and the thallus with four layers of cells except the marginal regions. The cortical cell lies upon each medullary cell in transverse section, and two cortical cells upon each medullary cell in longitudinal section. Tetrasporangium is observed for the first time, which is developed from cortical cell with stalk cell and singly scattered over the thallus surface, and has no indusia and paraphyses. The phylogenetic trees based on SSU, psaA, psbA, and rbcL gene sequences supported that Homoeostrichus species are closely related to Exallosorus species but distinctly different from Zonaria species.
References
Agardh CA: Synopsis algarum scandinaviae. Berling, Lund; 1817:1–135. I-xl + I-xl +
Agardh JG: Analecta alglogica. Continuatio I. Lunds universitets Års-skrift andra afdelningen, kongl. Fysiografiska sällskapets I. Lund Handlingar 1894,29(9):1–144.
Allender BM, Kraft GT: The marine algae of lord Howe island (New south Wales): the Dictyotales and Cutleriales (Phaeophyta). Brunonia 1983, 6: 73–130. 10.1071/BRU9830073
Børgesen F: Marine algae from the canary islands, especially from Teneriffe and Gran Canaria, II: Phaeophyceae . Biol Meddelelser 1926,6(2):1–112.
Chiang YM: Marine algae of northern Taiwan (Cyanophyta, Chlorophyta, Phaeophyta). Taiwania 1960, 7: 51–76.
Chun J Ph. D. Thesis. In Computer-assisted classification and identification of actinomycetes. University of Newcastle, UK; 1995.
Farrant PA, King RJ: The Dictyotales (algae: phaeophyta) of New South Wales. Proc Linnean Soc New South Wales 1989, 110: 369–406.
Gayral P Dion. In Les algues de côte francaises. Manche et Atlantique, Paris; 1966.
Hoshina R, Hasegawa K, Tanaka J, Hara Y: Molecular phylogeny of the dictyotaceae ( Phaeophyceae ) with emphasis on their morphology and its taxonomic implication. Jpn J Phycol 2004,52(suppl):189–194.
Hwang IK, Kim HS, Lee WJ: Confirmation on taxonomic status of Spatoglossum pacificum yendo (Dictyotaceae, Phaeophyceae ) based on morphology and plastid protein coding rbcL , rbcS , psaA , and psbA gene sequences. Algae 2004, 19: 161–174.
Hwang IK, Kim HS, Lee WJ: Polymorphism in the brown alga Dictyota dichotoma (Dictyotaceae, Dictyotales ) from Korea. Mar Biol 2005, 147: 999–1015. 10.1007/s00227-005-1623-8
Kitayama T, Lin S-M: Brown alga from Chaojing, Keelung City, Taiwan. Mem Natl Mus Sci, Tokyo 2012, 48: 149–157.
Lee WJ, Bae KS: Phylogenetic relationship among several genera of Dictyotaceae ( Dictyotales , Phaeophyceae ) based on 18S rRNA and rbcL gene sequences. Mar Biol 2002, 140: 1107–1115. 10.1007/s00227-002-0799-4
Lewis JE, Norris JN: A history and annotated account of the benthic marine algae of Taiwan. Smithson Contr Mar Sci 1987, 27: 38.
Okamura K: Nippon Kaiso-si. Uchi-da-ro-kaku-ho, Tokyo; 1936:158–185. pp. (In Japanese) pp. (In Japanese)
Papenfuss GF: Notes on algal nomenclature. II. Miscellaneous species of Chlorophyceae , Phaeophyceae and Rhaeophyta. Farlowia 1944, 1: 337–346.
Phillips JA: Genus and species concepts in Zonaria and Homoeostrichus ( Dictyotales , Phaeophyceae ), including the description of Exallosorus gen. nov. Eur J Phycol 1997, 32: 303–311. 10.1080/09670269710001737229
Phillips JA, Clayton MN: Comparative flagellar morphology of spermatozoids of the Dictyotales ( Phaeophyceae ). Eur J Phycol 1993,28(2):123–127. 10.1080/09670269300650201
Phillips JA, Clayton MN: Flagellate spores in Homoeostrichus olsenii Womersley ( Dictyotales , Phaeophyceae ): the largest know motile reproductive cells of marine macroalgae. Phycologia 1994,33(6):415–419. 10.2216/i0031-8884-33-6-415.1
Phillips JA, Clayton MN: Comparative studies on gametangial distribution and structure in species of Zonaria and Homoeostrichus ( Dictyotales , Phaeophyceae ) from Australia. Eur J Phycol 1997, 32: 25–34. 10.1080/09541449710001719345
Phillips JA, Nelson WA: Typification of the Australasian brown alga Zonaria turneriana J. Agardh ( Dictyotales ) and description of the endemic New Zealand species, Zonaria aureomarginata sp. nov. Bot Mar 1998, 41: 77–86.
Phillips JA, Clayton MN, Harvey AS: Comparative studies on sporangial distribution and structure in species of Zonaria , Lobophora and Homoeostrichus ( Dictyotales , Phaeophyceae ) from Australia. Eur J Phycol 1994, 29: 93–101. 10.1080/09670269400650541
Ribera MA, Gómez-Garreta A, Gallardo T, Comez-Garreta M, Gallardo T, Cormaci M, Furnari G, Giaccone G: Check-list of Mediterranean seaweeds. I. Fucophyceae (Warming 1884). Bot Mar 1992, 35: 109–130.
Seagarief SC: A catalogue of south African green, brown and red marine algae. Mem Bot Surv South Africa 1984, 47: 72.
Shen YF, Fan KC: Marine algae of Formosa. Taiwania 1950,1(2–4):317–345.
Silva PC: A review of nomenclatural conservation in the algae from the point of view of the type method. Univ Calif Pub Bot 1952, 25: 241–324.
Silva PC, Menez G, Moe RL: Catalog of benthic marine algae of the Philippines. Smithson Contr Mar Sci 1987, 27: 1–179.
Silva PC, Basson PW, Moe RL: Catalogue of the benthic marine algae of the Indian ocean. Univ California Publ Bot 1996, 79: 1–1259.
Simons RH: Notes on the species of Zonaria in south Africa. Bothalia 1964, 8: 195–197.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S: MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011, 28: 2731–2739. 10.1093/molbev/msr121
Taniguti M: Phytosociological study of marine algae in Taiwan. Bull Mie Univ 1976, 27: 51–57.
Taylor WR: Marine algae of the eastern tropical and subtropical coasts of the Americas. University of Michigan Press, America; 1960:214–238. 718–812 pp 718–812 pp
Wang WL, Chiang YM: The marine macroalgae of Lu Tao (green island), Taiwan. Taiwania 2001, 46: 49–61.
Womersley HBS: The marine benthic flora of southern Australia part II. South Australian Government Printing Division, Adelaide; 1987:187–259.
Yamada Y: Studien über die meeresalgen von der insel Formosa 2. Phaeophyceae Bot Mag (Tokyo) 1925, 39: 239–254.
Yoon HS, Hackett JD, Bhattacharya D: A single origin of the peridinin- and fucoxanthin-containing plastids in dinoflagellates through tertiary endosymbiosis. Proc Natl Acad Sci USA 2002, 99: 11724–11729. 10.1073/pnas.172234799
Yoshida T: Marine algae of Japan. Uchida Rokakuho Publishing, Tokyo, Japan; 1998:205–234.
Yoshida T, Nakajima Y, Nakata Y: Preliminary checklist of marine benthic algae of Japan-I. Chlorophyceae and Phaeophyceae . Jap J Phycol 1985, 33: 57–74.
Acknowledgments
We are very grateful to Dr. S.-M. Lin (National Taiwan Ocean University, Taiwan) for collecting the materials from the Pratas Island used in this study, to Mr. C.-K. Lin (National Museum of Natural Science, Taiwan) for loaning some Dictyotacean specimens, and to Dr. Lawrence Liao (Hiroshima University, Japan) for providing critical Dictyotacean references. We also thank to Mr. L.-C. Wang at Department of Biology, National Chunghua University of Education for the assistance of the field work and the map drawing. We also thank to Prof. Emerit. S. J. Fernando Mateos (Tien Educational Center, Taiwan) for translating the Latin of genus and species descriptions. This study was partly supported by a Taiwan National Science Council (NSC) grant (NSC89-2611-M-018-001) to W.-L. Wang, by a Korea Research Foundation grant (KRF2002-070-C00083) to W.J. Lee, and by a Taiwan NSC grant (NSC-101-2621-B-029-004) to S.-L. Liu.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
WL Wang and CS Lin carried out the morphological characteristics of this species and drafted the manuscript, while WJ Lee and SL Liu participated in the molecular genetic studies. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Wang, WL., Lin, CS., Lee, WJ. et al. Morphological and molecular characteristics of Homoeostrichus formosana sp. nov. (Dictyotaceae, Phaeophyceae) from Taiwan. Bot Stud 54, 13 (2013). https://doi.org/10.1186/1999-3110-54-13
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1999-3110-54-13